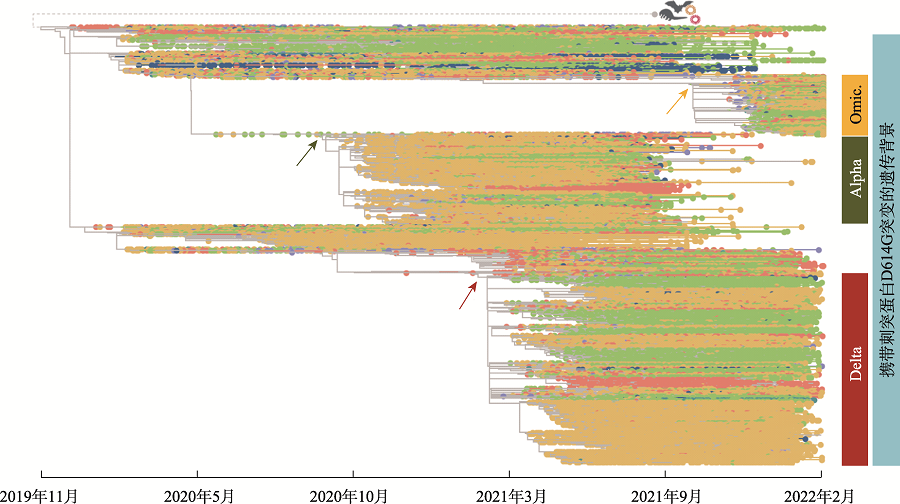
Hereditas(Beijing) ›› 2022, Vol. 44 ›› Issue (5): 442-446.doi: 10.16288/j.yczz.22-107
• Orginal Articles • Previous Articles
-
Received:2022-04-13Revised:2022-04-19Online:2022-05-20Published:2022-04-27 -
Supported by:Supported by the Strategic Priority Research Program of the Chinese Academy of Sciences(XDPB17)
Cite this article
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
| [1] |
Viana R, Moyo S, Amoako DG, Tegally H, Scheepers C, Althaus CL, Anyaneji UJ, Bester PA, Boni MF, Chand M, Choga WT, Colquhoun R, Davids M, Deforche K, Doolabh D, du PL, Engelbrecht S, Everatt J, Giandhari J, Giovanetti M, Hardie D, Hill V, Hsiao NY, Iranzadeh A, Ismail A, Joseph C, Joseph R, Koopile L, Kosakovsky Pond SL, Kraemer MUG, Kuate-Lere L, Laguda-Akingba O, Lesetedi-Mafoko O, Lessells R, Lockman S, Lucaci A, Maharaj A, Mahlangu B, Maponga T, Mahlakwane K, Makatini Z, Marais G, Maruapula D, Masupu K, Matshaba M, Mayaphi S, Mbhele N, Mbulawa MB, Mendes A, Mlisana K, Mnguni A, Mohale T, Moir M, Moruisi K, Mosepele M, Motsatsi G, Motswaledi MS, Mphoyakgosi T, Msomi N, Mwangi PN, Naidoo Y, Ntuli N, Nyaga M, Olubayo L, Pillay S, Radibe B, Ramphal Y, Ramphal U, San JE, Scott L, Shapiro R, Singh L, Smith-Lawrence P, Stevens W, Strydom A, Subramoney K, Tebeila N, Tshiabuila D, Tsui J, van Wyk S, Weaver S, Wibmer C, Wilkinson E, Wolter N, Zarebski AE, Zuze B, Goedhals D, Preiser W, Treurnicht F, Venter M, Williamson C, Pybus OG, Bhiman J, Glass A, Martin DP, Rambaut A, Gaseitsiwe S, von Gottberg A, de Oliveira T. Rapid epidemic expansion of the SARS-CoV-2 Omicron variant in southern Africa. Nature, 2022, 603(7902):679-686.
doi: 10.1038/s41586-022-04411-y |
| [2] |
Menni C, Valdes AM, Polidori L, Antonelli M, Penamakuri S, Nogal A, Louca P, May A, Figueiredo JC, Hu C, Molteni E, Canas L, Österdahl MF, Modat M, Sudre CH, Fox B, Hammers A, Wolf J, Capdevila J, Chan AT, David SP, Steves CJ, Ourselin S, Spector TD. Symptom prevalence, duration, and risk of hospital admission in individuals infected with SARS-CoV-2 during periods of omicron and delta variant dominance: a prospective observational study from the ZOE COVID study. Lancet, 2022, doi: 10.1016/s0140-6736(22)00327-0
doi: 10.1016/s0140-6736(22)00327-0 |
| [3] |
Chen JM, Chen YQ. China can prepare to end its zero-COVID policy. Nat Med, 2022, doi: 10.1038/s41591-022-01794-3
doi: 10.1038/s41591-022-01794-3 |
| [4] |
Wu F, Zhao S, Yu B, Chen YM, Wang W, Song ZG, Hu Y, Tao ZW, Tian JH, Pei YY, Yuan ML, Zhang YL, Dai FH, Liu Y, Wang QM, Zheng JJ, Xu L, Holmes EC, Zhang YZ. A new coronavirus associated with human respiratory disease in China. Nature, 2020, 579(7798):265-269.
doi: 10.1038/s41586-020-2008-3 |
| [5] |
Hu B, Guo H, Zhou P, Shi ZL. Characteristics of SARS- CoV-2 and COVID-19. Nat Rev Microbiol, 2021, 19(3):141-154.
doi: 10.1038/s41579-020-00459-7 |
| [6] | Wan Y, Shang J, Graham R, Baric RS, Li F. Receptor recognition by the novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS coronavirus. J Virol, 2020, 94(7):e00127-20. |
| [7] |
Zhang ZB, Xia YL, Shen JX, Du WW, Fu YX, Liu SQ. Mechanistic origin of different binding affinities of SARS-CoV and SARS-CoV-2 spike RBDs to human ACE2. Cells, 2022, 11(8):1274.
doi: 10.3390/cells11081274 |
| [8] |
Peng R, Wu LA, Wang Q, Qi J, Gao GF. Cell entry by SARS-CoV-2. Trends Biochem Sci, 2021, 46(10):848-860.
doi: 10.1016/j.tibs.2021.06.001 |
| [9] |
Yu D, Yang X, Tang BX, Pan YH, Yang JN, Duan GY, Zhu JW, Hao ZQ, Mu HL, Dai L, Hu WJ, Zhang MC, Cui Y, Jin T, Li CP, Ma L, Language translation team, Su X, Zhang G, Zhao W, Li H. Coronavirus GenBrowser for monitoring the transmission and evolution of SARS- CoV-2. Brief Bioinform, 2022, 23(2): bbab583.
doi: 10.1093/bib/bbab583 |
| [10] | Hartl DL, Clark AG. Principles of Population Genetics. Sinauer Associates, Inc., 1988. |
| [11] |
Lin K, Li H. Advances in detecting positive selection on genome. Hereditas (Beijing), 2009, 31(9):896-902.
doi: 10.3724/SP.J.1005.2009.00896 |
|
林拷, 李海鹏. DNA水平上检测正选择方法的研究进展. 遗传, 2009, 31(9):896-902
doi: 10.3724/SP.J.1005.2009.00896 |
|
| [12] |
Anderson RM, May RM. Coevolution of hosts and parasites. Parasitology, 1982, 85(Pt 2):411-426.
doi: 10.1017/S0031182000055360 |
| [13] |
Bull JJ. Virulence. Evolution, 1994, 48(5):1423-1437.
doi: 10.1111/j.1558-5646.1994.tb02185.x pmid: 28568406 |
| [14] |
Ebert D, Bull JJ. Challenging the trade-off model for the evolution of virulence: is virulence management feasible? Trends Microbiol, 2003, 11(1):15-20.
doi: 10.1016/S0966-842X(02)00003-3 |
| [15] |
Read AF. The evolution of virulence. Trends Microbiol, 1994, 2(3):73-76.
pmid: 8156274 |
| [16] |
Geoghegan JL, Holmes EC. The phylogenomics of evolving virus virulence. Nat Rev Genet, 2018, 19(12):756-769.
doi: 10.1038/s41576-018-0055-5 pmid: 30305704 |
| [17] |
Levin BR, Bull JJ. Short-sighted evolution and the virulence of pathogenic microorganisms. Trends Microbiol, 1994, 2(3):76-81.
pmid: 8156275 |
| [18] |
Read AF, Baigent SJ, Powers C, Kgosana LB, Blackwell L, Smith LP, Kennedy DA, Walkden-Brown SW, Nair VK. Imperfect vaccination can enhance the transmission of highly virulent pathogens. PLoS Biol, 2015, 13(7):e1002198.
doi: 10.1371/journal.pbio.1002198 |
| [19] |
Ma HL, Wang ZG, Zhao X, Han J, Zhang Y, Wang H, Chen C, Wang J, Jiang FC, Lei J, Song JD, Jiang SF, Zhu SL, Liu HH, Wang DY, Meng Y, Mao NY, Wang YH, Zhu Z, Chen ZX, Wang BL, Song QQ, Du HJ, Yuan Q, Xia D, Xia ZQ, Liu PP, Wu YC, Feng ZJ, Gao RQ, Gao GF, Xu WB. Long distance transmission of SARS-CoV-2 from contaminated cold chain products to humans -- Qingdao City, Shandong Province, China, September 2020. China CDC Weekly, 2021, 3(30):637-644.
doi: 10.46234/ccdcw2021.164 |
| [20] |
Yu D, Zhu JW, Yang JN, Pan YH, Mu HL, Cao RF, Tang BX, Duan GY, Hao ZQ, Dai L, Zhao GP, Zhang YP, Zhao W, Zhang G, Li H. Global cold-chain related SARS-CoV-2 transmission. Zenodo, 2022, doi: 10.5281/zenodo.6379310.
doi: 10.5281/zenodo.6379310 |
| [21] |
Liu PP, Yang MJ, Zhao X, Guo YY, Wang L, Zhang J, Lei WW, Han WF, Jiang FC, Liu WJ, Gao GF, Wu GZ. Cold-chain transportation in the frozen food industry may have caused a recurrence of COVID-19 cases in destination: successful isolation of SARS-CoV-2 virus from the imported frozen cod package surface. Biosaf Health, 2020, 2(4):199-201.
doi: 10.1016/j.bsheal.2020.11.003 |
| [22] |
Pang XH, Ren LL, Wu SS, Ma WT, Yang J, Di L, Li J, Xiao Y, Kang L, Du SC, Du J, Wang J, Li G, Zhai SG, Chen LJ, Zhou WX, Lai SJ, Gao L, Pan Y, Wang QY, Li MK, Wang JB, Huang YY, Wang JW, COVID-19 Field Response Group, COVID-19 Laboratory Testing Group. Cold-chain food contamination as the possible origin of COVID-19 resurgence in Beijing. Natl Sci Rev, 2020, 7(12):1861-1864.
doi: 10.1093/nsr/nwaa264 |
| [23] |
Bolze A, Basler T, White S, Rossi AD, Wyman D, Roychoudhury P, Greninger AL, Hayashibara K, Beatty M, Shah S, Stous S, Kil E, Dai H, Cassens T, Tsan K, Nguyen J, Ramirez J, Carter S, Cirulli ET, Barrett KS, Washington NL, Belda-Ferre P, Jacobs S, Sandoval E, Becker D, Lu JT, Isaksson M, Lee W, Luo SS. Evidence for SARS-CoV-2 Delta and Omicron co-infections and recombination. medRxiv, 2022, doi: 10.1101/2022.03.09.22272113.
doi: 10.1101/2022.03.09.22272113 |
| [24] |
Lacek KA, Rambo-Martin BL, Batra D, Zheng XY, Sakaguchi H, Peacock T, Keller M, Wilson MM, Sheth M, Davis ML, Borroughs M, Gerhart J, Hassell N, Shepard SS, Cook PW, Lee J, Wentworth DE, Barnes JR, Kondor R, Paden CR. Identification of a novel SARS-CoV-2 Delta-Omicron recombinant virus in the United States. bioRxiv, 2022, doi: 10.1101/2022.03.19.484981.
doi: 10.1101/2022.03.19.484981 |
| [25] |
Willyard C. What the Omicron wave is revealing about human immunity. Nature, 2022, 602(7895):22-25.
doi: 10.1038/d41586-022-00214-3 |
| [26] | Zhao SL, Sha T, Wu CI, Xue YB, Chen H. Will the large- scale vaccination succeed in containing the COVID-19 pandemic and how soon? Quantitative Biology, 2021, 3:304-316. |
| [27] |
Gong Z, Zhu JW, Li CP, Jiang S, Ma LN, Tang BX, Zou D, Chen ML, Sun YB, Song SH, Zhang Z, Xiao JF, Xue YB, Bao YM, Du ZL, Zhao WM. An online coronavirus analysis platform from the National Genomics Data Center. Zool Res, 2020, 41(6):705-708.
doi: 10.24272/j.issn.2095-8137.2020.065 |
| No related articles found! |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||