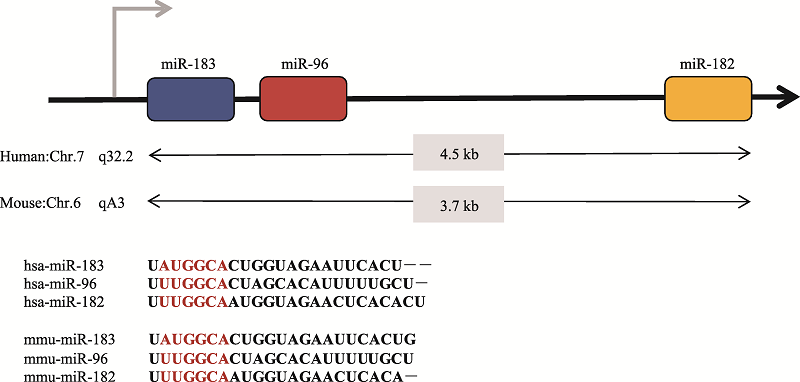
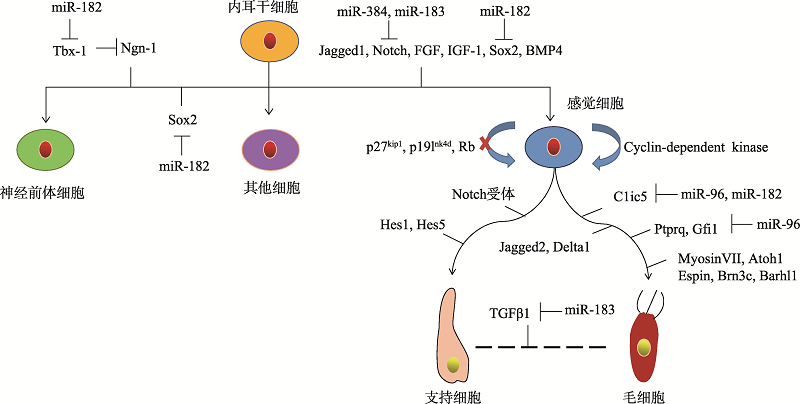
Hereditas(Beijing) ›› 2019, Vol. 41 ›› Issue (11): 994-1008.doi: 10.16288/j.yczz.19-119
• Review • Previous Articles Next Articles
Molecular mechanism of microRNA in regulating cochlear hair cell development
Lin Rao, Feilong Meng, Ran Fang, Chenyi Cai, Xiaoli Zhao( )
)
- Key Laboratory for Cell and Gene Engineering of Zhejiang Province, Institute of Genetics and Regenerative Biology, College of Life Sciences, Zhejiang University, Hangzhou 310058, China
-
Received:2019-07-20Revised:2019-09-26Online:2019-11-20Published:2019-10-18 -
Contact:Zhao Xiaoli E-mail:zhaoxiaoli@zju.edu.cn -
Supported by:Supported by the National Program on Key Basic Research Project (973 Program) No(2012CB967900)
Cite this article
Lin Rao, Feilong Meng, Ran Fang, Chenyi Cai, Xiaoli Zhao. Molecular mechanism of microRNA in regulating cochlear hair cell development[J]. Hereditas(Beijing), 2019, 41(11): 994-1008.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
Table 1
Target gene of miRNA in inner ear cochlea"
| miRNA | 靶基因 | 参考文献 |
|---|---|---|
| miR-9 | COL9A1 | [ |
| miR-29 | SIRT1 | [ |
| miR-34a | SIRT1、bcl-2和E2F-3 | [ |
| miR-96 | TRK和EGFR | [ |
| Slc26a5、Ocm、Gfi1、Ptprq和Pitpnm1 | [ | |
| miR-96/-182 | CLIC5 | [ |
| miR-124 | Sfrp4和Sfrp5 | [ |
| miR-135b | PSIP1-P75 | [ |
| miR-140 | NR2F1和Klf9 | [ |
| miR-182 | Sox2和Tbx1 | [ |
| miR-183 | Taok1和ltgA3 | [ |
| miR-194 | Fgf4和RhoB | [ |
| miR-200 | Zeb1和Zeb2 | [ |
| miR-204 | TMPRSS3 | [ |
| miR-224 | Ptx3 | [ |
| miR-376 | PRPS1 | [ |
| [1] |
Dhungel B, Ramlogan-Steel CA, Steel JC . MicroRNA- regulated gene delivery systems for research and therapeutic purposes. Molecules, 2018,23(7):E1500.
doi: 10.3390/molecules23071500 pmid: 29933586 |
| [2] |
Lau NC, Lim LP, Weinstein EG, Bartel DP . An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science, 2001,294(5543):858-862.
doi: 10.1126/science.1065062 pmid: 11679671 |
| [3] |
Tanzer A, Stadler PF . Molecular evolution of a microRNA cluster. J Mol Biol, 2004,339(2):327-335.
doi: 10.1016/j.jmb.2004.03.065 pmid: 15136036 |
| [4] |
Lagos-Quintana M, Rauhut R, Meyer J, Borkhardt A, Tuschl T . New microRNAs from mouse and human. RNA, 2003,9(2):175-179.
doi: 10.1261/rna.2146903 pmid: 12554859 |
| [5] |
Mittal R, Liu G, Polineni SP, Bencie N, Yan D, Liu XZ . Role of microRNAs in inner ear development and hearing loss. Gene, 2019,686:49-55.
doi: 10.1016/j.gene.2018.10.075 pmid: 30389561 |
| [6] |
Mahmoodian Sani RM, Hashemzadeh-Chaleshtori M, Saidijam M, Jami MS, Ghasemi-Dehkordi P . MicroRNA-183 family in inner ear: hair cell development and deafness. J Audiol Otol, 2016,20(3):131-138.
doi: 10.7874/jao.2016.20.3.131 pmid: 27942598 |
| [7] |
Fritzsch B, Elliott KL . Gene, cell, and organ multiplication drives inner ear evolution. Dev Biol, 2017,431(1):3-15.
doi: 10.1016/j.ydbio.2017.08.034 pmid: 28866362 |
| [8] |
Chan WX, Lee SH, Kim N, Shin CS, Yoon YJ . Mechanical model of an arched basilar membrane in the gerbil cochlea. Hear Res, 2017,345:1-9.
doi: 10.1016/j.heares.2016.12.003 pmid: 27986594 |
| [9] |
Ulfendahl M, Khanna SM, Decraemer WF . Acoustically induced vibrations of the Reissner's membrane in the guinea-pig inner ear. Acta Physiol Scand, 1996,158(3):275-285.
doi: 10.1046/j.1365-201X.1996.563313000.x pmid: 8931771 |
| [10] |
Mahmoudian-sani MR, Mehri-Ghahfarrokhi A, Ahmadinejad F, Hashemzadeh-Chaleshtori M, Saidijam M, Jami MS . MicroRNAs: effective elements in ear-related diseases and hearing loss. Eur Arch Oto-Rhino-L, 2017,274(6):2373-2380.
doi: 10.1007/s00405-017-4470-6 pmid: 28224282 |
| [11] |
Schrauwen I, Hasin-Brumshtein Y, Corneveaux JJ, Ohmen J, White C, Allen AN, Lusis AJ, Van Camp G, Huentelman MJ, Friedman RA . A comprehensive catalogue of the coding and non-coding transcripts of the human inner ear. Hear Res, 2016,333:266-274.
doi: 10.1016/j.heares.2015.08.013 pmid: 26341477 |
| [12] |
Mahmoodian-Sani MR, Mehri-Ghahfarrokhi A . The potential of miR-183 family expression in inner ear for regeneration, treatment, diagnosis and prognosis of hearing loss. J Otol, 2017,12(2):55-61.
doi: 10.1016/j.joto.2017.03.003 pmid: 29937838 |
| [13] |
Kwan KY . Single-cell transcriptome analysis of developing and regenerating spiral ganglion neurons. Curr Pharmacol Rep, 2016,2(5):211-220.
doi: 10.1007/s40495-016-0064-z pmid: 28758056 |
| [14] |
Matsunami T, Suzuki T, Hisa Y, Takata K, Takamatsu T, Oyamada M . Gap junctions mediate glucose transport between GLUT1-positive and -negative cells in the spiral limbus of the rat cochlea. Cell Commun Adhes, 2006,13(1-2):93-102.
doi: 10.1080/15419060600631805 pmid: 16613783 |
| [15] |
Lee SH, Ju HM, Choi JS, Ahn Y, Lee S, Seo YJ . Circulating serum miRNA-205 as a diagnostic biomarker for ototoxicity in mice treated with aminoglycoside antibiotics. Int J Mol Sci, 2018,19(9):2386.
doi: 10.3390/ijms19092836 pmid: 30235835 |
| [16] |
He WX, Kemp D, Ren TY . Timing of the reticular lamina and basilar membrane vibration in living gerbil cochleae. eLife Sciences, 2018,7.
doi: 10.7554/eLife.37625 pmid: 30183615 |
| [17] |
Monzack EL, Cunningham LL . Lead roles for supporting actors: critical functions of inner ear supporting cells. Hear Res, 2013,303:20-29.
doi: 10.1016/j.heares.2013.01.008 pmid: 23347917 |
| [18] |
Liu J, Liu W, Yang J . ATP-containing vesicles in stria vascular marginal cell cytoplasms in neonatal rat cochlea are lysosomes. Sci Rep, 2016,6:20903.
doi: 10.1038/srep20903 pmid: 26864824 |
| [19] |
Patel M, Hu BH . MicroRNAs in inner ear biology and pathogenesis. Hear Res, 2012,287(1-2):6-14.
doi: 10.1016/j.heares.2012.03.008 pmid: 22484222 |
| [20] |
Torres L, Juárez U, García L, Miranda-Ríos J, Frias S . External ear microRNA expression profiles during mouse development. Int J Dev Biol, 2015,59(10-12):497-503.
doi: 10.1387/ijdb.150124sf pmid: 26864490 |
| [21] |
Trujillo-Provencio C, Powers TR, Sultemeier DR, Ramirez-Gordillo D, Serrano EE . RNA extraction from xenopus auditory and vestibular organs for molecular cloning and expression profiling with RNA-seq and microarrays. Methods Mol Biol, 2016,1427:73-92.
doi: 10.1007/978-1-4939-3615-1_5 pmid: 27259922 |
| [22] |
Stenfelt S . Inner ear contribution to bone conduction hearing in the human. Hear Res, 2015,329:41-51.
doi: 10.1016/j.heares.2014.12.003 pmid: 25528492 |
| [23] |
Whitfield TT . Development of the inner ear. Curr Opin Genet Dev, 2015,32:112-118.
doi: 10.1016/j.gde.2015.02.006 pmid: 25796080 |
| [24] |
Pechriggl EJ, Bitsche M, Glueckert R, Rask-Andersen H, Blumer MJF, Schrott-Fischer A, Fritsch H . Development of the innervation of the human inner ear. Dev Neurobiol, 2015,75(7):683-702.
doi: 10.1002/dneu.22242 pmid: 25363666 |
| [25] |
Chadly DM, Best J, Ran C, Bruska M, Woźniak W, Kempisty B, Schwartz M , LaFleur B, Kerns BJ, Kessler JA, Matsuoka AJ. Developmental profiling of microRNAs in the human embryonic inner ear. PLoS One, 2018,13(1):e0191452.
doi: 10.1371/journal.pone.0191452 pmid: 29373586 |
| [26] |
Sai XR, Ladher RK . Early steps in inner ear development: induction and morphogenesis of the otic placode. Front Pharmacol, 2015,6.
doi: 10.3389/fphar.2015.00322 pmid: 26924983 |
| [27] |
McLean WJ, McLean DT, Eatock RA, Edge AS . Distinct capacity for differentiation to inner ear cell types by progenitor cells of the cochlea and vestibular organs. Development, 2016,143(23):4381-4393.
doi: 10.1242/dev.139840 pmid: 27789624 |
| [28] |
Corrales CE, Pan LY, Li HW, Liberman MC, Heller S, Edge AS . Engraftment and differentiation of embryonic stem cell-derived neural progenitor cells in the cochlear nerve trunk: Growth of processes into the organ of corti. J Neurobiol, 2006,66(13):1489-1500.
doi: 10.1002/neu.20310 pmid: 17013931 |
| [29] |
Varela-Nieto I, Palmero I, Magariños M . Complementary and distinct roles of autophagy, apoptosis and senescence during early inner ear development. Hear Res, 2019,376:86-96.
doi: 10.1016/j.heares.2019.01.014 pmid: 30711386 |
| [30] |
Robles L, Ruggero MA . Mechanics of the mammalian cochlea. Physiol Rev, 2001,81(3):1305-1352.
doi: 10.1152/physrev.2001.81.3.1305 pmid: 11427697 |
| [31] |
Hurd EA, Adams ME, Layman WS, Swiderski DL, Beyer LA, Halsey KE, Benson JM, Gong TW, Dolan DF, Raphael Y, Martin DM . Mature middle and inner ears express Chd7 and exhibit distinctive pathologies in a mouse model of CHARGE syndrome. Hear Res, 2011,282(1-2):184-195.
doi: 10.1016/j.heares.2011.08.005 pmid: 21875659 |
| [32] |
Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T . Identification of tissue-specific microRNAs from mouse. Curr Biol, 2002,12(9):735-739.
doi: 10.1016/S0960-9822(02)00809-6 |
| [33] |
Berghaus A, Nicoló MS . Milestones in the history of ear reconstruction. Facial Plast Surg, 2015,31(6):563-566.
doi: 10.1055/s-0035-1567886 pmid: 26667630 |
| [34] |
Lee S, Shin JO, Sagong B, Kim UK, Bok J . Spatiotemporal expression patterns of clusterin in the mouse inner ear. Cell Tissue Res, 2017,370(1):89-97.
doi: 10.1007/s00441-017-2650-8 pmid: 28687930 |
| [35] |
Roser AE, Gomes LC, Halder R, Jain G, Maass F, Tönges L, Tatenhorst L, Bähr M, Fischer A , Lingor P. miR-182-5p and miR-183-5p Act as GDNF mimics in dopaminergic midbrain neurons. Mol Ther-Nucl Acids, 2018,11:9-22.
doi: 10.1016/j.omtn.2018.01.005 pmid: 29858093 |
| [36] |
Bai YS, Li L, Wei HX, Zhu C, Zhang CQ . The effect of microRNAs on the regulatory network of pluripotency in embryonic stem cells. Hereditas(Beijing), 2013,35(10):1153-1166.
doi: 10.3724/SP.J.1005.2013.01153 |
|
白银山, 李莉, 卫恒习, 朱翠, 张守全 . MicroRNA对胚胎干细胞的多能性网络调控. 遗传, 2013,35(10):1153-1166.
doi: 10.3724/SP.J.1005.2013.01153 |
|
| [37] |
Weston MD, Tarang S, Pierce ML, Pyakurel U, Rocha-Sanchez SM, McGee J, Walsh EJ, Soukup GA. A mouse model of miR-96, miR-182 and miR-183 misexpression implicates miRNAs in cochlear cell fate and homeostasis. Sci Rep, 2018,8(1):3569.
doi: 10.1038/s41598-018-21811-1 pmid: 29476110 |
| [38] |
Weston MD, Pierce ML, Rocha-Sanchez S, Beisel KW, Soukup GA . MicroRNA gene expression in the mouse inner ear. Brain Res, 2006,1111(1):95-104.
doi: 10.1016/j.brainres.2006.07.006 pmid: 16904081 |
| [39] |
Li HQ, Kloosterman W, Fekete DM . MicroRNA-183 family members regulate sensorineural fates in the inner ear. J Neurosci, 2010,30(9):3254-3263.
doi: 10.1523/JNEUROSCI.4948-09.2010 pmid: 20203184 |
| [40] |
Xiang L, Chen XJ, Wu KC, Zhang CJ, Zhou GH, Lv JN, Sun LF, Cheng FF, Cai XB, Jin ZB, . miR-183/96 plays a pivotal regulatory role in mouse photoreceptor maturation and maintenance. Proc Natl Acad Sci USA, 2017,114(24):6376-6381.
doi: 10.1073/pnas.1618757114 pmid: 28559309 |
| [41] |
Sacheli R, Nguyen L, Borgs L, Vandenbosch R, Bodson M, Lefebvre P, Malgrange B . Expression patterns of miR-96, miR-182 and miR-183 in the development inner ear. Gene Expr Patterns, 2009,9(5):364-370.
doi: 10.1016/j.gep.2009.01.003 pmid: 19602392 |
| [42] |
Fettiplace R . Hair cell transduction, tuning, and synaptic transmission in the mammalian cochlea. Compr Physiol, 2017,7(4):1197-1227.
doi: 10.1002/cphy.c160049 pmid: 28915323 |
| [43] |
Cunningham LL, Tucci DL . Hearing loss in adults. N Engl J Med, 2017,377(25):2465-2473.
doi: 10.1056/NEJMra1616601 pmid: 29262274 |
| [44] |
Liberman MC, Epstein MJ, Cleveland SS, Wang HB, Maison SF . Toward a differential diagnosis of hidden hearing loss in humans. PLoS One, 2016,11(9):e0162726.
doi: 10.1371/journal.pone.0162726 pmid: 27618300 |
| [45] |
Berrettini S, De Vito A, Bruschini L, Fortunato S, Forli F . Idiopathic sensorineural hearing loss in the only hearing ear. Acta Otorhinolaryngol Ital, 2016,36(2):119-126.
doi: 10.14639/0392-100X-587 pmid: 27196076 |
| [46] |
Leung MA, Flaherty A, Zhang JA, Hara J, Barber W, Burgess L . Sudden sensorineural hearing loss: primary care update. Hawaii J Med Public Health, 2016,75(6):172-174.
pmid: 27413627 |
| [47] |
Bermingham-McDonogh O, Reh TA . Regulated reprogramming in the regeneration of sensory receptor cells. Neuron, 2011,71(3):389-405.
doi: 10.1016/j.neuron.2011.07.015 |
| [48] |
Sekine K, Matsumura T, Takizawa T, Kimura Y, Saito S, Shiiba K, Shindo S, Okubo K, Ikezono T . Expression profiling of MicroRNAs in the inner ear of elderly people by real-time PCR quantification. Audiol Neuro- Otol, 2017,22(3):135-145.
doi: 10.1159/000479724 pmid: 28968605 |
| [49] |
Pierce ML, Weston MD, Fritzsch B, Gabel HW, Ruvkun G, Soukup GA . MicroRNA-183 family conservation and ciliated neurosensory organ expression. Evol Dev, 2008,10(1):106-113.
doi: 10.1111/j.1525-142X.2007.00217.x pmid: 18184361 |
| [50] |
Dambal S, Baumann B, McCray T, Williams L, Richards Z, Deaton R, Prins GS, Nonn L . The miR-183 family cluster alters zinc homeostasis in benign prostate cells, organoids and prostate cancer xenografts. Sci Rep, 2017,7(1):7704.
doi: 10.1038/s41598-017-07979-y pmid: 28794468 |
| [51] |
Xu S, Witmer PD, Lumayag S, Kovacs B, Valle D . MicroRNA (miRNA) transcriptome of mouse retina and identification of a sensory organ-specific miRNA cluster. J Biol Chem, 2007,282(34):25053-25066.
doi: 10.1074/jbc.M700501200 pmid: 17597072 |
| [52] |
Li JY, Ling YH, Huang WH, Sun LM, Li YY, Wang CH, Zhang YH, Wang XD, Dahlgren RA, Wang HL . Regulatory mechanisms of miR-96 and miR-184 abnormal expressions on otic vesicle development of zebrafish following exposure to β-diketone antibiotics. Chemosphere, 2019,214:228-238.
doi: 10.1016/j.chemosphere.2018.09.118 pmid: 30265930 |
| [53] |
Raymond M, Walker E, Dave I, Dedhia K . Genetic testing for congenital non-syndromic sensorineural hearing loss. Int J Pediatr Otorhinolaryngol, 2019,124:68-75.
doi: 10.1016/j.ijporl.2019.05.038 pmid: 31163360 |
| [54] |
Li HQ, Fekete DM . MicroRNAs in hair cell development and deafness. Curr Opin Otolaryngo, 2010,18(5):459-465.
doi: 10.1097/MOO.0b013e32833e0601 pmid: 20717030 |
| [55] |
Chen J, Johnson SL, Lewis MA, Hilton JM, Huma A, Marcotti W, Steel KP . A reduction in Ptprq associated with specific features of the deafness phenotype of the miR-96 mutant mouse diminuendo. Eur J Neurosci, 2014,39(5):744-756.
doi: 10.1111/ejn.12484 |
| [56] |
Sánchez-Mora C, Ramos-Quiroga JA, Garcia-Martínez I, Fernàndez-Castillo N, Bosch R, Richarte V, Palomar G, Nogueira M, Corrales M, Daigre C, Martínez-Luna N, Grau-Lopez L, Toma C, Cormand B, Roncero C, Casas M, Ribasés M . Evaluation of single nucleotide polymorphisms in the miR-183-96-182 cluster in adulthood attention-deficit and hyperactivity disorder (ADHD) and substance use disorders (SUDs). Eur Neuropsychopharmacol, 2013,23(11):1463-1473.
doi: 10.1016/j.euroneuro.2013.07.002 pmid: 23906647 |
| [57] |
Mencía A, Modamio-Høybjør S, Redshaw N, Morín M, Mayo-Merino F, Olavarrieta L, Aguirre LA, del Castillo I, Steel KP, Dalmay T, Moreno F, Moreno-Pelayo MA . Mutations in the seed region of human miR-96 are responsible for nonsyndromic progressive hearing loss. Nat Genet, 2009,41(5):609-613.
doi: 10.1038/ng.355 pmid: 19363479 |
| [58] |
Lewis MA, Quint E, Glazier AM, Fuchs H, De Angelis MH, Langford C, van Dongen S, Abreu-Goodger C, Piipari M, Redshaw N, Dalmay T, Moreno-Pelayo MA, Enright AJ, Steel KP . An ENU-induced mutation of miR-96 associated with progressive hearing loss in mice. Nat Genet, 2009,41(5):614-618.
doi: 10.1038/ng.369 pmid: 19363478 |
| [59] |
Kuhn S, Johnson SL, Furness DN, Chen J, Ingham N, Hilton JM, Steffes G, Lewis MA, Zampini V, Hackney CM, Masetto S, Holley MC, Steel KP, Marcotti W. miR-96 regulates the progression of differentiation in mammalian cochlear inner and outer hair cells. Proc Natl Acad Sci USA, 2011,108(6):2355-2360.
doi: 10.1073/pnas.1016646108 pmid: 21245307 |
| [60] |
Li YM, Li A, Wu JF, He YZ, Yu HQ, Chai RJ . MiR- 182-5p protects inner ear hair cells from cisplatin- induced apoptosis by inhibiting FOXO3a. Cell Death Dis, 2016,7(9):e2362.
doi: 10.1038/cddis.2016.246 pmid: 27607577 |
| [61] |
Patel M, Cai QF, Ding DL, Salvi R, Hu ZH, Hu BH . The miR-183/Taok1 target pair is implicated in cochlear responses to acoustic trauma. PLoS One, 2013,8(3):e58471.
doi: 10.1371/journal.pone.0058471 pmid: 23472202 |
| [62] |
MacFarlane LA, Murphy PR . MicroRNA: biogenesis, function and role in cancer. Curr Genomics, 2010,11(7):537-561.
doi: 10.2174/138920210793175895 pmid: 21532838 |
| [63] |
Giraldez AJ, Cinalli RM, Glasner ME, Enright AJ, Thomson JM, Baskerville S, Hammond SM, Bartel DP, Schier AF . MicroRNAs regulate brain morphogenesis in zebrafish. Science, 2005,308(5723):833-838.
doi: 10.1126/science.1109020 pmid: 15774722 |
| [64] |
Weston MD, Pierce ML, Jensen-Smith HC, Fritzsch B, Rocha-Sanchez S, Beisel KW, Soukup GA . MicroRNA- 183 family expression in hair cell development and requirement of MicroRNAs for hair cell maintenance and survival. Dev Dyn, 2011,240(4):808-819.
doi: 10.1002/dvdy.22591 pmid: 21360794 |
| [65] |
Hildebrand MS, Witmer PD, Xu S, Newton SS, Kahrizi K, Najmabadi H, Valle D, Smith RJ. miRNA mutations are not a common cause of deafness. Am J Med Genet A, 2010,152A(3):646-652.
doi: 10.1002/ajmg.a.33299 pmid: 20186779 |
| [66] |
Wang XR, Zhang XM, Du JT, Jiang HY . MicroRNA-182 regulates otocyst-derived cell differentiation and targets T-box1 gene. Hear Res, 2012,286(1-2):55-63.
doi: 10.1016/j.heares.2012.02.005 pmid: 22381690 |
| [67] |
Schellenberg GD, Dawson G, Sung YJ, Estes A, Munson J, Rosenthal E, Rothstein J, Flodman P, Smith M, Coon H, Leong L, Yu CE, Stodgell C, Rodier PM, Spence MA, Minshew N, McMahon WM, Wijsman EM . Evidence for multiple loci from a genome scan of autism kindreds. Mol Psychiatr, 2006,11(11):1049-1060.
doi: 10.1038/sj.mp.4001874 pmid: 16880825 |
| [68] |
Soukup GA, Fritzsch B, Pierce ML, Weston MD, Jahan I, McManus MT, Harfe BD, . Residual microRNA expression dictates the extent of inner ear development in conditional dicer knockout mice. Dev Biol, 2009,328(2):328-341.
doi: 10.1016/j.ydbio.2009.01.037 pmid: 19389351 |
| [69] |
Bhattacharya A, Cui Y . Knowledge-based analysis of functional impacts of mutations in microRNA seed regions. J Biosci, 2015,40(4):791-798.
doi: 10.1007/s12038-015-9560-2 pmid: 26564979 |
| [70] |
Kim CW, Han JH, Wu L, Choi JY. microRNA-183 is essential for hair cell regeneration after neomycin injury in zebrafish. Yonsei Med J, 2018,59(1):141-147.
doi: 10.3349/ymj.2018.59.1.141 pmid: 29214789 |
| [71] |
Van den Ackerveken P, Mounier A, Huyghe A, Sacheli R, Vanlerberghe PB, Volvert ML, Delacroix L, Nguyen L, Malgrange B . The miR-183/ItgA3 axis is a key regulator of prosensory area during early inner ear development. Cell Death Differ, 2017,24(12):2054-2065.
doi: 10.1038/cdd.2017.127 pmid: 28777373 |
| [72] |
Sivakumaran TA, Resendes BL, Robertson NG, Giersch ABS, Morton CC . Characterization of an abundant COL9A1 transcript in the cochlea with a novel 3ʹ UTR: Expression studies and detection of miRNA target sequence. Jaro-J Assoc Res Oto, 2006,7(2):160-172.
doi: 10.1007/s10162-006-0032-0 |
| [73] |
Gu CH, Li XD, Tan Q, Wang Z, Chen LM, Liu YM . MiR-183 family regulates chloride intracellular channel 5 expression in inner ear hair cells. Toxicol in Vitro, 2013,27(1):486-491.
doi: 10.1016/j.tiv.2012.07.008 pmid: 22889583 |
| [74] |
Jiang D, Du JT, Zhang XM, Zhou W, Zong L, Dong C, Chen K, Chen Y, Chen XH, Jiang HY . miR-124 promotes the neuronal differentiation of mouse inner ear neural stem cells. Int J Mol Med, 2016,38(5):1367-1376.
doi: 10.3892/ijmm.2016.2751 pmid: 28025992 |
| [75] |
Rudnicki A, Isakov O, Ushakov K, Shivatzki S, Weiss I, Friedman LM, Shomron N, Avraham KB . Next-generation sequencing of small RNAs from inner ear sensory epithelium identifies microRNAs and defines regulatory pathways. Bmc Genomics, 2014,15:484.
doi: 10.1186/1471-2164-15-484 pmid: 24942165 |
| [76] |
Du JT, Zhang XM, Cao H, Jiang D, Wang XR, Zhou W, Chen KT, Zhou J, Jiang HY, Ba L . MiR-194 is involved in morphogenesis of spiral ganglion neurons in inner ear by rearranging actin cytoskeleton via targeting RhoB. Int J Dev Neurosci, 2017,63:16-26.
doi: 10.1016/j.ijdevneu.2017.09.004 pmid: 28941704 |
| [77] |
Park SM, Gaur AB, Lengyel E, Peter ME . The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev, 2008,22(7):894-907.
doi: 10.1101/gad.1640608 pmid: 18381893 |
| [78] |
Yan D, Xing YZ, Ouyang XM, Zhu JH, Chen ZY, Lang HN, Liu XZ . Analysis of miR-376 RNA cluster members in the mouse inner ear. Int J Exp Pathol, 2012,93(6):450-457.
doi: 10.1111/j.1365-2613.2012.00840.x |
| [79] |
Elkan-Miller T, Ulitsky I, Hertzano R, Rudnicki A, Dror AA, Lenz DR, Elkon R, Irmler M, Beckers J, Shamir R, Avraham KB . Integration of transcriptomics, proteomics, and microRNA analyses reveals novel microRNA regulation of targets in the mammalian inner ear. PLoS One, 2011,6(4):e18195.
doi: 10.1371/journal.pone.0018195 pmid: 21483685 |
| [80] |
Kurtz CL, Fannin EE, Toth CL, Pearson DS, Vickers KC, Sethupathy P . Inhibition of miR-29 has a significant lipid-lowering benefit through suppression of lipogenic programs in liver. Sci Rep, 2015,5:12911.
doi: 10.1038/srep12911 pmid: 26246194 |
| [81] |
Pang JQ, Xiong H, Yang HD, Ou YK, Xu YD, Huang QH, Lai L, Chen SJ, Zhang ZG, Cai YX, Zheng YQ . Circulating miR-34a levels correlate with age-related hearing loss in mice and humans. Exp Gerontol, 2016,76:58-67.
doi: 10.1016/j.exger.2016.01.009 pmid: 26802970 |
| [82] |
Ling H, Fabbri M, Calin GA . MicroRNAs and other non-coding RNAs as targets for anticancer drug development. Nat Rev Drug Discov, 2013,12(11):847-865.
doi: 10.1038/nrd4140 |
| [83] |
Chiang DY, Cuthbertson DW, Ruiz FR, Li N, Pereira FA . A coregulatory network of NR2F1 and microRNA-140. PLoS One, 2013,8(12):e83358.
doi: 10.1371/journal.pone.0083358 pmid: 24349493 |
| [84] |
Lee YJ, Bernstock JD, Klimanis D, Hallenbeck JM . Akt protein kinase, miR-200/miR-182 expression and epithelial- mesenchymal transition proteins in hibernating ground squirrels. Front Mol Neurosci, 2018,11:22.
doi: 10.3389/fnmol.2018.00022 pmid: 29440989 |
| [85] |
Burk U, Schubert J, Wellner U, Schmalhofer O, Vincan E, Spaderna S, Brabletz T . A reciprocal repression between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cells. Embo Rep, 2008,9(6):582-589.
doi: 10.1038/embor.2008.74 pmid: 18483486 |
| [86] |
Li YZ, Peng AQ, Ge SL, Wang Q, Liu JJ . miR-204 suppresses cochlear spiral ganglion neuron survival in vitro by targeting TMPRSS3. Hear Res, 2014,314:60-64.
doi: 10.1016/j.heares.2014.05.002 pmid: 24924414 |
| [87] |
Rudnicki A, Shivatzki S, Beyer LA, Takada Y, Raphael Y, Avraham KB . microRNA-224 regulates Pentraxin 3, a component of the humoral arm of innate immunity, in inner ear inflammation. Hum Mol Genet, 2014,23(12):3138-3146.
doi: 10.1093/hmg/ddu023 |
| [88] |
Kiernan AE, Xu JX, Gridley T . The Notch ligand JAG1 is required for sensory progenitor development in the mammalian inner ear. PLoS Genet, 2006,2(1):e4.
doi: 10.1371/journal.pgen.0020004 pmid: 16410827 |
| [89] |
Pan BF, Akyuz N, Liu XP, Asai Y, Nist-Lund C, Kurima K, Derfler BH, Gyorgy B, Limapichat W, Walujkar S, Wimalasena LN, Sotomayor M, Corey DP, Holt JR. TMC1 forms the pore of mechanosensory transduction channels in vertebrate inner ear hair cells. Neuron, 2018, 99(4): 736-753. e6.
doi: 10.1016/j.neuron.2018.07.033 pmid: 30138589 |
| [90] |
Müller U, Barr-Gillespie PG . New treatment options for hearing loss. Nat Rev Drug Discov, 2015,14(5):346-385.
doi: 10.1038/nrd4533 pmid: 25792261 |
| [91] |
Bermingham NA, Hassan BA, Price SD, Vollrath MA, Ben-Arie N, Eatock RA, Bellen HJ, Lysakowski A, Zoghbi HY . Math1: an essential gene for the generation of inner ear hair cells. Science, 1999,284(5421):1837-1841.
doi: 10.1126/science.284.5421.1837 pmid: 10364557 |
| [92] |
Fan QQ, Meng FL, Fang R, Li GP, Zhao XL . Functions of Wnt signaling pathway in hair cell differentiation and regeneration. Hereditas(Beijing), 2017,39(10):897-907.
doi: 10.16288/j.yczz.17-037 pmid: 29070485 |
|
范晴晴, 孟飞龙, 房冉, 李高鹏, 赵小立 . Wnt信号通路在毛细胞分化和再生过程中的作用. 遗传, 2017,39(10):897-907.
doi: 10.16288/j.yczz.17-037 pmid: 29070485 |
|
| [93] |
Slowik AD , Bermingham-McDonogh O. Hair cell generation by notch inhibition in the adult mammalian cristae. Jaro-J Assoc Res Oto, 2013,14(6):813-828.
doi: 10.1007/s10162-013-0414-z |
| [94] |
Tateya T, Imayoshi I, Tateya I, Hamaguchi K, Torii H, Ito J, Kageyama R . Hedgehog signaling regulates prosensory cell properties during the basal-to-apical wave of hair cell differentiation in the mammalian cochlea. Development, 2013,140(18):3848-3857.
doi: 10.1242/dev.095398 |
| [95] |
Mansour SL, Noyes CA, Li CY, Wang XF, Hatch E, Twigg S, WIlkie AOM, Urness L . FGF signaling in inner ear development. FASEB J, 2009,23.
doi: 10.1096/fj.09-1201ufm pmid: 19948522 |
| [96] |
Kim HJ, Kang KY, Baek JG, Jo HC, Kim H . Expression of TGFβ family in the developing internal ear of rat embryos. J Korean Med Sci, 2006,21(1):136-142.
doi: 10.3346/jkms.2006.21.1.136 pmid: 16479080 |
| [97] |
Geng RS, Noda T, Mulvaney JF, Lin VYW, Edge ASB, Dabdoub A . Comprehensive expression of wnt signaling pathway genes during development and maturation of the mouse cochlea. PLoS One, 2016,11(2):e0148339.
doi: 10.1371/journal.pone.0148339 pmid: 26859490 |
| [98] |
Chen C, Xiang H, Peng YL, Peng J, Jiang SW . Mature miR-183, negatively regulated by transcription factor GATA3, promotes 3T3-L1 adipogenesis through inhibition of the canonical Wnt/β-catenin signaling pathway by targeting LRP6. Cell Signal, 2014,26(6):1155-1165.
doi: 10.1016/j.cellsig.2014.02.003 |
| [99] |
Tang XL, Zheng D, Hu P, Zeng ZY, Li M, Tucker L, Monahan R, Resnick MB, Liu M, Ramratnam B . Glycogen synthase kinase 3 beta inhibits microRNA- 183-96-182 cluster via the β-Catenin/TCF/LEF-1 pathway in gastric cancer cells. Nucleic Acids Res, 2014,42(5):2988-2998.
doi: 10.1093/nar/gkt1275 pmid: 24335145 |
| [100] |
Hartman BH, Reh TA , Bermingham-McDonogh O. Notch signaling specifies prosensory domains via lateral induction in the developing mammalian inner ear. Proc Natl Acad Sci USA, 2010,107(36):15792-15797.
doi: 10.1073/pnas.1002827107 pmid: 20798046 |
| [101] |
Chen ZB Pu MM, Yao J, Cao X, Cheng L . Screening of microRNAs targeting Notch signaling pathway implicated in inner ear development and the role of microRNA-384-5p. Chin J Otorhinol Head Neck Surg, 2018,53(11):830-837.
doi: 10.3760/cma.j.issn.1673-0860.2018.11.007 pmid: 30453402 |
| [102] |
Zhou W, Du JT, Jiang D, Wang XR, Chen KT, Tang HC, Zhang XM, Cao H, Zong L, Dong C , Jiang HY. microRNA-183 is involved in the differentiation and regeneration of Notch signaling-prohibited hair cells from mouse cochlea. Mol Med Rep, 2018,18(2):1253-1262.
doi: 10.3892/mmr.2018.9127 pmid: 29901127 |
| [103] |
Yang Z, Yao J, Cao X . Roles of the FGF signaling pathway in regulating inner ear development and hair cell regeneration. Hereditas(Beijing), 2018,40(7):515-524
doi: 10.16288/j.yczz.17-407 pmid: 30021715 |
|
杨志, 姚俊, 曹新 . FGF信号通路在内耳发育调控和毛细胞再生中的作用. 遗传, 2018,40(7):515-524.
doi: 10.16288/j.yczz.17-407 pmid: 30021715 |
|
| [104] |
Brennecke J, Hipfner DR, Stark A, Russell RB, Cohen SM . Bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell, 2003,113(1):25-36.
doi: 10.1016/s0092-8674(03)00231-9 pmid: 12679032 |
| [105] |
Tan PX, Du SS, Ren C, Yao QW, Zheng R, Li R, Yuan YW . MicroRNA-207 enhances radiation-induced apoptosis by directly targeting akt3 in cochlea hair cells. Cell Death Dis, 2014,5:e1433.
doi: 10.1038/cddis.2014.407 pmid: 25275594 |
| [106] |
Yamashita H, Takahashi M, Bagger-Sjöbäck D . Expression of epidermal growth factor, epidermal growth factor receptor and transforming growth factor-alpha in the human fetal inner ear. Eur Arch Oto-Rhino-L, 1996,253(8):494-497.
doi: 10.1007/bf00179956 pmid: 8950550 |
| [107] |
Lu YY, Zheng JY, Liu J, Huang CL, Zhang W, Zeng Y. miR-183 induces cell proliferation, migration, and invasion by regulating PDCD4 expression in the SW1990 pancreatic cancer cell line. Biomed Pharmacother, 2015,70:151-157.
doi: 10.1016/j.biopha.2015.01.016 pmid: 25776494 |
| [108] |
Mendell JT, Olson EN . MicroRNAs in stress signaling and human disease. Cell, 2012,148(6):1172-1187.
doi: 10.1016/j.cell.2012.02.005 |
| [109] |
Cui J, Zhou B, Ross SA, Zempleni J . Nutrition, microRNAs, and human health. Adv Nutr, 2017,8(1):105-112.
doi: 10.3945/an.116.013839 pmid: 28096131 |
| [110] |
Miguel V, Cui JY, Daimiel L, Espinosa-Díez C, Fernández-Hernando C, Kavanagh TJ, Lamas S . The role of microRNAs in environmental risk factors, noise- induced hearing loss, and mental stress. Antioxid Redox Sign, 2018,28(9):773-796.
doi: 10.1089/ars.2017.7175 pmid: 28562070 |
| [111] |
Bardin P, Sonneville F, Corvol H, Tabary O . Emerging microRNA therapeutic approaches for cystic fibrosis. Front Pharmacol, 2018,9:1113.
doi: 10.3389/fphar.2018.01113 pmid: 30349480 |
| [112] |
Takeda H, Dondzillo A, Randall JA, Gubbels SP . Challenges in cell-based therapies for the treatment of hearing loss. Trends Neurosci, 2018,41(11):823-837.
doi: 10.1016/j.tins.2018.06.008 pmid: 30033182 |
| [113] |
Simoni E, Orsini G, Chicca M, Bettini S, Franceschini V, Martini A, Astolfi L . Regenerative medicine in hearing recovery. Cytotherapy, 2017,19(8):909-915.
doi: 10.1016/j.jcyt.2017.04.008 pmid: 28532627 |
| [114] |
Shrestha BR, Chia C, Wu L, Kujawa SG, Liberman MC, Goodrich LV. Sensory neuron diversity in the inner ear is shaped by activity. Cell, 2018, 174(5): 1229-1246. e17.
doi: 10.1016/j.cell.2018.07.007 pmid: 30078709 |
| [115] |
Sennett R, Rendl M . Mesenchymal-epithelial interactions during hair follicle morphogenesis and cycling. Semin Cell Dev Biol, 2012,23(8):917-927.
doi: 10.1016/j.semcdb.2012.08.011 pmid: 22960356 |
| [116] |
Chen H, Sun YM, Dong RQ, Yang SS, Pan CY, Xiang D, Miao MY, Jiao BH . Mir-34a is upregulated during liver regeneration in rats and is associated with the supperssion of hepatocyte proliferation. PLoS One, 2011,6(5):e20238.
doi: 10.1371/journal.pone.0020238 pmid: 21655280 |
| [117] |
Liang DD, Li J, Wu YH, Zhen LX, Li CM, Qi M, Wang LJ, Deng FF, Huang J, Lv F, Liu Y, Ma C, Yu ZR, Zhang YZ, Chen YH. miRNA-204 drives cardiomyocyte proliferation via targeting Jarid2. Int J Cardiol, 2015,201:38-48.
doi: 10.1016/j.ijcard.2015.06.163 pmid: 26298346 |
| [118] |
Vos T, Abajobir AA, Abate KH, Abbafati C, Abbas KM, Abd-Allah F, Abdulkader RS, Abdulle AM, Abebo TA, Abera SF, Aboyans V, Abu-Raddad LJ, Ackerman IN, Adamu AA, Adetokunboh O, Afarideh M, Afshin A, Agarwal SK, Aggarwal R, Agrawal A, Agrawal S, Ahmadieh H, Ahmed MB, Aichour MTE, Aichour AN, Aichour I, Aiyar S, Akinyemi RO, Akseer N, Al Lami FH, Alahdab F, Al-Aly Z, Alam K, Alam N, Alam T, Alasfoor D, Alene KA, Ali R, Alizadeh-Navaei R, Alkerwi A, Alla F, Allebeck P, Allen C, Al-Maskari F, Al-Raddadi R, Alsharif U, Alsowaidi S, Altirkawi KA, Amare AT, Amini E, Ammar W, Amoako YA, Andersen HH, Antonio CAT, Anwari P, Ärnlöv J, Artaman A, Aryal KK, Asayesh H, Asgedom SW, Assadi R, Atey TM, Atnafu NT, Atre SR, Avila-Burgos L, Avokphako EFGA, Awasthi A, Bacha U, Badawi A, Balakrishnan K, Banerjee A, Bannick MS, Barac A, Barber RM, Barker- Collo SL, Bärnighausen T, Barquera S, Barregard L, Barrero LH, Basu S, Battista B, Battle KE, Baune BT, Bazargan-Hejazi S, Beardsley J, Bedi N, Beghi E, Béjot Y, Bekele BB, Bell ML, Bennett DA, Bensenor IM, Benson J, Berhane A, Berhe DF, Bernabé E, Betsu BD, Beuran M, Beyene AS, Bhala N, Bhansali A, Bhatt S, Bhutta ZA, Biadgilign S, Bicer BK, Bienhoff K, Bikbov B, Birungi C, Biryukov S, Bisanzio D, Bizuayehu HM, Boneya DJ, Boufous S, Bourne RRA, Brazinova A, Brugha TS, Buchbinder R, Bulto LNB, Bumgarner BR, Butt ZA, Cahuana-Hurtado L, Cameron E, Car M, Carabin H, Carapetis JR, Cárdenas R, Carpenter DO, Carrero JJ, Carter A, Carvalho F, Casey DC, Caso V, Castañeda-Orjuela CA, Castle CD, Catalá-López F, Chang HY, Chang JC, Charlson FJ, Chen H, Chibalabala M, Chibueze CE, Chisumpa VH, Chitheer AA, Christopher DJ, Ciobanu LG, Cirillo M, Colombara D, Cooper C, Cortesi PA, Criqui MH, Crump JA, Dadi AF, Dalal K, Dandona L, Dandona R, das Neves J, Davitoiu DV, de Courten B, De Leo D, Defo BK, Degenhardt L, Deiparine S, Dellavalle RP, Deribe K, Des Jarlais DC, Dey S, Dharmaratne SD, Dhillon PK, Dicker D, Ding EL, Djalalinia S, Do HP, Dorsey ER, Dos Santos KPB, Douwes-Schultz D, Doyle KE, Driscoll TR, Dubey M, Duncan BB, El-Khatib ZZ, Ellerstrand J, Enayati A, Endries AY, Ermakov SP, Erskine HE, Eshrati B, Eskandarieh S, Esteghamati A, Estep K, Fanuel FBB, Farinha CSES, Faro A, Farzadfar F, Fazeli MS, Feigin VL, Fereshtehnejad SM, Fernandes JC, Ferrari AJ, Feyissa TR, Filip I, Fischer F, Fitzmaurice C, Flaxman AD, Flor LS, Foigt N, Foreman KJ, Franklin RC, Fullman N, Fürst T, Furtado JM, Futran ND, Gakidou E, Ganji M, Garcia-Basteiro AL, Gebre T, Gebrehiwot TT, Geleto A, Gemechu BL, Gesesew HA, Gething PW, Ghajar A, Gibney KB, Gill PS, Gillum RF, Ginawi IAM, Giref AZ, Gishu MD, Giussani G, Godwin WW, Gold AL, Goldberg EM, Gona PN, Goodridge A, Gopalani SV, Goto A, Goulart AC, Griswold M, Gugnani HC, Gupta R, Gupta R, Gupta T, Gupta V, Hafezi-Nejad N, Hailu GB, Hailu AD, Hamadeh RR, Hamidi S, Handal AJ, Hankey GJ, Hanson SW, Hao Y, Harb HL, Hareri HA, Haro JM, Harvey J, Hassanvand MS, Havmoeller R, Hawley C, Hay SI, Hay RJ, Henry NJ, Heredia-Pi IB, Hernandez JM, Heydarpour P, Hoek HW, Hoffman HJ, Horita N, Hosgood HD, Hostiuc S, Hotez PJ, Hoy DG, Htet AS, Hu G, Huang H, Huynh C, Iburg KM, Igumbor EU, Ikeda C, Irvine CMS, Jacobsen KH, Jahanmehr N, Jakovljevic MB, Jassal SK, Javanbakht M, Jayaraman SP, Jeemon P, Jensen PN, Jha V, Jiang G, John D, Johnson SC, Johnson CO, Jonas JB, Jürisson M, Kabir Z, Kadel R, Kahsay A, Kamal R, Kan H, Karam NE, Karch A, Karema CK, Kasaeian A, Kassa GM, Kassaw NA, Kassebaum NJ, Kastor A, Katikireddi SV, Kaul A, Kawakami N, Keiyoro PN, Kengne AP, Keren A, Khader YS, Khalil IA, Khan EA, Khang YH, Khosravi A, Khubchandani J, Kiadaliri AA, Kieling C, Kim YJ, Kim D, Kim P, Kimokoti RW, Kinfu Y, Kisa A, Kissimova-Skarbek KA, Kivimaki M, Knudsen AK, Kokubo Y, Kolte D, Kopec JA, Kosen S, Koul PA, Koyanagi A, Kravchenko M, Krishnaswami S, Krohn KJ, Kumar GA, Kumar P, Kumar S, Kyu HH, Lal DK, Lalloo R, Lambert N, Lan Q, Larsson A, Lavados PM, Leasher JL, Lee PH, Lee JT, Leigh J, Leshargie CT, Leung J, Leung R, Levi M, Li Y, Li Y, Li Kappe D, Liang X, Liben ML, Lim SS, Linn S, Liu PY, Liu A, Liu S, Liu Y, Lodha R, Logroscino G, London SJ, Looker KJ, Lopez AD, Lorkowski S, Lotufo PA, Low N, Lozano R, Lucas TCD, Macarayan ERK, Magdy Abd El Razek H, Magdy Abd El Razek M, Mahdavi M, Majdan M, Majdzadeh R, Majeed A, Malekzadeh R, Malhotra R, Malta DC, Mamun AA, Manguerra H, Manhertz T, Mantilla A, Mantovani LG, Mapoma CC, Marczak LB, Martinez-Raga J, Martins-Melo FR, Martopullo I, März W, Mathur MR, Mazidi M, McAlinden C, McGaughey M, McGrath JJ, McKee M, McNellan C, Mehata S, Mehndiratta MM, Mekonnen TC, Memiah P, Memish ZA, Mendoza W, Mengistie MA, Mengistu DT, Mensah GA, Meretoja TJ, Meretoja A, Mezgebe HB, Micha R, Millear A, Miller TR, Mills EJ, Mirarefin M, Mirrakhimov EM, Misganaw A, Mishra SR, Mitchell PB, Mohammad KA, Mohammadi A, Mohammed KE, Mohammed S, Mohanty SK, Mokdad AH, Mollenkopf SK, Monasta L, Montico M, Moradi-Lakeh M, Moraga P, Mori R, Morozoff C, Morrison SD, Moses M, Mountjoy-Venning C, Mruts KB, Mueller UO, Muller K, Murdoch ME, Murthy GVS, Musa KI, Nachega JB, Nagel G, Naghavi M, Naheed A, Naidoo KS, Naldi L, Nangia V, Natarajan G, Negasa DE, Negoi RI, Negoi I, Newton CR, Ngunjiri JW, Nguyen TH, Nguyen QL, Nguyen CT, Nguyen G, Nguyen M, Nichols E, Ningrum DNA, Nolte S, Nong VM, Norrving B, Noubiap JJN, O'Donnell MJ, Ogbo FA, Oh IH, Okoro A, Oladimeji O, Olagunju TO, Olagunju AT, Olsen HE, Olusanya BO, Olusanya JO, Ong K, Opio JN, Oren E, Ortiz A, Osgood-Zimmerman A, Osman M, Owolabi MO, Pa M, Pacella RE, Pana A, Panda BK, Papachristou C, Park EK, Parry CD, Parsaeian M, Patten SB, Patton GC, Paulson K, Pearce N, Pereira DM, Perico N, Pesudovs K, Peterson CB, Petzold M, Phillips MR, Pigott DM, Pillay JD, Pinho C, Plass D, Pletcher MA, Popova S, Poulton RG, Pourmalek F, Prabhakaran D, Prasad NM, Prasad N, Purcell C, Qorbani M, Quansah R, Quintanilla BPA, Rabiee RHS, Radfar A, Rafay A, Rahimi K, Rahimi-Movaghar A, Rahimi-Movaghar V, Rahman MHU, Rahman M, Rai RK, Rajsic S, Ram U, Ranabhat CL, Rankin Z, Rao PC, Rao PV, Rawaf S, Ray SE, Reiner RC, Reinig N, Reitsma MB, Remuzzi G, Renzaho AMN, Resnikoff S, Rezaei S, Ribeiro AL, Ronfani L, Roshandel G, Roth GA, Roy A, Rubagotti E, Ruhago GM, Saadat S, Sadat N, Safdarian M, Safi S, Safiri S, Sagar R, Sahathevan R, Salama J, Saleem HOB, Salomon JA, Salvi SS, Samy AM, Sanabria JR, Santomauro D, Santos IS, Santos JV, Santric Milicevic MM, Sartorius B, Satpathy M, Sawhney M, Saxena S, Schmidt MI, Schneider IJC, Schöttker B, Schwebel DC, Schwendicke F, Seedat S, Sepanlou SG, Servan-Mori EE, Setegn T, Shackelford KA, Shaheen A, Shaikh MA, Shamsipour M, Shariful Islam SM, Sharma J, Sharma R, She J, Shi P, Shields C, Shifa GT, Shigematsu M, Shinohara Y, Shiri R, Shirkoohi R, Shirude S, Shishani K, Shrime MG, Sibai AM, Sigfusdottir ID, Silva DAS, Silva JP, Silveira DGA, Singh JA, Singh NP, Sinha DN, Skiadaresi E, Skirbekk V, Slepak EL, Sligar A, Smith DL, Smith M, Sobaih BHA, Sobngwi E, Sorensen RJD, Sousa TCM, Sposato LA, Sreeramareddy CT, Srinivasan V, Stanaway JD, Stathopoulou V, Steel N, Stein MB, Stein DJ, Steiner TJ, Steiner C, Steinke S, Stokes MA, Stovner LJ, Strub B, Subart M, Sufiyan MB, Sunguya BF, Sur PJ, Swaminathan S, Sykes BL, Sylte DO, Tabarés-Seisdedos R, Taffere GR, Takala JS, Tandon N, Tavakkoli M, Taveira N, Taylor HR, Tehrani-Banihashemi A, Tekelab T, Terkawi AS, Tesfaye DJ, Tesssema B, Thamsuwan O, Thomas KE, Thrift AG, Tiruye TY, Tobe-Gai R, Tollanes MC, Tonelli M, Topor-Madry R, Tortajada M, Touvier M, Tran BX, Tripathi S, Troeger C, Truelsen T, Tsoi D, Tuem KB, Tuzcu EM, Tyrovolas S, Ukwaja KN, Undurraga EA, Uneke CJ, Updike R, Uthman OA, Uzochukwu BSC, van Boven JFM, Varughese S, Vasankari T, Venkatesh S, Venketasubramanian N, Vidavalur R, Violante FS, Vladimirov SK, Vlassov VV, Vollset SE, Wadilo F, Wakayo T, Wang YP, Weaver M, Weichenthal S, Weiderpass E, Weintraub RG, Werdecker A, Westerman R, Whiteford HA, Wijeratne T, Wiysonge CS, Wolfe CDA, Woodbrook R, Woolf AD, Workicho A, Xavier D, Xu G, Yadgir S, Yaghoubi M, Yakob B, Yan LL, Yano Y, Ye P, Yimam HH, Yip P, Yonemoto N, Yoon SJ, Yotebieng M, Younis MZ, Zaidi Z, Zaki MES, Zegeye EA, Zenebe ZM, Zhang X, Zhou M, Zipkin B, Zodpey S, Zuhlke LJ, Murray CJL . Global, regional, and national incidence, prevalence, and years lived with disability for 328 diseases and injuries for 195 countries, 1990-2016: a systematic analysis for the Global Burden of Disease Study 2016. Lancet, 2017,390(10100):1211-1259.
doi: 10.1016/S0140-6736(17)32154-2 pmid: 28919117 |
| [119] |
He P, Luo YN, Hu XY, Gong R, Wen X, Zheng XY . Association of socioeconomic status with hearing loss in Chinese working-aged adults: A population-based study. PLoS One, 2018,13(3):e0195227.
doi: 10.1371/journal.pone.0195227 pmid: 29596478 |
| [120] |
Soukup GA . Little but loud: Small RNAs have a resounding affect on ear development. Brain Res, 2009,1277:104-114.
doi: 10.1016/j.brainres.2009.02.027 pmid: 19245798 |
| [1] | Huijie Yang, De Li, Huiling Bai, Ming Zhang, Jun Huang, Xiaoqing Yuan. Diagnosis, treatment and genetic analysis of a case of Alstrom syndrome caused by compoud heterozygous mutation of ALMS1 [J]. Hereditas(Beijing), 2022, 44(12): 1148-1157. |
| [2] | Yan Zhu, Ming Wei, Xiao Zhou, Linhua Deng, Jian Qiu, Guo Li, Shiqiang Zhou, Hao Xie, Desheng Li, Chengdong Wang. Progress on miRNA in giant panda (Ailuropoda melanoleuca) [J]. Hereditas(Beijing), 2021, 43(9): 849-857. |
| [3] | Yong Wei, Yulan He, Xueli Zheng. Research progress in RNA interference against the infection of mosquito-borne viruses [J]. Hereditas(Beijing), 2020, 42(2): 153-160. |
| [4] | Wenquan Liang,Yu Hou,Cunyou Zhao. Schizophrenia-associated single nucleotide polymorphisms affecting microRNA function [J]. Hereditas(Beijing), 2019, 41(8): 677-685. |
| [5] | Xia Mengmeng,Shen Xueyi,Niu Changmin,Xia Jing,Sun Hongya,Zheng Ying. MicroRNA regulates Sertoli cell proliferation and adhesion [J]. Hereditas(Beijing), 2018, 40(9): 724-732. |
| [6] | Hailong Liu, Yang Shen, Yang Gao, Ling Zhou, Xiaosong Han, Changzhi Zhao, Gaojuan Yang, Yilong Chen, Hui Yang, Shengsong Xie. Assessing abundance and specificity of different types of sgRNA targeting miRNA precursors [J]. Hereditas(Beijing), 2018, 40(7): 561-571. |
| [7] | Juan Xiao, Xun Wang, Yi Luo, Xiaokai Li, Xuewei Li. Research progress in sRNAs and functional proteins in epididymosomes [J]. Hereditas(Beijing), 2018, 40(3): 197-206. |
| [8] | Cuicui Wang,Huijun Yuan. Application and progress of high-throughput sequencing technologies in the research of hereditary hearing loss [J]. Hereditas(Beijing), 2017, 39(3): 208-219. |
| [9] | Xinyun Li, Liangliang Fu, Huijun Cheng, Shuhong Zhao. Advances on microRNA in regulating mammalian skeletal muscle development [J]. Hereditas(Beijing), 2017, 39(11): 1046-1053. |
| [10] | Chendong Liu, Lu Yang, Hongzhou Pu, Qiong Yang, Wenyao Huang, Xue Zhao, Li Zhu, Shunhua Zhang. Epigenetics regulates gene expression patterns of skeletal muscle induced by physical exercise [J]. Hereditas(Beijing), 2017, 39(10): 888-896. |
| [11] | Jun Wei,Xiujun Lu,Xiaolin Zhang,Mei Mei,Xiaoli Huang. Functions of microRNA in seed development, dormancy and germination processes [J]. Hereditas(Beijing), 2017, 39(1): 14-21. |
| [12] | Ke Zhang, Guangde Feng, Baoyun Zhang, Wei Xiang, Long Chen, Fang Yang, Mingxing Chu, Pingqing Wang. Application of epigenetic markers in molecular breeding of the swine [J]. Hereditas(Beijing), 2016, 38(7): 634-643. |
| [13] | Mei Fu, Kehui Xu, Wenming Xu. Research advances of Dicer in regulating reproductive function [J]. HEREDITAS(Beijing), 2016, 38(7): 612-622. |
| [14] | Long Chen, Baoyun Zhang, Guangde Feng, Wei Xiang, Yunxia Ma, Hang Chen, Mingxing Chu, Pingqing Wang. The mechanism of miRNA-mediated PGR signaling pathway in regulating female reproduction [J]. HEREDITAS(Beijing), 2016, 38(1): 40-51. |
| [15] | Xue Zhou, Yilan Du, Ping Jin, Fei Ma. Bioinformatic analysis of cancer-related microRNAs and their target genes [J]. HEREDITAS(Beijing), 2015, 37(9): 855-864. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||