Hereditas(Beijing) ›› 2024, Vol. 46 ›› Issue (5): 421-430.doi: 10.16288/j.yczz.24-036
• Research Article • Previous Articles Next Articles
Comparison of genomic prediction methods for early growth traits of Inner Mongolia cashmere goats based on multi trait models
Linyu Gao1( ), Qi Xu1, Yuxiao He1, Haijiao Xi1, Yifan Liu1, Tao Zhang1, Jinquan Li1,2, Yanjun Zhang1, Ruijun Wang1, Qi Lü1, Bujun Mei4, Rui Su1, Zhiying Wang1,3(
), Qi Xu1, Yuxiao He1, Haijiao Xi1, Yifan Liu1, Tao Zhang1, Jinquan Li1,2, Yanjun Zhang1, Ruijun Wang1, Qi Lü1, Bujun Mei4, Rui Su1, Zhiying Wang1,3( )
)
- 1. College of Animal Science, Inner Mongolia Agricultural University, Hohhot 010018, China
2. Key Laboratory of Meat Sheep Genetic Breeding of the Ministry of Agriculture and Rural Affairs, Hohhot 010018, China
3. Key Laboratory of Sheep Genetic Breeding and Reproduction in Inner Mongolia Autonomous Region, Hohhot 010018, China
4. Department of Agriculture, Hetao University, Bayannur 015000, China
-
Received:2024-01-31Revised:2024-04-25Online:2024-05-06Published:2024-05-06 -
Contact:Zhiying Wang E-mail:gaolinyu0629@163.com;wzhy0321@126.com -
Supported by:National Key Research and Development Program(2022YFE0113300);National Key Research and Development Program(2022YFD1300201);National Key Research and Development Program(2022YFD1300204);Inner Mongolia Autonomous Region Science and Technology Research Project(2021GG0086);Inner Mongolia Autonomous Region Higher Education Youth Science and Technology Talent Support Program(NJYT22038);Ministry of Finance and the Ministry of Agriculture and Rural Affairs, the National Technical System for Woolen Sheep Industry(CARS-39);High Level Achievement Cultivation Project of Inner Mongolia Agricultural University(QT202201);Inner Mongolia Autonomous Region Higher Education Innovation Team Development Program(NMGIRT2322);Basic Research Expenses of Universities Directly of Inner Mongolia Autonomous Region(BR220112)
Cite this article
Linyu Gao, Qi Xu, Yuxiao He, Haijiao Xi, Yifan Liu, Tao Zhang, Jinquan Li, Yanjun Zhang, Ruijun Wang, Qi Lü, Bujun Mei, Rui Su, Zhiying Wang. Comparison of genomic prediction methods for early growth traits of Inner Mongolia cashmere goats based on multi trait models[J]. Hereditas(Beijing), 2024, 46(5): 421-430.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
Table 1
Basic descriptive statistics on early growth traits of Inner Mongolia cashmere goat"
| 性状 | 个体数 | 父本数 | 母本数 | 最大值 | 最小值 | 平均值 | 标准差(sd) | 变异系数(C.V) |
|---|---|---|---|---|---|---|---|---|
| 初生重 | 1811 | 87 | 829 | 4.53 | 1.00 | 2.76 | 0.47 | 16.96% |
| 断乳重 | 1809 | 115 | 1351 | 34.77 | 13.60 | 21.97 | 3.34 | 15.42% |
| 日增重 | 967 | 85 | 789 | 0.27 | 0.09 | 0.17 | 0.03 | 17.79% |
| 周岁重 | 1953 | 116 | 1419 | 80.50 | 20.50 | 37.20 | 7.68 | 21.04% |
Table 2
Estimation of variance components of birth weight in Inner Mongolia cashmere goats using different methods"
| 方法 | 方差组分 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| ha2±SE | hm2 | C2 | hT2 | ra,m | |||||||
| ABLUP | 0.03 | 0.01 | -3.87×10-3 | 0.02 | 0.14 | 0.20 | 0.15±0.07 | 0.05 | 0.10 | 0.15 | -0.22 |
| GBLUP | 0.02 | 0.02 | -4.31×10-3 | 0.02 | 0.12 | 0.18 | 0.11±0.05 | 0.11 | 0.11 | 0.13 | -0.22 |
| ssGBLUP | 0.02 | 0.02 | -4.63×10-3 | 0.02 | 0.12 | 0.18 | 0.11±0.06 | 0.11 | 0.11 | 0.13 | -0.23 |
Table 3
Estimation of variance components of weaning weight in Inner Mongolia cashmere goats using different methods"
| 方法 | 方差组分 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| ha2±SE | hm2 | C2 | hT2 | ra,m | |||||||
| ABLUP | 1.93 | 0.67 | -0.91 | 0.13 | 3.12 | 4.94 | 0.39±0.13 | 0.14 | 0.03 | 0.18 | -0.80 |
| GBLUP | 1.62 | 0.90 | -0.83 | 6.46×10-5 | 3.85 | 5.54 | 0.29±0.10 | 0.16 | 1.17×10-5 | 0.20 | -0.69 |
| ssGBLUP | 1.82 | 0.94 | -1.07 | 6.94×10-3 | 3.68 | 5.38 | 0.34±0.05 | 0.17 | 1.29×10-3 | 0.13 | -0.82 |
Table 4
Estimation of variance components and genetic parameters of average daily weight gain in Inner Mongolia cashmere goats using different methods"
| 方法 | 方差组分 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| ha2±SE | hm2 | C2 | hT2 | ra,m | |||||||
| ABLUP | 3.31×10-5 | 2.77×10-5 | -1.95×10-6 | 2.93×10-6 | 2.51×10-4 | 3.13×10-4 | 0.11±0.05 | 0.09 | 9.36×10-3 | 0.14 | -0.06 |
| GBLUP | 5.50×10-5 | 2.96×10-5 | -1.49×10-5 | 1.26×10-5 | 3.16×10-4 | 3.98×10-4 | 0.14±0.04 | 0.07 | 0.03 | 0.12 | -0.37 |
| ssGBLUP | 5.43×10-5 | 3.14×10-5 | -1.42×10-5 | 3.68×10-6 | 3.85×10-4 | 4.73×10-4 | 0.11±0.05 | 0.07 | 7.78×10-3 | 0.11 | -0.34 |
Table 5
Estimation of variance components and genetic parameters of yearling weight in Inner Mongolia cashmere goats using different methods"
| 方法 | 方差组分 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| ha2±SE | hm2 | C2 | hT2 | ra,m | |||||||
| ABLUP | 6.51 | 4.98 | -4.46 | 3.35×10-4 | 17.32 | 24.35 | 0.27±0.09 | 0.20 | 1.38×10-5 | 0.09 | -0.78 |
| GBLUP | 4.87 | 2.38 | -2.33 | 3.80×10-4 | 15.52 | 20.44 | 0.24±0.07 | 0.12 | 1.86×10-5 | 0.13 | -0.68 |
| ssGBLUP | 5.75 | 2.87 | -2.64 | 3.67×10-4 | 17.04 | 23.02 | 0.25±0.03 | 0.12 | 1.59×10-5 | 0.14 | -0.65 |
Table 9
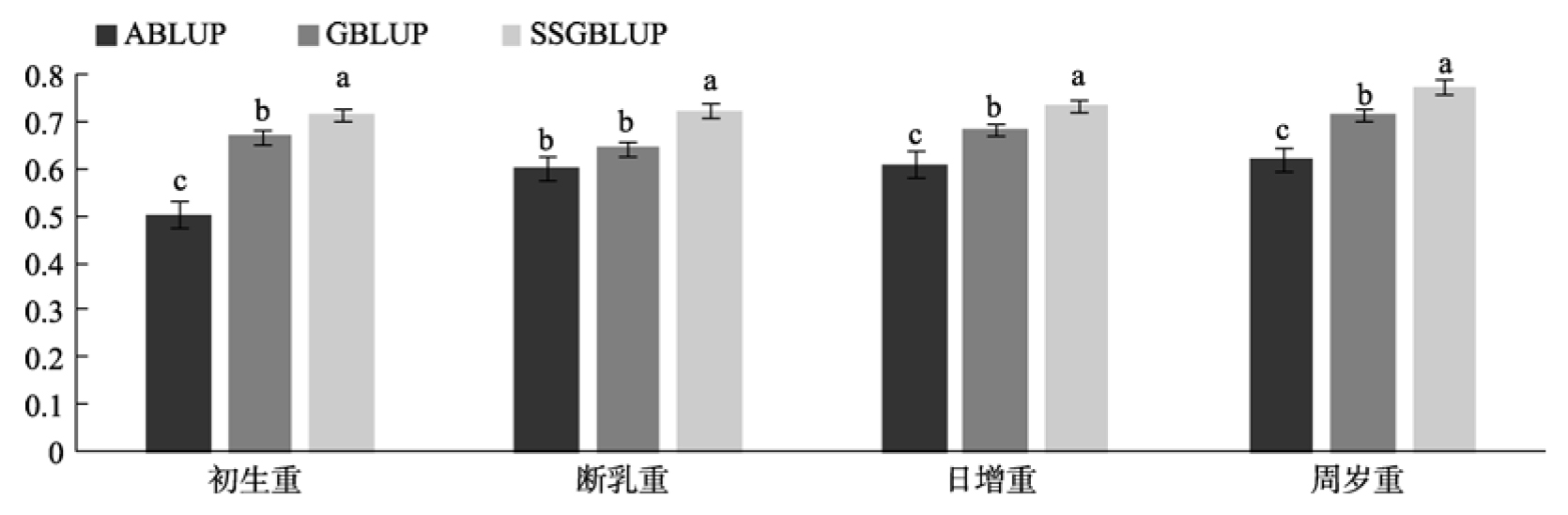
Accuracy and reliability evaluation of genomic breeding value of early growth traits with multi trait animal model"
| 性状 | 方法 | 准确性 | 准确性误差 | 可靠性 |
|---|---|---|---|---|
| 初生重 | ABLUP | 0.5047 (0.5209) | 0.1063 (0.1276) | 0.2547 (0.2713) |
| GBLUP | 0.6694 (0.5556) | 0.0097 (0.0079) | 0.4481 (0.3087) | |
| ssGBLUP | 0.7156 (0.6057) | 0.0068 (0.0058) | 0.5121 (0.3669) | |
| 断乳重 | ABLUP | 0.6207 (0.6831) | 0.1021 (0.1651) | 0.3853 (0.4666) |
| GBLUP | 0.6456 (0.7274) | 0.0719 (0.0110) | 0.4168 (0.5291) | |
| ssGBLUP | 0.7254 (0.7577) | 0.0658 (0.0081) | 0.5262 (0.5741) | |
| 日增重 | ABLUP | 0.6110 (0.5857) | 0.1146 (0.1604) | 0.3733 (0.3430) |
| GBLUP | 0.6855 (0.6630) | 0.0529 (0.0202) | 0.4699 (0.4396) | |
| ssGBLUP | 0.7357 (0.7021) | 0.0268 (0.0080) | 0.5413 (0.4929) | |
| 周岁重 | ABLUP | 0.6209 (0.5965) | 0.1231 (0.1494) | 0.3855 (0.3558) |
| GBLUP | 0.7155 (0.6381) | 0.0082 (0.0209) | 0.5119 (0.4072) | |
| ssGBLUP | 0.7756 (0.6683) | 0.0048 (0.0046) | 0.6016 (0.4466) |
| [1] |
Su R, Gong G, Zhang LT, Yan XC, Wang FH, Zhang L, Qiao X, Li XK, Li JQ. Screening the key genes of hair follicle growth cycle in Inner Mongolian cashmere goat based on RNA sequencing. Arch Anim Breed, 2020, 63(1): 155-164.
doi: 10.5194/aab-63-155-2020 pmid: 32490151 |
| [2] |
Meuwissen TH, Hayes BJ, Goddard ME. Prediction of total genetic value using genome-wide dense marker maps. Genetics, 2001, 157(4): 1819-1829.
doi: 10.1093/genetics/157.4.1819 pmid: 11290733 |
| [3] | Sandhu KS, Mihalyov PD, Lewien MJ, Pumphrey MO, Carter AH. Combining genomic and phenomic information for predicting grain protein content and grain yield in spring wheat. Front Plant Sci, 2021, 12: 613300. |
| [4] | Villar-Hernández BJ, Pérez-Elizalde S, Martini JWR, Toledo F, Perez-Rodriguez P, Krzuse M, García-Calvillo ID, Covarrubias-Pazaran G, Crossa J. Corrigendum to: application of multi-trait bayesian decision theory for parental genomic selection. G3 (Bethesda), 2021, 11(3): jkab034. |
| [5] |
Henderson CR. Best linear unbiased estimation and prediction under a selection model. Biometrics, 1975, 31(2): 423-447.
pmid: 1174616 |
| [6] |
Nilforooshan MA, Jakobsen JH, Fikse WF, Berglund B, Jorjani H. Application of a multiple-trait, multiple-country genetic evaluation model for female fertility traits. J Dairy Sci, 2010, 93(12): 5977-5986.
doi: 10.3168/jds.2010-3437 pmid: 21094772 |
| [7] | Galluzzo F, Visentin G, van Kaam JBCHM, Finocchiaro R, Biffani S, Costa A, Marusi M, Cassandro M. Genetic evaluation of gestation length in Italian Holstein breed. J Anim Breed Genet, 2024, 141(2): 113-123. |
| [8] | Adekale D, Alkhoder H, Liu ZT, Segelke D, Tetens J. Single-step SNPBLUP evaluation in six German beef cattle breeds. J Anim Breed Genet, 2023, 140(5): 496-507. |
| [9] | Lee SH, Lee SH, Park HB, Kim JM. Estimation of genetic parameters for pork belly traits. Anim Biosci, 2023, 36(8): 1156-1166. |
| [10] | Zhang X, Tsuruta S, Andonov S, Lourenco DAL, Sapp RL, Wang C, Misztal I. Relationships among mortality, performance, and disorder traits in broiler chickens: a genetic and genomic approach. Poult Sci, 2018, 97(5): 1511-1518. |
| [11] | Butler DG, Cullis BR, Gilmour AR, Gogel BG, Thompson R.ASReml-R Reference Manual Version 4. VSN International Ltd, Hemel Hempstead, HP1 1ES, UK. 2017. |
| [12] |
Christensen OF, Madsen P, Nielsen B, Ostersen T, Su G. Single-step methods for genomic evaluation in pigs. Animal, 2012, 6(10): 1565-1571.
doi: 10.1017/S1751731112000742 pmid: 22717310 |
| [13] | Kong J, Luan S, Tan J, Sui J, Luo K, Li XP, Dai P, Meng XH, Lu X, Chen BL, Cao JW, Cao BX. Progress of study on penaeid shrimp selective breeding. J Ocean Univ China(Nat Sci Ed), 2020, 50(9): 81-97. |
| 孔杰, 栾生, 谭建, 隋娟, 罗坤, 李旭鹏, 代平, 孟宪红, 卢霞, 陈宝龙, 曹家旺, 曹宝祥. 对虾选择育种研究进展. 中国海洋大学学报(自然科学版), 2020, 50 (9): 81-97. | |
| [14] |
Aguilar I, Misztal I, Tsuruta S, Wiggans GR, Lawlor TJ. Multiple trait genomic evaluation of conception rate in Holsteins. J Dairy Sci, 2011, 94(5): 2621-2624.
doi: 10.3168/jds.2010-3893 pmid: 21524554 |
| [15] | Tsuruta S, Misztal I, Aguilar I, Lawlor TJ. Multiple-trait genomic evaluation of linear type traits using genomic and phenotypic data in US Holsteins. J Dairy Sci, 2011, 94(8): 4198-4204. |
| [16] |
Guo G, Zhao FP, Wang YC, Zhang Y, Du LX, Su GS. Comparison of single-trait and multiple-trait genomic prediction models. BMC Genet, 2014, 15: 30.
doi: 10.1186/1471-2156-15-30 pmid: 24593261 |
| [17] |
Roy R, Mandal A, Notter DR. Estimates of (co)variance components due to direct and maternal effects for body weights in Jamunapari goats. Animal, 2008, 2(3): 354-359.
doi: 10.1017/S1751731107001218 pmid: 22445036 |
| [18] |
Magotra A, Bangar YC, Chauhan A, Malik BS, Malik ZS. Influence of maternal and additive genetic effects on offspring growth traits in Beetal goat. Reprod Domest Anim, 2021, 56(7): 983-991.
doi: 10.1111/rda.13940 pmid: 33884683 |
| [19] | Di J, Zhang Y, Tian KC, Lazate, Liu JF, Xu XM, Zhang YJ, Zhang TH. Estimation of (co)variance components and genetic parameters for growth and wool traits of Chinese superfine merino sheep with the use of a multi-trait animal model. Livest Sci, 2011, 138(1-3): 278-288. |
| [20] |
Gowane GR, Chopra A, Prakash V, Arora AL. Estimates of (co)variance components and genetic parameters for growth traits in Sirohi goat. Trop Anim Health Prod, 2011, 43(1): 189-198.
doi: 10.1007/s11250-010-9673-4 pmid: 20711812 |
| [21] |
Barazandeh A, Moghbeli SM, Vatankhah M, Mohammadabadi M. Estimating non-genetic and genetic parameters of pre-weaning growth traits in Raini cashmere goat. Trop Anim Health Prod, 2012, 44(4): 811-817.
doi: 10.1007/s11250-011-9971-5 pmid: 21901301 |
| [22] | Tesema Z, Alemayehu K, Getachew T, Kebede D, Deribe B, Taye M, Tilahun M, Lakew M, Kefale A, Belayneh N, Zegeye A, Yizengaw L. Estimation of genetic parameters for growth traits and Kleiber ratios in Boer × Central Highland goat. Trop Anim Health Prod, 2020, 52(6): 3195-3205. |
| [23] | 张沅. 家畜育种学. 北京: 中国农业出版社, 2001. |
| [24] |
Bangar YC, Magotra A, Yadav AS. Estimates of covariance components and genetic parameters for growth, average daily gain and Kleiber ratio in Harnali sheep. Trop Anim Health Prod, 2020, 52(5): 2291-2296.
doi: 10.1007/s11250-020-02248-z pmid: 32144658 |
| [25] | Wang RJ, Liu Y, Shi Y, Qi YP, Li YB, Wang ZY, Zhang YJ, Zhao YH, Su R, Li JQ. Study of genetic parameters for pre-weaning growth traits in inner Mongolia white Arbas cashmere goats. Front Vet Sci, 2023, 9: 1026528. |
| [26] | 王大广, 赵志辉, 赵玉民, 鲍志鸿. 萨福克羊表型测定值遗传参数的估计. 中国草食动物, 2003, (S1): 67-68. |
| [27] |
Baloche G, Legarra A, Sallé G, Larroque H, Astruc J-M, Robert-Granié C, Barillet F. Assessment of accuracy of genomic prediction for French Lacaune dairy sheep. J Dairy Sci, 2014, 97(2): 1107-1116.
doi: 10.3168/jds.2013-7135 pmid: 24315320 |
| [28] | Negro A, Cesarani A, Cortellari M, Bionda A, Fresi P, Macciotta NPP, Grande S, Biffani S, Crepaldi P. A comparison of genetic and genomic breeding values in Saanen and Alpine goats. Animal, 2024, 18(4): 101118. |
| [29] | Wang FH.Design of goat SNP chip and genome-wide association analysis and genome selection of important economic traits in Inner Mongolia cashmere goats[Dissertation]. Inner Mongolia Agricultural University, 2021. |
| 王凤红.山羊SNP芯片设计与内蒙古绒山羊重要经济性状全基因组关联分析及基因组选择研究[学位论文]. 内蒙古农业大学, 2021. |
| No related articles found! |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||