Hereditas(Beijing) ›› 2021, Vol. 43 ›› Issue (9): 849-857.doi: 10.16288/j.yczz.21-209
• Review • Previous Articles Next Articles
Progress on miRNA in giant panda (Ailuropoda melanoleuca)
Yan Zhu, Ming Wei, Xiao Zhou, Linhua Deng, Jian Qiu, Guo Li, Shiqiang Zhou, Hao Xie, Desheng Li, Chengdong Wang( )
)
- State Forestry and Grassland Administration Key Laboratory of Conservation Biology for Rare Animals of the Giant Panda State Park, China Conservation and Research Center for the Giant Panda, Chengdu 611830, China
-
Received:2021-06-11Revised:2021-07-28Online:2021-09-20Published:2021-08-23 -
Contact:Wang Chengdong E-mail:wolongpanda@qq.com -
Supported by:Supported by Giant Panda International Cooperation Fund Project No(GH201710)
Cite this article
Yan Zhu, Ming Wei, Xiao Zhou, Linhua Deng, Jian Qiu, Guo Li, Shiqiang Zhou, Hao Xie, Desheng Li, Chengdong Wang. Progress on miRNA in giant panda (Ailuropoda melanoleuca)[J]. Hereditas(Beijing), 2021, 43(9): 849-857.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
Table 1
Regulatory effects of miRNA in giant panda"
| 生物学功能 | 组织 | 研究策略 | 参考文献 |
|---|---|---|---|
| 调控免疫反应,包括血液免疫、母乳免疫 | 血液、乳汁 | 对不同年龄大熊猫的血液进行miRNA-seq;对接种犬瘟热病毒疫苗后的大熊猫血液进行miRNA-seq;对不同哺乳期的大熊猫母乳外泌体进行miRNA-seq | [ |
| 调控乳腺发育及新生幼崽生长发育 | 血液、乳汁 | 对怀孕后期和哺乳早期大熊猫血液进行miRNA-seq;对不同哺乳期母乳外泌体进行miRNA-seq | [ |
| 调控精子冷冻耐受 | 精子 | 对大熊猫新鲜和冷冻的精子外泌体进行miRNA-seq | [ |
| 调控其他多种生物学过程 | 内脏(心、肝、脾、肺、肾) | 对不同内脏组织进行miRNA-seq;对新生和成年大熊猫的脾脏进行miRNA-seq | [ |
| [1] |
Li Y, Viña A, Yang W, Chen XD, Zhang JD, Ouyang ZY, Liang Z, Liu JG. Effects of conservation policies on forest cover change in giant panda habitat regions, China. Land Use Policy, 2013, 33:42-53.
doi: 10.1016/j.landusepol.2012.12.003 |
| [2] |
Huang GP, Wang X, Hu YB, Wu Q, Nie RG, Dong JH, Ding Y, Yan L, Wei FW. Diet drives convergent evolution of gut microbiomes in bamboo-eating species. Sci China Life Sci, 2021, 64(1):88-95.
doi: 10.1007/s11427-020-1750-7 |
| [3] |
Heiderer M, Westenberg C, Li DS, Zhang HM, Preininger D, Dungl E. Giant panda twin rearing without assistance requires more interactions and less rest of the mother-A case study at Vienna Zoo. PLoS One, 2018, 13(11):e0207433.
doi: 10.1371/journal.pone.0207433 |
| [4] |
Wei W, Swaisgood RR, Owen MA, Pilfold NW, Han H, Hong MS, Zhou H, Wei FW, Nie YG, Zhang ZJ. The role of den quality in giant panda conservation. Biol Conserv, 2019, 231:189-196.
doi: 10.1016/j.biocon.2018.12.031 |
| [5] |
Li T, Luo P, Luo C, Yang H, Li YJ, Zuo DD, Xiong QL, Mo L, Mu CX, Gu XD, Zhou SQ, Huang JY, Li HL, Wu SJ, Cao WQ, Zhang YB, Wang MJ, Li JL, Liu Y, Gou PJ, Zhu ZF, Wang DY, Liang Y, Bai S, Zou Y. Long-term empirical monitoring indicates the tolerance of the giant panda habitat to climate change under contemporary conservation policies. Ecol Indic, 2020, 110:105886.
doi: 10.1016/j.ecolind.2019.105886 |
| [6] |
Han H, Wei W, Hu YB, Nie YG, Ji XP, Yan L, Zhang ZJ, Shi XX, Zhu LF, Luo YB, Chen WC, Wei FW. Diet evolution and habitat contraction of giant pandas via stable isotope analysis. Curr Biol, 2019, 29(4):664-669.
doi: S0960-9822(19)30004-1 pmid: 30713107 |
| [7] |
Figueirido B, Palmqvist P, Pérez-Claros JA, Wei D. Cranial shape transformation in the evolution of the giant panda ( Ailuropoda melanoleuca). Naturwissenschaften, 2011, 98(2):107-116.
doi: 10.1007/s00114-010-0748-x pmid: 21132275 |
| [8] |
Hu YD, Pang HZ, Li DS, Ling SS, Lan D, Wang Y, Zhu Y, Li DY, Wei RP, Zhang HM, Wang CD. Analysis of the cytochrome c oxidase subunit 1 ( COX1) gene reveals the unique evolution of the giant panda. Gene, 2016, 592(2):303-307.
doi: 10.1016/j.gene.2016.07.029 |
| [9] |
Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell, 1993, 75(5):843.
pmid: 8252621 |
| [10] |
Kozomara A, Birgaoanu M, Griffiths-Jones S. miRBase: from microRNA sequences to function. Nucleic Acids Res, 2019, 47(D1):D155-D162.
doi: 10.1093/nar/gky1141 |
| [11] |
Gupta V, Markmann K, Pedersen CNS, Stougaard J, Andersen SU. Shortran: a pipeline for small RNA-seq data analysis. Bioinformatics, 2012, 28(20):2698-2700.
doi: 10.1093/bioinformatics/bts496 |
| [12] |
Yang F, Yi F, Cao HQ, Du Q, Liang ZC. The emerging landscape of long non-coding RNAs. Hereditas(Beijing), 2014, 36(5):456-468.
pmid: 24846995 |
|
杨峰, 易凡, 曹慧青, 杜权, 梁子才. 长链非编码RNA研究进展. 遗传, 2014, 36(5):456-468.
pmid: 24846995 |
|
| [13] |
Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature, 2000, 403(6772):901-906.
doi: 10.1038/35002607 |
| [14] |
Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem, 2010, 79(1):351-379.
doi: 10.1146/annurev-biochem-060308-103103 |
| [15] |
Nahvi A, Shoemaker CJ, Green R. An expanded seed sequence definition accounts for full regulation of the hid 3' UTR by bantam miRNA. RNA, 2009, 15(5):814-822.
doi: 10.1261/rna.1565109 pmid: 19286629 |
| [16] |
Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell, 2009, 136(2):215-233.
doi: 10.1016/j.cell.2009.01.002 pmid: 19167326 |
| [17] |
Jones-Rhoades MW, Bartel DP. Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol Cell, 2004, 14(6):787-799.
pmid: 15200956 |
| [18] |
Brennecke J, Stark A, Russell RB, Cohen SM. Principles of microRNA-target recognition. PLoS Biol, 2005, 3(3):e85.
doi: 10.1371/journal.pbio.0030085 |
| [19] |
Krek A, Grün D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, Macmenamin P, da Piedade I, Gunsalus KC, Stoffel M, Rajewsky N. Combinatorial microRNA target predictions. Nat Genet, 2005, 37(5):495-500.
doi: 10.1038/ng1536 |
| [20] |
Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell, 2005, 120(1):15-20.
doi: 10.1016/j.cell.2004.12.035 |
| [21] |
Shimono Y, Zabala M, Cho RW, Lobo N, Dalerba P, Qian DL, Diehn M, Liu HP, Panula SP, Chiao E, Dirbas FM, Somlo G, Reijo Pera RA, Lao KQ, Clarke MF. Downregulation of miRNA-200c links breast cancer stem cells with normal stem cells. Cell, 2009, 138(3):592-603.
doi: 10.1016/j.cell.2009.07.011 pmid: 19665978 |
| [22] |
Zhao Y, Ransom JF, Li AK, Vedantham V, von Drehle M, Muth AN, Tsuchihashi T, McManus MT, Schwartz RJ, Srivastava D. Dysregulation of cardiogenesis, cardiac conduction, and cell cycle in mice lacking miRNA-1-2. Cell, 2007, 129(2):303-317.
doi: 10.1016/j.cell.2007.03.030 |
| [23] | Zhang QH, Chang B, Zheng GZ, Du SX, Li XD. Quercetin stimulates osteogenic differentiation of bone marrow stromal cells through miRNA-206/connexin 43 pathway. Am J Transl Res, 2020, 12(5):2062-2070. |
| [24] |
Ma T, Li JY, Li JP, Wu SF, Ba X, Jiang HZ, Zhang QL. Expression of miRNA-203 and its target gene in hair follicle cycle development of cashmere goat. Cell Cycle, 2021, 20(2):1-7.
doi: 10.1080/15384101.2020.1864941 |
| [25] |
Gong ZD, Niu YH, Yuan ZF, Yang J, Wei SC. MiRNA-let-7b decreases proliferation activities and development of follicular cells via targeting MAP3K1 gene. Vet Arhiv, 2021, 91(2):149-158.
doi: 10.24099/vet.arhiv |
| [26] | Zhao Y, Cao XY, Zhou HT, Song LY, Tu HQ, Huang SY, Zhao JL. Analysis of miRNA transcriptome in early developmental stage and identification of growth-related miRNA of siniperca chuatsi. Biotechnol Bull, 2018, 34(8):181-189. |
| 赵岩, 曹晓颖, 周昊天, 宋凌元, 涂翰卿, 黄思颖, 赵金良. 鳜不同孵化时期miRNA转录组分析及生长相关miRNA鉴定. 生物技术通报, 2018, 34(8):181-189. | |
| [27] |
Wang LM, Zhu WB, Dong ZJ, Song FB, Dong JJ, Fu JJ. Comparative microRNA-seq analysis depicts candidate miRNAs involved in skin color differentiation in red tilapia. Int J Mol Sci, 2018, 19(4):1209.
doi: 10.3390/ijms19041209 |
| [28] |
Zhou J, Zhao H, Zhang L, Liu C, Feng SY, Ma JD, Li Q, Ke HY, Wang XY, Liu LY, Liu C, Su XT, Liu YK, Yang S. Integrated analysis of RNA-seq and microRNA-seq depicts miRNA-mRNA networks involved in stripe patterns of Botia superciliaris skin. Funct Integr Genomics, 2019, 19(5):827-838.
doi: 10.1007/s10142-019-00683-2 |
| [29] |
Wang N, Wang RK, Wang RQ, Chen SL. RNA-seq and microRNA-seq analysis of Japanese flounder ( Paralichthys olivaceus) larvae treated by thyroid hormones. Fish Physiol Biochem, 2019, 45(4):1233-1244.
doi: 10.1007/s10695-019-00654-1 pmid: 31115741 |
| [30] | Ma Y. Transcriptome sequencing and analysis of milk exosomal miRNA of columba livia[Dissertation]. Sichuan Agricultural University, 2018. |
| 马瑶. 鸽乳exosomal miRNA转录组测序与分析[学位论文]. 四川农业大学, 2018. | |
| [31] | Yuan M, Jiang FM, Xu YO, Lin YQ, Jiang XS, Yang CW, Yu CL, Li ZX. Analysis of transcriptome and microRNA in leg muscle of tibetan chicken at different developmental stages. Acta Vet Et Zootech Sin, 2019, 50(12): 2400-2412. |
| 袁茂, 江明锋, 徐亚欧, 林亚秋, 蒋小松, 杨朝武, 余春林, 李志雄. 藏鸡不同发育阶段腿部肌肉组织转录组及microRNA联合分析. 畜牧兽医学报, 2019, 50(12):2400-2412. | |
| [32] | Long KR. The miRNA transcriptome level reveals the mechanism of multi-species adaptation to high altitude [Dissertation]. Sichuan Agricultural University, 2018. |
| 龙科任. 从miRNA转录组层面揭示多物种的高海拔适应性机制[学位论文]. 四川农业大学, 2018. | |
| [33] | Zhang SF. Identification and characterization of miRNA transcriptome in sheep[Dissertation]. Chinese Academy of Agricultural Sciences, 2013. |
| 张世芳. 绵羊miRNA转录组的鉴定与特征分析[学位论文]. 中国农业科学院, 2013. | |
| [34] | Xie LL, Li Y, Huang WL, Zhang XX, Miao XY. Identification and analysis of miRNA in ovary of sheep at different developmental stages. Acta Vet Et Zootech Sin, 2019, 50(7):1396-1404. |
| 解领丽, 李嫒, 黄万龙, 张秀秀, 苗向阳. 湖羊卵巢不同发育阶段的miRNA鉴定与分析. 畜牧兽医学报, 2019, 50(7):1396-1404. | |
| [35] | Bao LY, Cao BY, Liu YH, Ma Y, An XP, Zhang Y, Zhang M, Wang JG, Du B, Li G. Regulation of miR-92a on proliferation and apoptosis of mammary epithelial cells of dairy goats. Acta Vet Et Zootech Sin, 2020, 51(1):137-149. |
| 包黎娟, 曹斌云, 刘育含, 马毅, 安小鹏, 张月, 张梦, 王建刚, 堵斌, 李广. miR-92a对奶山羊乳腺上皮细胞增殖及凋亡的调控分析. 畜牧兽医学报, 2020, 51(1):137-149. | |
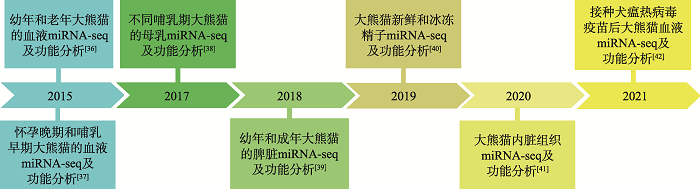
| [36] |
Yang MY, Du LM, Li WJ, Shen FJ, Fan ZX, Jian ZY, Hou R, Shen YM, Yue BS, Zhang XY. Profile of microRNA in giant panda blood: a resource for immune-related and novel microRNAs. PLoS One, 2015, 10(11):e0143242.
doi: 10.1371/journal.pone.0143242 |
| [37] |
Wang CD, Long K, Jin L, Huang S, Li DH, Ma XP, Wei M, Gu Y, Ma JD, Zhang H. Identification of conserved microRNAs in peripheral blood from giant panda: expression of mammary gland-related microRNAs during late pregnancy and early lactation. Genet Mol Res, 2015, 14(4):14216-14228.
doi: 10.4238/2015.November.13.5 pmid: 26600479 |
| [38] |
Ma JD, Wang CD, Long KR, Zhang HM, Zhang JW, Jin L, Tang QZ, Jiang AA, Wang X, Tian SL, Chen L, He DF, Li DS, Huang S, Jiang Z, Li MZ. Exosomal microRNAs in giant panda (Ailuropoda melanoleuca) breast milk: potential maternal regulators for the development of newborn cubs. Sci Rep, 2017, 7(1):3507.
doi: 10.1038/s41598-017-03707-8 |
| [39] |
Peng R, Liu YL, Cai ZG, Shen FJ, Chen JS, Hou R, Zou FD. Characterization and analysis of whole transcriptome of giant panda spleens: implying critical roles of long non-coding RNAs in immunity. Cell Physiol Biochem, 2018, 46(3):1065-1077.
doi: 10.1159/000488837 pmid: 29669315 |
| [40] |
Ran MX, Zhou YM, Liang K, Wang WC, Zhang Y, Zhang M, Yang JD, Zhou GB, Wu K, Wang CD, Huang Y, Luo B, Qazi IH, Zhang HM, Zeng CJ. Comparative analysis of microRNA and mRNA profiles of sperm with different freeze tolerance capacities in boar (Sus scrofa) and giant panda (Ailuropoda melanoleuca). Biomolecules, 2019, 9(9):432.
doi: 10.3390/biom9090432 |
| [41] |
Wang CD, Li F, Deng LH, Li MZ, Wei M, Zeng B, Wu K, Xu ZX, Wei RP, Wei LM, Liu WP, Zhang SY, Xu L, Huang Y, Li DS, Li Y, Zhang HM. Identification and characterization of miRNA expression profiles across five tissues in giant panda. Gene, 2020, 769(5):145206.
doi: 10.1016/j.gene.2020.145206 |
| [42] |
Sun J, Shen FJ, Zhang L, Luo L, Fan ZX, Hou R, Yue BS, Zhang XY. Changes in the microRNA profile of the giant panda after canine distemper vaccination and the integrated analysis of microRNA-messenger RNA. DNA Cell Biol, 2021, 40(4):595-605.
doi: 10.1089/dna.2020.5942 |
| [43] |
Sonkoly E, Ståhle M, Pivarcsi A. MicroRNAs and immunity: novel players in the regulation of normal immune function and inflammation. Semin Cancer Biol, 2008, 18(2):131-140.
doi: 10.1016/j.semcancer.2008.01.005 pmid: 18291670 |
| [44] |
Du LM, Li WJ, Fan ZX, Shen FJ, Yang MY, Wang ZL, Jian ZY, Hou R, Yue BS, Zhang XY. First insights into the giant panda (Ailuropoda melanoleuca) blood transcriptome: a resource for novel gene loci and immunogenetics. Mol Ecol Resour, 2015, 15(4):1001-1013.
doi: 10.1111/1755-0998.12367 |
| [45] | Du LM, Liu Q, Shen FJ, Fan ZX, Hou R, Yue BS, Zhang XY. Transcriptome analysis reveals immune-related gene expression changes with age in giant panda (Ailuropoda melanoleuca) blood. Aging (Albany NY), 2019, 11(1):249-262. |
| [46] | Gartner LM, Morton J, Lawrence RA, Naylor AJ, O'Hare D, Schanler RJ, Eidelman AI. Breastfeeding and the use of human milk. Pediatrics, 2005, 129(3):496-506. |
| [47] |
Kosaka N, Izumi H, Sekine K, Ochiya T. MicroRNA as a new immune-regulatory agent in breast milk. Silence, 2010, 1(1):7.
doi: 10.1186/1758-907X-1-7 |
| [48] |
Izumi H, Kosaka N, Shimizu T, Sekine K, Ochiya T, Takase M. Bovine milk contains microRNA and messenger RNA that are stable under degradative conditions. J Dairy Sci, 2012, 95(9):4831-4841.
doi: S0022-0302(12)00497-3 pmid: 22916887 |
| [49] |
Gu YR, Liang Y, Gong JJ, Zeng K, Li ZQ, Lei YF, He ZP, Lv XB. Suitable internal control microRNA genes for measuring miRNA abundance in pig milk during different lactation periods. Genet Mol Res, 2012, 11(3):2506-2512.
doi: 10.4238/2012.June.18.3 pmid: 22782633 |
| [50] | Jin XL, Yang JX, Li Z. Liu YH, Liu JX. Progress on the miRNA related with mammary gland development and lactation. Hereditas(Beijing), 2013, 35(6):695-702. |
| 金晓露, 杨建香, 李真, 刘红云, 刘建新. 乳腺发育及泌乳相关miRNA研究进展. 遗传, 2013, 35(6):695-702. | |
| [51] | Oatley JM, Brinster RL. Spermatogonial stem cells. Methods Enzymol, 2006, 419:259-282. |
| [52] | Ran ML, Chen B, Yang QA, Jiang M, Yin J. Advances in miRNA research related to testis development and spermatogenesis. Hereditas(Beijing), 2014, 36(7):646-654. |
| 冉茂良, 陈斌, 杨岸奇, 蒋明, 尹杰. 睾丸发育和精子生成相关miRNA研究进展. 遗传, 2014, 36(7):646-654. | |
| [53] | Tong MH, Mitchell D, Evanoff R, Griswold MD. Expression of Mirlet7 family microRNAs in response to retinoic acid-induced spermatogonial differentiation in mice. Biol Reprod, 2011, 85(1):189-197. |
| [54] | Tong MH, Mitchell DA, McGowan SD, Evanoff R, Griswold MD. Two miRNA clusters, Mir-17-92 (Mirc1) and Mir-106b-25 (Mirc3), are involved in the regulation of spermatogonial differentiation in mice. Biol Reprod, 2012, 86(3):72. |
| [55] |
Lichner Z, Páll E, Kerekes A, Pállinger E, Maraghechi P, Bosze Z, Gócza E. The miR-290-295 cluster promotes pluripotency maintenance by regulating cell cycle phase distribution in mouse embryonic stem cells. Differentiation, 2011, 81(1):11-24.
doi: 10.1016/j.diff.2010.08.002 |
| [56] |
Rosa A, Brivanlou AH. A regulatory circuitry comprised of miR-302 and the transcription factors OCT4 and NR2F2 regulates human embryonic stem cell differentiation. EMBO J, 2011, 30(2):237-248.
doi: 10.1038/emboj.2010.319 |
| [57] |
Jung YH, Gupta MK, Shin JY, Uhm SJ, Lee HT. MicroRNA signature in testes-derived male germ-line stem cells. Mol Hum Reprod, 2010, 16(11):804-810.
doi: 10.1093/molehr/gaq058 |
| [58] |
Ucar A, Erikci E, Ucar O, Chowdhury K. miR-212 and miR-132 are dispensable for mouse mammary gland development. Nat Genet, 2014, 46(8):802-804.
doi: 10.1038/ng.2990 |
| [59] |
Phua YW, Nguyen A, Roden DL, Elsworth B, Deng NT, Nikolic I, Yang J, Mcfarland A, Russell R, Kaplan W, Cowley MJ, Nair R, Zotenko E, O’Toole S, Tan SX, James DE, Clark SJ, Kouros-Mehr H, Swarbrick A. MicroRNA profiling of the pubertal mouse mammary gland identifies miR-184 as a candidate breast tumour suppressor gene. Breast Cancer Res, 2015, 17(1):83.
doi: 10.1186/s13058-015-0593-0 |
| [60] | Zhang M, Hou R, Liu YL, Zheng HP, Zhu Q, Zhang ZH, Xian H. Effects of epidermal growth factor and insulin on biological characteristics of giant panda (Ailuropoda melanoleuca) cutaneous fibroblast in vitro. Zoolog Res, 2005, 26(5):499-505. |
| 张明, 侯蓉, 刘玉良, 郑鸿培, 朱庆, 张志和, 鲜红. 表皮生长因子和胰岛素对大熊猫体外培养皮肤成纤维细胞生物学特性的影响. 动物学研究, 2005, 26(5):499-505. | |
| [61] | Zhang M, Hou R, Zheng HP, Zhu Q, Xian H, Li J. Studies on biological characteristic of giant panda (Ailuropoda melanoleuca) skin fibroblasts in the different media. J Yangzhou Univ (Agric Life Sci Ed), 2006, 27(3):35-39. |
| 张明, 侯蓉, 郑鸿培, 朱庆, 鲜红, 李俊. 大熊猫皮肤成纤维细胞在不同培养液中的生物学特性研究. 扬州大学学报(农业与生命科学版), 2006, 27(3):35-39. | |
| [62] |
Yu FJ, Zeng CJ, Zhang Y, Wang CD, Xiong TY, Fang SG, Zhang HM. Establishment and cryopreservation of a giant panda skeletal muscle-derived cell line. Biopreserv Biobank, 2015, 13(3):195-199.
doi: 10.1089/bio.2014.0073 pmid: 26035009 |
| [63] |
Mali P, Yang LH, Esvelt KM, Aach J, Guell M, Dicarlo JE, Norville JE, Church GM. RNA-guided human genome engineering via Cas9. Science, 2013, 339(6121):823-826.
doi: 10.1126/science.1232033 |
| [64] |
Chang NN, Sun CH, Gao L, Zhu D, Xu XF, Zhu XJ, Xiong JW, Xi JJ. Genome editing with RNA-guided Cas9 nuclease in zebrafish embryos. Cell Res, 2013, 23(4):465-472.
doi: 10.1038/cr.2013.45 |
| [65] |
Wu YX, Liang D, Wang YH, Bai MZ, Tang W, Bao SM, Yan ZQ, Li DS, Li JS. Correction of a genetic disease in mouse via use of CRISPR-Cas9. Cell Stem Cell, 2013, 13(6):653-658.
doi: 10.1016/j.stem.2013.11.002 |
| [66] |
Hai T, Teng F, Guo RF, Li W, Zhou Q. One-step generation of knockout pigs by zygote injection of CRISPR/Cas system. Cell Res, 2014, 24(3):327-375.
doi: 10.1016/0014-4827(61)90435-9 |
| [67] |
Han HB, Ma YH, Wang T, Lian L, Tian XZ, Hu R, Deng SL, Li KP, Wang F, Li N. One-step generation of myostatin gene knockout sheep via the CRISPR/Cas9 system. Front Agr Sci Eng, 2014, 1(1):2-5.
doi: 10.15302/J-FASE-2014007 |
| [68] |
Clop A, Marcq F, Takeda H, Pirottin D, Tordoir X, Bibé B, Bouix J, Caiment F, Elsen JM, Eychenne F, Larzul C, Laville E, Meish F, Milenkovic D, Tobin J, Charlier C, Georges M. A mutation creating a potential illegitimate microRNA target site in the myostatin gene affects muscularity in sheep. Nat Genet, 2006, 38(7):813-818.
doi: 10.1038/ng1810 |
| [69] | Li C, Cao GW. Advances in CRISPR/Cas9-mediated gene editing. Chin J Biotechnol, 2015, 31(11):1531-1542. |
| 李聪, 曹文广. CRISPR/Cas9介导的基因编辑技术研究进展. 生物工程学报, 2015, 31(11):1531-1542. |
| [1] | Jie Kou, Yan Li, Peng Wang, Hong Liu, Jiawen Liu, Juan Wang, Ye Wang, Liang Zhang, Fujun Shen. Optimization of microsatellite genotyping system used for genetic diversity evaluation of Ailuropoda melanoleuca [J]. Hereditas(Beijing), 2022, 44(3): 253-266. |
| [2] | Yong Wei, Yulan He, Xueli Zheng. Research progress in RNA interference against the infection of mosquito-borne viruses [J]. Hereditas(Beijing), 2020, 42(2): 153-160. |
| [3] | Xin Li, Chengming Qu, Yinglun Han, Xin Liu, Qingwei Li. Identification, recombinant expression and immunological study of Lja-SHP2 in Lampetra japonica [J]. Hereditas(Beijing), 2020, 42(2): 183-193. |
| [4] | Wenquan Liang,Yu Hou,Cunyou Zhao. Schizophrenia-associated single nucleotide polymorphisms affecting microRNA function [J]. Hereditas(Beijing), 2019, 41(8): 677-685. |
| [5] | Wenwen Deng, Caiwu Li, Siyue Zhao, Rengui Li, Yongguo He, Daifu Wu, Shengzhi Yang, Yan Huang, Hemin Zhang, Likou Zou. Whole genome sequencing reveals the distribution of resistance and virulence genes of pathogenic Escherichia coli CCHTP from giant panda [J]. Hereditas(Beijing), 2019, 41(12): 1138-1147. |
| [6] | Lin Rao, Feilong Meng, Ran Fang, Chenyi Cai, Xiaoli Zhao. Molecular mechanism of microRNA in regulating cochlear hair cell development [J]. Hereditas(Beijing), 2019, 41(11): 994-1008. |
| [7] | Xia Mengmeng,Shen Xueyi,Niu Changmin,Xia Jing,Sun Hongya,Zheng Ying. MicroRNA regulates Sertoli cell proliferation and adhesion [J]. Hereditas(Beijing), 2018, 40(9): 724-732. |
| [8] | Hailong Liu, Yang Shen, Yang Gao, Ling Zhou, Xiaosong Han, Changzhi Zhao, Gaojuan Yang, Yilong Chen, Hui Yang, Shengsong Xie. Assessing abundance and specificity of different types of sgRNA targeting miRNA precursors [J]. Hereditas(Beijing), 2018, 40(7): 561-571. |
| [9] | Huawei Zhang, Xingyu Meng, anfeng Li, Yuying Yang, Huaji Qiu. Long non-coding RNAs: Emerging regulators of antiviral innate immune responses [J]. Hereditas(Beijing), 2018, 40(7): 525-533. |
| [10] | Juan Xiao, Xun Wang, Yi Luo, Xiaokai Li, Xuewei Li. Research progress in sRNAs and functional proteins in epididymosomes [J]. Hereditas(Beijing), 2018, 40(3): 197-206. |
| [11] | Xinyun Li, Liangliang Fu, Huijun Cheng, Shuhong Zhao. Advances on microRNA in regulating mammalian skeletal muscle development [J]. Hereditas(Beijing), 2017, 39(11): 1046-1053. |
| [12] | Chendong Liu, Lu Yang, Hongzhou Pu, Qiong Yang, Wenyao Huang, Xue Zhao, Li Zhu, Shunhua Zhang. Epigenetics regulates gene expression patterns of skeletal muscle induced by physical exercise [J]. Hereditas(Beijing), 2017, 39(10): 888-896. |
| [13] | Jun Wei,Xiujun Lu,Xiaolin Zhang,Mei Mei,Xiaoli Huang. Functions of microRNA in seed development, dormancy and germination processes [J]. Hereditas(Beijing), 2017, 39(1): 14-21. |
| [14] | Ke Zhang, Guangde Feng, Baoyun Zhang, Wei Xiang, Long Chen, Fang Yang, Mingxing Chu, Pingqing Wang. Application of epigenetic markers in molecular breeding of the swine [J]. Hereditas(Beijing), 2016, 38(7): 634-643. |
| [15] | Mei Fu, Kehui Xu, Wenming Xu. Research advances of Dicer in regulating reproductive function [J]. HEREDITAS(Beijing), 2016, 38(7): 612-622. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||