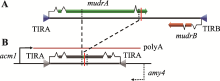
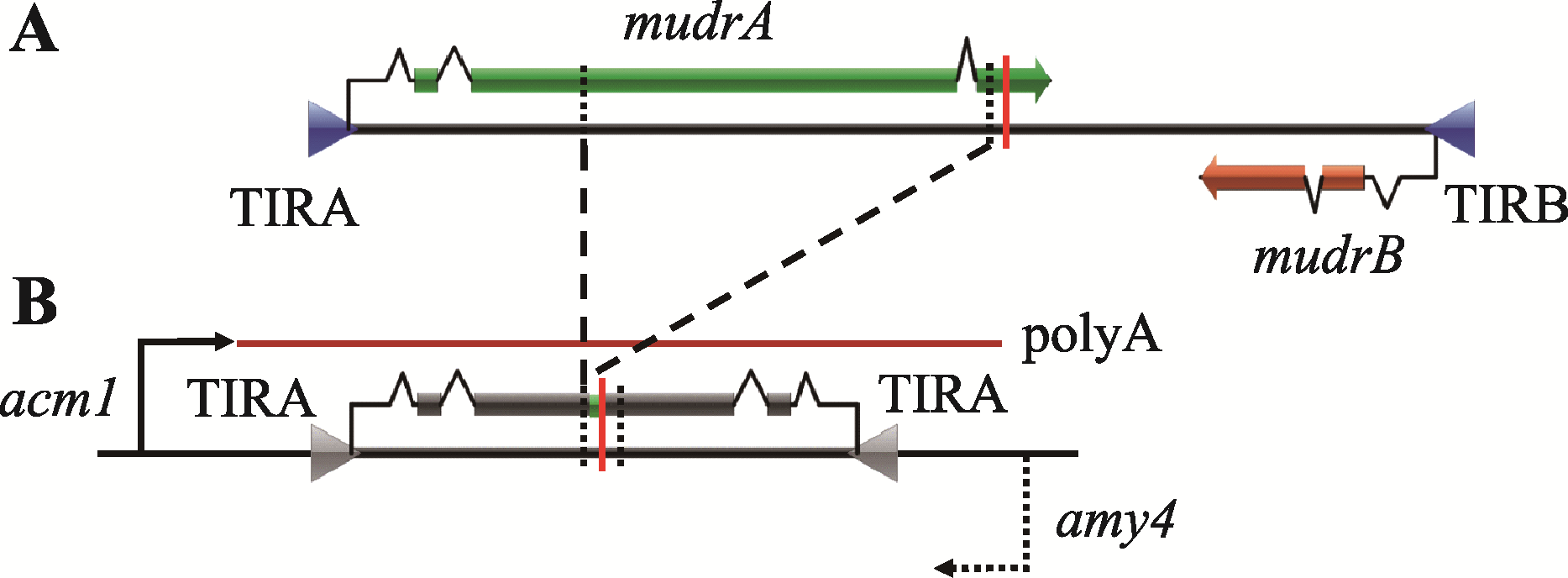
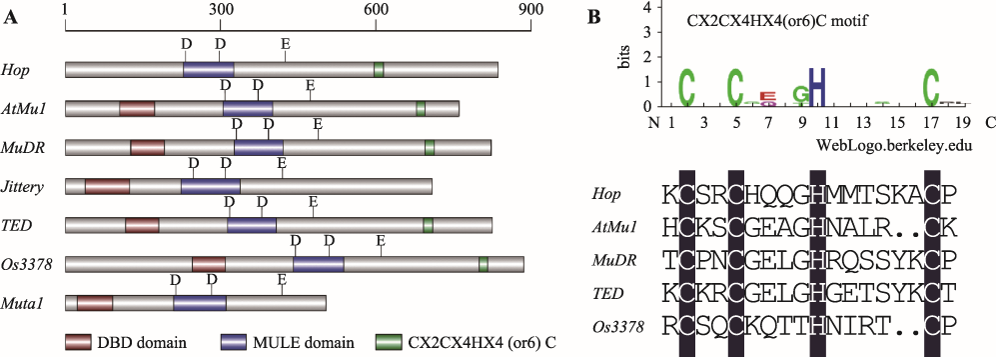
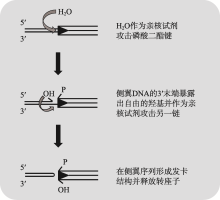
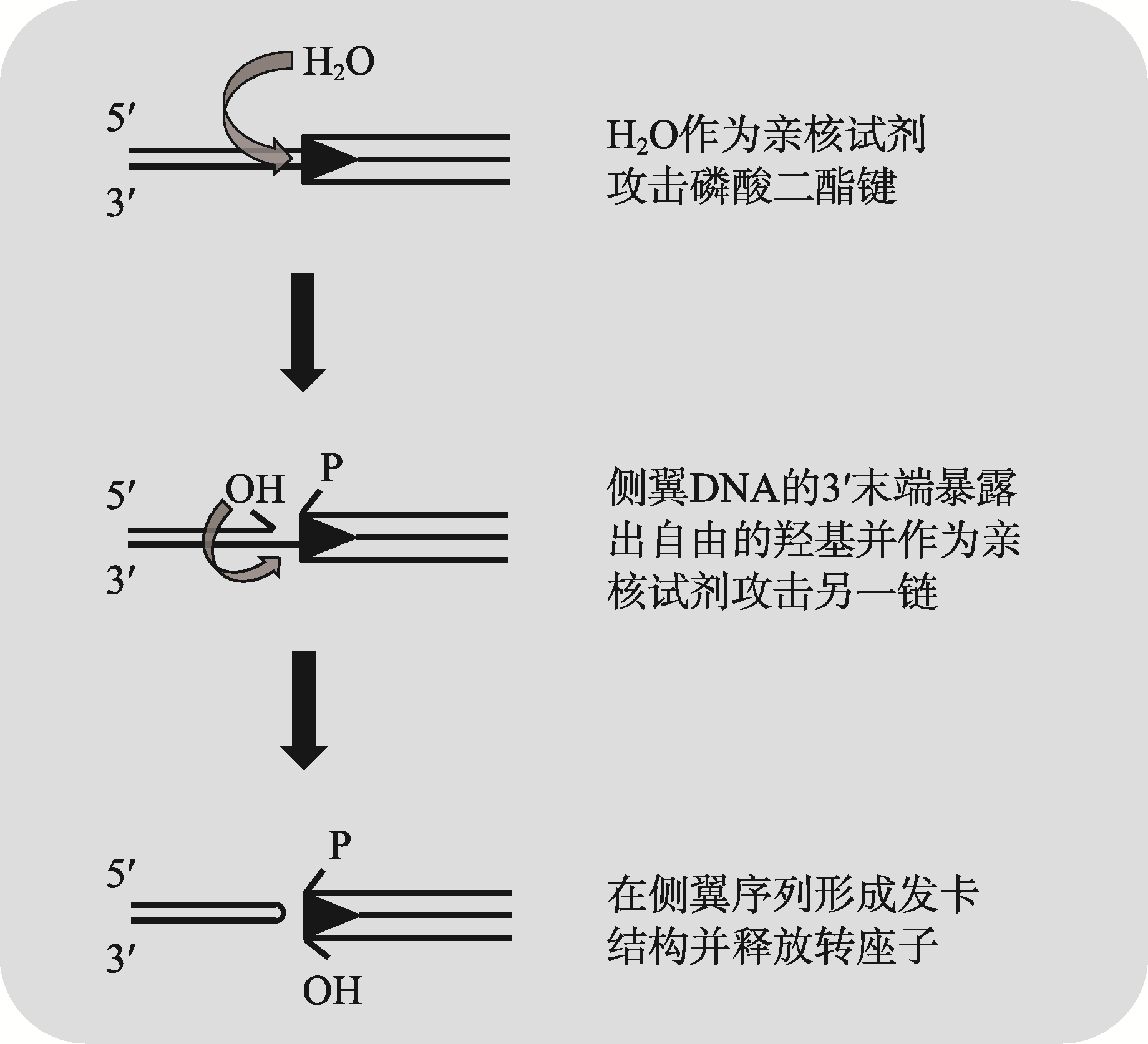
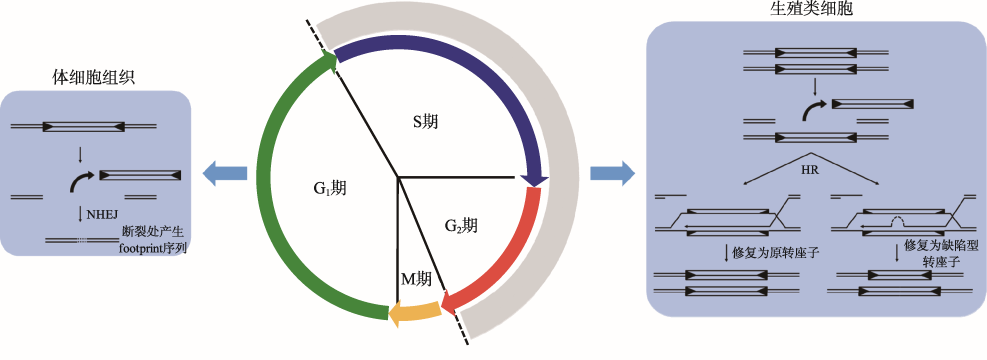
Hereditas(Beijing) ›› 2020, Vol. 42 ›› Issue (2): 131-144.doi: 10.16288/j.yczz.19-301
• Review • Next Articles
Progress on Mutator superfamily
Chunsheng Cong1( ), Yubin Li1,2(
), Yubin Li1,2( )
)
- 1. Biotechnology Research Institute, Chinese Academy of Agricultural Sciences, Beijing 100081, China
2. College of Agronomy, Qingdao Agricultural University, Qingdao 266109, China
-
Received:2019-09-27Revised:2019-12-17Online:2020-01-02Published:2019-12-27 -
Supported by:the National Natural Science Foundation of China(31871642)
Cite this article
Chunsheng Cong, Yubin Li. Progress on Mutator superfamily[J]. Hereditas(Beijing), 2020, 42(2): 131-144.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
| [1] |
Ravindran S . Barbara McClintock and the discovery of jumping genes. Proc Natl Acad Sci USA, 2012,109(50):20198-20199.
doi: 10.1073/pnas.1219372109 pmid: 23236127 |
| [2] |
Bourque G, Burns KH, Gehring M, Gorbunova V, Seluanov A, Hammell M, Imbeault M, Izsvák Z, Levin HL, Macfarlan TS, Mager DL, Feschotte C . Ten things you should know about transposable elements. Genome Biol, 2018,19(1):199.
doi: 10.1186/s13059-018-1577-z pmid: 30454069 |
| [3] |
Huang CR, Burns KH, Boeke JD . Active transposition in genomes. Annu Rev Genet, 2012,46:651-675.
doi: 10.1146/annurev-genet-110711-155616 |
| [4] |
Liu Z, Xu JH . The application of the high throughput sequencing technology in the transposable elements. Hereditas(Beijing), 2015,37(9):885-898.
doi: 10.16288/j.yczz.15-140 pmid: 26399528 |
|
刘振, 徐建红 . 高通量测序技术在转座子研究中的应用. 遗传, 2015,37(9):885-898.
doi: 10.16288/j.yczz.15-140 pmid: 26399528 |
|
| [5] | Li SF, Li S, Deng CL, Lu LD, Gao WJ . Role of transposons in origin and evolution of plant XY sex chromosomes. Hereditas(Beijing), 2015,37(2):157-164. |
| 李书粉, 李莎, 邓传良, 卢龙斗, 高武军 . 转座子在植物XY性染色体起源与演化过程中的作用. 遗传, 2015,37(2):157-164. | |
| [6] |
Li MM, Zhang DF, Gao Q, Luo YF, Zhang H, Ma B, Chen CH, Whibley A, Zhang YE, Cao YH, Li Q, Guo H, Li JH, Song YZ, Zhang Y, Copsey L, Li Y, Li XX, Qi M, Wang JW, Chen Y, Wang D, Zhao JY, Liu GC, Wu B, Yu LL, Xu CY, Li J, Zhao SC, Zhang YJ, Hu SN, Liang CZ, Yin Y, Coen E, Xue YB . Genome structure and evolution of Antirrhinum majus L. Nat Plants, 2019,5(2):174-183.
doi: 10.1038/s41477-018-0349-9 pmid: 30692677 |
| [7] |
Mascher M, Gundlach H, Himmelbach A, Beier S, Twardziok SO, Wicker T, Radchuk V, Dockter C, Hedley PE, Russell J, Bayer M, Ramsay L, Liu H, Haberer G, Zhang XQ, Zhang Q, Barrero RA, Li L, Taudien S, Groth M, Felder M, Hastie A, Šimková H, Staňková H, Vrána J, Chan S, Muñoz-Amatriaín M, Ounit R, Wanamaker S, Bolser D, Colmsee C, Schmutzer T, Aliyeva-Schnorr L, Grasso S, Tanskanen J, Chailyan A, Sampath D, Heavens D, Clissold L, Cao S, Chapman B, Dai F, Han Y, Li H, Li X, Lin C, Mccooke JK, Tan C, Wang P, Wang S, Yin S, Zhou G, Poland JA, Bellgard MI, Borisjuk L, Houben A, Dolezel J, Ayling S, Lonardi S, Kersey P, Langridge P, Muehlbauer GJ, Clark MD, Caccamo M, Schulman AH, Mayer FXK, Platzer M, Close TJ, Scholz U, Hansson M, Zhang G, Braumann I, Spannagl M, Li C, Waugh R, Stein N . A chromosome conformation capture ordered sequence of the barley genome. Nature, 2017,544(7651):427-433.
doi: 10.1038/nature22043 pmid: 28447635 |
| [8] |
Clavijo BJ, Venturini L, Schudoma C, Accinelli GG, Kaithakottil G, Wright J, Borrill P, Kettleborough G, Heavens D, Chapman H, Lipscombe J, Barker T, Lu FH, Mckenzie N, Raats D, Ramirez-Gonzalez RH, Coince A, Peel N, Percival-Alwyn L, Duncan O, Trösch J, Yu G, Bolser DM, Namaati G, Kerhornou A, Spannagl M, Gundlach H, Haberer G, Davey RP, Fosker C, Palma FD, Phillips AL, Millar AH, Kersey PJ, Uauy C, Krasileva KV, Swarbreck D, Bevan MW, Clark MD . An improved assembly and annotation of the allohexaploid wheat genome identifies complete families of agronomic genes and provides genomic evidence for chromosomal translocations. Genome Res, 2017,27(5):885-896.
doi: 10.1101/gr.217117.116 pmid: 28420692 |
| [9] |
Li FG, Fan GY, Lu CR, Xiao GH, Zou CS, Kohel RJ, Ma ZY, Shang HH, Ma XF, Wu JY, Liang XM, Huang G, Percy RG, Liu K, Yang WH, Chen WB, Du XM, Shi CC, Yuan YL, Ye WW, Liu X, Zhang XY, Liu WQ, Wei HL, Wei SJ, Huang GD, Zhang XL, Zhu SJ, Zhang H, Sun FM, Wang XF, Liang J, Wang JH, He Q, Huang LH, Wang J, Cui JJ, Song GL, Wang KB, Xu X, Yu JZ, Zhu YX, Yu SX . Genome sequence of cultivated upland cotton (Gossypium hirsutum TM-1) provides insights into genome evolution. Nat Biotechnol, 2015,33(5):524-530.
doi: 10.1038/nbt.3208 pmid: 25893780 |
| [10] |
Schmutz J, Cannon SB, Schlueter J, Ma J, Mitros T, Nelson W, Hyten DL, Song Q, Thelen JJ, Cheng J, Xu D, Hellsten U, May GD, Yu Y, Sakurai T, Umezawa T, Bhattacharyya MK, Sandhu D, Valliyodan B, Lindquist E, Peto M, Grant D, Shu S, Goodstein D, Barry K, Futrell-Griggs M, Abernathy B, Du J, Tian Z, Zhu L, Gill N, Joshi T, Libault M, Sethuraman A, Zhang XC, Shinozaki K, Nguyen HT, Wing RA, Cregan P, Specht J, Grimwood J, Rokhsar D, Stacey G, Shoemaker RC, Jackson SA . Genome sequence of the palaeopolyploid soybean. Nature, 2010,463(7278):178-183.
doi: 10.1038/nature08670 pmid: 20075913 |
| [11] |
Schnable PS, Ware D, Fulton RS, Stein JC, Wei F, Pasternak S, Liang C, Zhang J, Fulton L, Graves TA, Minx P, Reily AD, Courtney L, Kruchowski SS, Tomlinson C, Strong C, Delehaunty K, Fronick C, Courtney B, Rock SM, Belter E, Du F, Kim K, Abbott RM, Cotton M, Levy A, Marchetto P, Ochoa K, Jackson SM, Gillam B, Chen W, Yan L, Higginbotham J, Cardenas M, Waligorski J, Applebaum E, Phelps L, Falcone J, Kanchi K, Thane T, Scimone A, Thane N, Henke J, Wang T, Ruppert J, Shah N, Rotter K, Hodges J, Ingenthron E, Cordes M, Kohlberg S, Sgro J, Delgado B, Mead K, Chinwalla A, Leonard S, Crouse K, Collura K, Kudrna D, Currie J, He R, Angelova A, Rajasekar S, Mueller T, Lomeli R, Scara G, Ko A, Delaney K, Wissotski M, Lopez G, Campos D, Braidotti M, Ashley E, Golser W, Kim H, Lee S, Lin J, Dujmic Z, Kim W, Talag J, Zuccolo A, Fan C, Sebastian A, Kramer M, Spiegel L, Nascimento L, Zutavern T, Miller B, Ambroise C, Muller S, Spooner W, Narechania A, Ren L, Wei S, Kumari S, Faga B, Levy MJ, Mcmahan L, Van Buren P, Vaughn MW, Ying K, Yeh CT, Emrich SJ, Jia Y, Kalyanaraman A, Hsia AP, Barbazuk WB, Baucom RS, Brutnell TP, Carpita NC, Chaparro C, Chia JM, Deragon JM, Estill JC, Fu Y, Jeddeloh JA, Han Y, Lee H, Li P, Lisch DR, Liu S, Liu Z, Nagel DH, Mccann MC, Sanmiguel P, Myers AM, Nettleton D, Nguyen J, Penning BW, Ponnala L, Schneider KL, Schwartz DC, Sharma A, Soderlund C, Springer NM, Sun Q, Wang H, Waterman M, Westerman R, Wolfgruber TK, Yang L, Yu Y, Zhang L, Zhou S, Zhu Q, Bennetzen JL, Dawe RK, Jiang J, Jiang N, Presting GG, Wessler SR, Aluru S, Martienssen RA, Clifton SW, Mccombie WR, Wing RA, Wilson RK . The B73 maize genome: complexity, diversity, and dynamics. Science, 2009,326(5956):1112-1115.
doi: 10.1126/science.1178534 pmid: 19965430 |
| [12] |
Paterson AH, Bowers JE, Bruggmann R, Dubchak I, Grimwood J, Gundlach H, Haberer G, Hellsten U, Mitros T, Poliakov A, Schmutz J, Spannagl M, Tang H, Wang X, Wicker T, Bharti AK, Chapman J, Feltus FA, Gowik U, Grigoriev IV, Lyons E, Maher CA, Martis M, Narechania A, Otillar RP, Penning BW, Salamov AA, Wang Y, Zhang L, Carpita NC, Freeling M, Gingle AR, Hash CT, Keller B, Klein P, Kresovich S, Mccann MC, Ming R, Peterson DG, Mehboob-Ur-Rahman, Ware D, Westhoff P, Mayer KF, Messing J, Rokhsar DS . The Sorghum bicolor genome and the diversification of grasses. Nature, 2009,457(7229):551-556.
doi: 10.1038/nature07723 pmid: 19189423 |
| [13] |
Studer A, Zhao Q, Ross-Ibarra J, Doebley J . Identification of a functional transposon insertion in the maize domestication gene tb1. Nat Genet, 2011,43(11):1160-1163.
doi: 10.1038/ng.942 pmid: 21946354 |
| [14] |
Yang Q, Li Z, Li WQ, Ku LX, Wang C, Ye JR, Li K, Yang N, Li YP, Zhong T, Li JS, Chen YH, Yan JB, Yang XH, Xu ML . CACTA-like transposable element in ZmCCT attenuated photoperiod sensitivity and accelerated the postdomestication spread of maize. Proc Natl Acad Sci USA, 2013,110(42):16969-16974.
doi: 10.1073/pnas.1310949110 pmid: 24089449 |
| [15] |
Castelletti S, Tuberosa R, Pindo M, Salvi S . A MITE transposon insertion is associated with differential methylation at the maize flowering time QTL Vgt1. G3 (Bethesda), 2014,4(5):805-812.
doi: 10.1534/g3.114.010686 pmid: 24607887 |
| [16] |
Wang C, Yang Q, Wang WX, Li YP, Guo YL, Zhang DF, Ma XM, Song W, Zhao JR, Xu ML . A transposon- directed epigenetic change in ZmCCT underlies quantitative resistance to Gibberella stalk rot in maize. New Phytol, 2017,215(4):1503-1515.
doi: 10.1111/nph.14688 pmid: 28722229 |
| [17] |
Zhang ZH, Zhang X, Lin ZL, Wang J, Liu HQ, Zhou LN, Zhong SY, Li Y, Zhu C, Lai JS, Li XR, Yu JM, Lin ZW . A large transposon insertion in the stiff1 promoter increases stalk strength in maize. Plant Cell, 2019, DOI: 10.1105/tpc.19.00486.
doi: 10.1105/tpc.19.00486 pmid: 31690654 |
| [18] | Kojima KK . Structural and sequence diversity of eukaryotic transposable elements. Genes Genet Syst, 2018, DOI: 10.1266/ggs.18-00024 |
| [19] |
Finnegan DJ . Eukaryotic transposable elements and genome evolution. Trends Genet, 1989,5(4):103-107.
doi: 10.1016/0168-9525(89)90039-5 pmid: 2543105 |
| [20] |
Kapitonov VV, Jurka J . A universal classification of eukaryotic transposable elements implemented in Repbase. Nat Rev Genet, 2008,9(5):411-412, 414.
doi: 10.1038/nrg2165-c1 pmid: 18421312 |
| [21] |
Wicker T, Sabot F, Hua-Van A, Bennetzen JL, Capy P, Chalhoub B, Flavell A, Leroy P, Morgante M, Panaud O, Paux E, Sanmiguel P, Schulman AH . A unified classification system for eukaryotic transposable elements. Nat Rev Genet, 2007,8(12):973-982.
doi: 10.1038/nrg2165 pmid: 17984973 |
| [22] |
Bao WD, Kojima KK, Kohany O . Repbase update, a database of repetitive elements in eukaryotic genomes. Mob DNA, 2015,6:11.
doi: 10.1186/s13100-015-0041-9 pmid: 26045719 |
| [23] |
Xu HE, Zhang HH, Han MJ, Sheng YH, Huang XZ, Xiang ZH, Zhang Z . Computational approaches for identification and classification of transposable elements in eukaryotic genomes. Hereditas(Beijing), 2012,34(8):1009-1019.
doi: 10.3724/SP.J.1005.2012.01009 |
|
许红恩, 张化浩, 韩民锦, 沈以红, 黄先智, 向仲怀, 张泽 . 真核生物转座子鉴定和分类计算方法. 遗传, 2012,34(8):1009-1019.
doi: 10.3724/SP.J.1005.2012.01009 |
|
| [24] |
Hou XG, Zhang X, Guo DL . Identification and analysis methods of plant LTR retrotransposon sequences. Hereditas (Beijing), 2012,34(11):1491-1500.
doi: 10.3724/sp.j.1005.2012.01491 pmid: 23208147 |
|
侯小改, 张曦, 郭大龙 . 植物LTR类反转录转座子序列分析识别方法. 遗传, 2012,34(11):1491-1500.
doi: 10.3724/sp.j.1005.2012.01491 pmid: 23208147 |
|
| [25] | Shen D, Chen C, Wang SS, Chen W, Gao B, Song CY . Research progress of Tc1/Mariner superfamily. Hereditas (Beijing), 2017,39(1):1-13. |
| 沈丹, 陈才, 王赛赛, 陈伟, 高波, 宋成义 . Tc1/Mariner转座子超家族的研究进展. 遗传, 2017,39(1):1-13. | |
| [26] | Robertson DS . Characterization of a mutator system in maize. Mutat Res, 1978,51(1):21-28. |
| [27] |
Robertson DS . Genetic studies on the loss of Mu mutator activity in maize. Genetics, 1986,113(3):765-773.
pmid: 17246337 |
| [28] |
Strommer JN, Hake S, Bennetzen J, Taylor WC, Freeling M . Regulatory mutants of the maize Adh1 gene caused by DNA insertions. Nature, 1982,300(5892):542-544.
doi: 10.1038/300539a0 pmid: 7144906 |
| [29] |
Bennetzen JL . Transposable element Mu1 is found in multiple copies only in Robertson's Mutator maize lines. J Mol Appl Genet, 1984,2(6):519-524.
pmid: 6099399 |
| [30] |
Taylor LP, Walbot V . Isolation and characterization of a 1.7-kb transposable element from a mutator line of maize. Genetics, 1987,117(2):297-307.
pmid: 2444493 |
| [31] | Lisch D, Jiang N. Mutator and MULE transposons. In: Bennetzen JL, Hake S, eds. Handbook of Maize: Genetics and Genomics. New York, NY: Springer New York, 2009, 277-306. |
| [32] |
Hershberger RJ, Warren CA, Walbot V . Mutator activity in maize correlates with the presence and expression of the Mu transposable element Mu9. Proc Natl Acad Sci USA, 1991,88(22):10198-10202.
doi: 10.1073/pnas.88.22.10198 pmid: 1719548 |
| [33] |
Chomet P, Lisch D, Hardeman KJ, Chandler VL, Freeling M . Identification of a regulatory transposon that controls the Mutator transposable element system in maize. Genetics, 1991,129(1):261-270.
pmid: 1657702 |
| [34] |
Qin MM, Robertson DS, Ellingboe AH . Cloning of the Mutator transposable element MuA2, a putative regulator of somatic mutability of the a1-Mum2 allele in maize. Genetics, 1991,129(3):845-854.
pmid: 1661256 |
| [35] |
Mccarty DR, Settles AM, Suzuki M, Tan BC, Latshaw S, Porch T, Robin K, Baier J, Avigne W, Lai J, Messing J, Koch KE, Hannah LC . Steady-state transposon mutagenesis in inbred maize. Plant J, 2005,44(1):52-61.
doi: 10.1111/j.1365-313X.2005.02509.x pmid: 16167895 |
| [36] |
May BP, Liu H, Vollbrecht E, Senior L, Rabinowicz PD, Roh D, Pan XK, Stein L, Freeling M, Alexander D, Martienssen R . Maize-targeted mutagenesis: A knockout resource for maize. Proc Natl Acad Sci USA, 2003,100(20):11541-11546.
doi: 10.1073/pnas.1831119100 pmid: 12954979 |
| [37] |
Raizada MN . RescueMu protocols for maize functional genomics. Methods Mol Biol, 2003,236:37-58.
doi: 10.1385/1-59259-413-1:37 pmid: 14501057 |
| [38] |
Qian YX, Cheng X, Liu Y, Jiang HY, Zhu SW, Cheng BJ . Reactivation of a silenced minimal Mutator transposable element system following low-energy nitrogen ion implantation in maize. Plant Cell Rep, 2010,29(12):1365-1376.
doi: 10.1007/s00299-010-0922-9 |
| [39] |
Lisch D, Chomet P, Freeling M . Genetic characterization of the Mutator system in maize: Behavior and regulation of Mu transposons in a minimal line. Genetics, 1995,139(4):1777-1796.
pmid: 7789777 |
| [40] | Lisch D, Freeling M . Loss of Mutator activity in a minimal line. Maydica, 1994,39(4):289-300. |
| [41] |
Slotkin RK, Freeling M, Lisch D . Mu killer causes the heritable inactivation of the Mutator family of transposable elements in Zea mays. Genetics, 2003,165(2):781-797.
pmid: 14573488 |
| [42] |
Slotkin RK, Freeling M, Lisch D . Heritable transposon silencing initiated by a naturally occurring transposon inverted duplication. Nat Genet, 2005,37(6):641-644.
doi: 10.1038/ng1576 pmid: 15908951 |
| [43] |
Stawujak K, Startek M, Gambin A, Grzebelus D . MuTAnT: A family of Mutator-like transposable elements targeting TA microsatellites in Medicago truncatula. Genetica, 2015,143(4):433-440.
doi: 10.1007/s10709-015-9842-5 pmid: 25981486 |
| [44] |
Yan L, Gu YH, Tao X, Lai XJ, Zhang YZ, Tan XM, Wang HY . Scanning of transposable elements and analyzing expression of transposase genes of sweet potato [Ipomoea batatas]. PLoS One, 2014,9(3):e90895.
doi: 10.1371/journal.pone.0090895 pmid: 24608103 |
| [45] |
Gbadegesin MA, Wills MA, Beeching JR . Diversity of LTR-retrotransposons and enhancer/suppressor mutator- like transposons in cassava(Manihot esculenta Crantz). Mol Genet Genomics, 2008,280(4):305-317.
doi: 10.1007/s00438-008-0366-x pmid: 18636276 |
| [46] |
van Leeuwen H, Monfort A, Puigdomenech P . Mutator- like elements identified in melon, Arabidopsis and rice contain ULP1 protease domains. Mol Genet Genomics, 2007,277(4):357-364.
doi: 10.1007/s00438-006-0194-9 pmid: 17136348 |
| [47] |
Rossi M , Araujo PG, de Jesus EM, Varani AM, Van Sluys MA. Comparative analysis ofMutator-like transposases in sugarcane. Mol Genet Genomics, 2004,272(2):194-203.
doi: 10.1007/s00438-004-1036-2 pmid: 15338280 |
| [48] |
Neuvéglise C, Chalvet F, Wincker P, Gaillardin C, Casaregola S . Mutator-like element in the yeast Yarrowia lipolytica displays multiple alternative splicings. Eukaryot Cell, 2005,4(3):615-624.
doi: 10.1128/EC.4.3.615-624.2005 pmid: 15755923 |
| [49] |
Chalvet F, Grimaldi C, Kaper F, Langin T, Daboussi MJ . Hop, an active Mutator-like element in the genome of the fungus Fusarium oxysporum. Mol Biol Evol, 2003,20(8):1362-1375.
doi: 10.1093/molbev/msg155 pmid: 12777515 |
| [50] |
Lopes FR, Silva JC, Benchimol M, Costa GG, Pereira GA, Carareto CM . The protist Trichomonas vaginalis harbors multiple lineages of transcriptionally active Mutator-like elements. BMC Genomics, 2009,10:330.
doi: 10.1186/1471-2164-10-330 pmid: 19622157 |
| [51] |
Pritham EJ, Feschotte C, Wessler SR . Unexpected diversity and differential success of DNA transposons in four species of Entamoeba protozoans. Mol Biol Evol, 2005,22(9):1751-1763.
doi: 10.1093/molbev/msi169 pmid: 15901838 |
| [52] |
Jacinto DS, Muniz Hdos S, Venancio TM, Wilson RA, Verjovski-Almeida S, Demarco R . Curupira-1 and Curupira-2, two novel Mutator-like DNA transposons from the genomes of human parasites Schistosoma mansoni and Schistosoma japonicum. Parasitology, 2011,138(9):1124-1133.
doi: 10.1017/S0031182011000886 |
| [53] |
Marquez CP, Pritham EJ . Phantom, a new subclass of Mutator DNA transposons found in insect viruses and widely distributed in animals. Genetics, 2010,185(4):1507-1517.
doi: 10.1534/genetics.110.116673 pmid: 20457878 |
| [54] |
Singer T, Yordan C, Martienssen RA . Robertson's Mutator transposons in A. thaliana are regulated by the chromatin-remodeling gene Decrease in DNA Methylation (DDM1). Genes Dev, 2001,15(5):591-602.
doi: 10.1101/gad.193701 pmid: 11238379 |
| [55] |
Xu NZ, Yan XH, Maurais S, Fu HH , O'Brien DG, Mottinger J, Dooner HK Jittery, a Mutator distant relative with a paradoxical mobile behavior: excision without reinsertion. Plant Cell, 2004,16(5):1105-1114.
doi: 10.1105/tpc.019802 pmid: 15075398 |
| [56] |
Li YB, Harris L, Dooner HK . TED, an autonomous and rare maize transposon of the Mutator superfamily with a high gametophytic excision frequency. Plant Cell, 2013,25(9):3251-3265.
doi: 10.1105/tpc.113.116517 |
| [57] |
Gao DY . Identification of an active Mutator-like element (MULE) in rice (Oryza sativa). Mol Genet Genomics, 2012,287(3):261-271.
doi: 10.1007/s00438-012-0676-x |
| [58] |
Liu K, Wessler SR . Functional characterization of the active Mutator-like transposable element, Muta1 from the mosquito aedes aegypti. Mob DNA, 2017,8:1.
doi: 10.1186/s13100-016-0084-6 pmid: 28096902 |
| [59] |
Ming R, Vanburen R, Liu Y, Yang M, Han Y, Li LT, Zhang Q, Kim MJ, Schatz MC, Campbell M, Li J, Bowers JE, Tang H, Lyons E, Ferguson AA, Narzisi G, Nelson DR, Blaby-Haas CE, Gschwend AR, Jiao Y, Der JP, Zeng F, Han J, Min XJ, Hudson KA, Singh R, Grennan AK, Karpowicz SJ, Watling JR, Ito K, Robinson SA, Hudson ME, Yu Q, Mockler TC, Carroll A, Zheng Y, Sunkar R, Jia R, Chen N, Arro J, Wai CM, Wafula E, Spence A, Han Y, Xu L, Zhang J, Peery R, Haus MJ, Xiong W, Walsh JA, Wu J, Wang ML, Zhu YJ, Paull RE, Britt AB, Du C, Downie SR, Schuler MA, Michael TP, Long SP, Ort DR, Schopf JW, Gang DR, Jiang N, Yandell M, Depamphilis CW, Merchant SS, Paterson AH, Buchanan BB, Li S, Shen-Miller J . Genome of the long-living sacred lotus (Nelumbo nucifera Gaertn.). Genome Biol, 2013,14(5):R41.
doi: 10.1186/gb-2013-14-5-r41 pmid: 23663246 |
| [60] |
Ferguson AA, Jiang N . Mutator-like elements with multiple long terminal inverted repeats in plants. Comp Funct Genomics, 2012,2012:695827.
doi: 10.1155/2012/695827 pmid: 22474413 |
| [61] |
Holligan D, Zhang XY, Jiang N, Pritham EJ, Wessler SR . The transposable element landscape of the model legume Lotus japonicus. Genetics, 2006,174(4):2215-2228.
doi: 10.1534/genetics.106.062752 pmid: 17028332 |
| [62] |
Hoen DR, Park KC, Elrouby N, Yu Z, Mohabir N, Cowan RK, Bureau TE . Transposon-mediated expansion and diversification of a family of ULP-like genes. Mol Biol Evol, 2006,23(6):1254-1268.
doi: 10.1093/molbev/msk015 pmid: 16581939 |
| [63] |
Jiang N, Bao ZR, Zhang XY, Eddy SR, Wessler SR . Pack-MULE transposable elements mediate gene evolution in plants. Nature, 2004,431(7008):569-573.
doi: 10.1038/nature02953 pmid: 15457261 |
| [64] |
Lisch D . Mutator transposons. Trends Plant Sci, 2002,7(11):498-504.
doi: 10.1016/s1360-1385(02)02347-6 pmid: 12417150 |
| [65] |
Yu Z, Wright SI, Bureau TE . Mutator-like elements in Arabidopsis thaliana: structure, diversity and evolution. Genetics, 2000,156(4):2019-2031.
pmid: 11102392 |
| [66] |
Wang J, Yu Y, Tao F, Zhang JW, Copetti D, Kudrna D, Talag J, Lee S, Wing RA, Fan CZ . DNA methylation changes facilitated evolution of genes derived from Mutator-like transposable elements. Genome Biol, 2016,17(1):92.
doi: 10.1186/s13059-016-0954-8 pmid: 27154274 |
| [67] |
Ferguson AA, Zhao D, Jiang N . Selective acquisition and retention of genomic sequences by Pack-Mutator- like elements based on guanine-cytosine content and the breadth of expression. Plant Physiol, 2013,163(3):1419-1432.
doi: 10.1104/pp.113.223271 |
| [68] |
Jiang N, Ferguson AA, Slotkin RK, Lisch D . Pack-Mutator-like transposable elements (Pack-MULEs) induce directional modification of genes through biased insertion and DNA acquisition. Proc Natl Acad Sci USA, 2011,108(4):1537-1542.
doi: 10.1073/pnas.1010814108 pmid: 21220310 |
| [69] |
Zhao D, Hamilton JP, Vaillancourt B, Zhang W, Eizenga GC, Cui Y, Jiang J, Buell CR, Jiang N . The unique epigenetic features of Pack-MULEs and their impact on chromosomal base composition and expression spectrum. Nucleic Acids Res, 2018,46(5):2380-2397.
doi: 10.1093/nar/gky025 pmid: 29365184 |
| [70] |
Benito MI, Walbot V . Characterization of the maizeMutator transposable element MURA transposase as a DNA-binding protein. Mol Cell Biol, 1997,17(9):5165-5175.
doi: 10.1128/mcb.17.9.5165 pmid: 9271394 |
| [71] |
Zhao D, Ferguson A, Jiang N . Transposition of a rice Mutator-like element in the yeast Saccharomyces cerevisiae. Plant Cell, 2015,27(1):132-148.
doi: 10.1105/tpc.114.128488 pmid: 25587002 |
| [72] |
Raizada MN, Benito MI, Walbot V . The MuDR transposon terminal inverted repeat contains a complex plant promoter directing distinct somatic and germinal programs. Plant J, 2001,25(1):79-91.
doi: 10.1046/j.1365-313x.2001.00939.x pmid: 11169184 |
| [73] | Lisch D . Mutator and MULE transposons. Microbiol Spectr, 2015,3(2):A3-A32. |
| [74] | Tan BC, Chen ZL, Shen Y, Zhang YF, Lai JS, Sun SSM . Identification of an active new Mutator transposable element in maize. G3 (Bethesda), 2011,1(4):293-302. |
| [75] |
Dietrich CR, Cui F, Packila ML, Li J, Ashlock DA, Nikolau BJ, Schnable PS . Maize Mu transposons are targeted to the 5' untranslated region of the gl8 gene and sequences flanking Mu target-site duplications exhibit nonrandom nucleotide composition throughout the genome. Genetics, 2002,160(2):697-716.
pmid: 11861572 |
| [76] | Bennetzen JL, Springer P, Cresse AD, Hendrickx M . Specificity and regulation of the Mutator transposable element system in maize. Crit Rev Plant Sci, 1993,12(1-2):57. |
| [77] |
Liu K, Wessler SR . Transposition of Mutator-like transposable elements (MULEs) resembles hAT and Transib elements and V(D)J recombination. Nucleic Acids Res, 2017,45(11):6644-6655.
doi: 10.1093/nar/gkx357 pmid: 28482040 |
| [78] |
Wang QH, Dooner HK . Remarkable variation in maize genome structure inferred from haplotype diversity at the bz locus. Proc Natl Acad Sci USA, 2006,103(47):17644-17649.
doi: 10.1073/pnas.0603080103 pmid: 17101975 |
| [79] |
Eisen JA, Benito MI, Walbot V . Sequence similarity of putative transposases links the maize Mutator autonomous element and a group of bacterial insertion sequences. Nucleic Acids Res, 1994,22(13):2634-2636.
doi: 10.1093/nar/22.13.2634 pmid: 8041625 |
| [80] |
Hua-Van A, Capy P . Analysis of the DDE motif in the Mutator superfamily. J Mol Evol, 2008,67(6):670-681.
doi: 10.1007/s00239-008-9178-1 |
| [81] |
Raizada MN, Walbot V . The late developmental pattern of Mu transposon excision is conferred by a cauliflower mosaic virus 35S-driven MURA cDNA in transgenic maize. Plant Cell, 2000,12(1):5-21.
doi: 10.1105/tpc.12.1.5 pmid: 10634904 |
| [82] |
Lisch D, Girard L, Donlin M, Freeling M . Functional analysis of deletion derivatives of the maize transposon MuDR delineates roles for the MURA and MURB proteins. Genetics, 1999,151(1):331-341.
pmid: 9872971 |
| [83] |
Aziz RK, Breitbart M, Edwards RA . Transposases are the most abundant, most ubiquitous genes in nature. Nucleic Acids Res, 2010,38(13):4207-4217.
doi: 10.1093/nar/gkq140 pmid: 20215432 |
| [84] |
Yuan YW, Wessler SR . The catalytic domain of all eukaryotic cut-and-paste transposase superfamilies. Proc Natl Acad Sci USA, 2011,108(19):7884-7889.
doi: 10.1073/pnas.1104208108 pmid: 21518873 |
| [85] |
Hickman AB, Ewis HE, Li X, Knapp JA, Laver T, Doss AL, Tolun G, Steven AC, Grishaev A, Bax A, Atkinson PW, Craig NL, Dyda F . Structural basis of hAT transposon end recognition by Hermes, an octameric DNA transposase from Musca domestica. Cell, 2014,158(2):353-367.
doi: 10.1016/j.cell.2014.05.037 |
| [86] |
Zhou L, Mitra R, Atkinson PW, Hickman AB, Dyda F, Craig NL . Transposition of hAT elements links transposable elements and V(D)J recombination. Nature, 2004,432(7020):995-1001.
doi: 10.1038/nature03157 pmid: 15616554 |
| [87] |
Babu MM, Iyer LM, Balaji S, Aravind L . The natural history of the WRKY-GCM1 zinc fingers and the relationship between transcription factors and transposons. Nucleic Acids Res, 2006,34(22):6505-6520.
doi: 10.1093/nar/gkl888 pmid: 17130173 |
| [88] | Mccarty DR, Meeley RB. Transposon resources for forward and reverse genetics in maize. In: Bennetzen JL, Hake S, eds. Handbook of Maize: Genetics and Genomics.New York, NY: Springer New York, 2009, 561-584. |
| [89] |
Liang L, Zhou L, Tang Y, Li N, Song T, Shao W, Zhang Z, Cai P, Feng F, Ma Y, Yao D, Feng Y, Ma Z, Zhao H, Song R . A Sequence-Indexed mutator insertional library for maize functional genomics study. Plant Physiol, 2019,181(4):1404-1414.
doi: 10.1104/pp.19.00894 pmid: 31636104 |
| [90] |
Hickman AB, Chandler M, Dyda F . Integrating prokaryotes and eukaryotes: DNA transposases in light of structure. Crit Rev Biochem Mol Biol, 2010,45(1):50-69.
doi: 10.3109/10409230903505596 pmid: 20067338 |
| [91] |
Hsia AP, Schnable PS . DNA sequence analyses support the role of interrupted gap repair in the origin of internal deletions of the maize transposon, MuDR. Genetics, 1996,142(2):603-618.
pmid: 8852857 |
| [92] |
Donlin MJ, Lisch D, Freeling M . Tissue-specific accumulation of MURB, a protein encoded by MuDR, the autonomous regulator of the Mutator transposable element family. Plant Cell, 1995,7(12):1989-2000.
doi: 10.1105/tpc.7.12.1989 pmid: 8718617 |
| [93] |
Britt AB, Walbot V . Germinal and somatic products of Mu1 excision from the Bronze-1 gene of Zea mays. Mol Gen Genet, 1991,227(2):267-276.
doi: 10.1007/bf00259680 pmid: 1648169 |
| [94] |
Doseff A, Martienssen R, Sundaresan V . Somatic excision of the Mu1 transposable element of maize. Nucleic Acids Res, 1991,19(3):579-584.
doi: 10.1093/nar/19.3.579 pmid: 1849263 |
| [95] |
Raizada MN, Nan GL, Walbot V . Somatic and germinal mobility of the RescueMu transposon in transgenic maize. Plant Cell, 2001,13(7):1587-1608.
doi: 10.1105/tpc.010002 pmid: 11449053 |
| [96] |
Li J, Harper LC, Golubovskaya I, Wang CR, Weber D, Meeley RB, Mcelver J, Bowen B, Cande WZ, Schnable PS . Functional analysis of maize RAD51 in meiosis and double-strand break repair. Genetics, 2007,176(3):1469-1482.
doi: 10.1534/genetics.106.062604 pmid: 17507687 |
| [97] |
Franklin AE, Mcelver J, Sunjevaric I, Rothstein R, Bowen B, Cande WZ . Three-dimensional microscopy of the Rad51 recombination protein during meiotic prophase. Plant Cell, 1999,11(5):809-824.
doi: 10.1105/tpc.11.5.809 pmid: 10330467 |
| [98] |
Loessner I, Dietrich K, Dittrich D, Hacker J, Ziebuhr W . Transposase-dependent formation of circular IS 256 derivatives in Staphylococcus epidermidis and Staphylococcus aureus. J Bacteriol, 2002,184(17):4709-4714.
doi: 10.1128/jb.184.17.4709-4714.2002 pmid: 12169594 |
| [99] |
Sundaresan V, Freeling M . An extrachromosomal form of the Mu transposons of maize. Proc Natl Acad Sci USA, 1987,84(14):4924-4928.
doi: 10.1073/pnas.84.14.4924 pmid: 3037528 |
| [100] |
Gorbunova V, Levy AA . Analysis of extrachromosomal Ac/Ds transposable elements. Genetics, 2000,155(1):349-359.
pmid: 10790408 |
| [101] |
Gorbunova V, Levy AA . Circularized Ac/Ds transposons: formation, structure and fate. Genetics, 1997,145(4):1161-1169.
pmid: 9093867 |
| [102] |
Settles AM, Holding DR, Tan BC, Latshaw SP, Liu J, Suzuki M, Li L , O'Brien BA, Fajardo DS, Wroclawska E, Tseung CW, Lai J, Hunter CT, Avigne WT, Baier J, Messing J, Hannah LC, Koch KE, Becraft PW, Larkins BA, Mccarty DR. Sequence-indexed mutations in maize using the UniformMu transposon-tagging population. BMC Genomics, 2007,8:116.
doi: 10.1186/1471-2164-8-116 pmid: 17490480 |
| [103] |
Fernandes J, Dong QF, Schneider B, Morrow DJ, Nan GL, Brendel V, Walbot V . Genome-wide mutagenesis of Zea mays L. using RescueMu transposons. Genome Biol, 2004,5(10):R82.
doi: 10.1186/gb-2004-5-10-r82 pmid: 15461800 |
| [104] |
Cresse AD, Hulbert SH, Brown WE, Lucas JR, Bennetzen JL . Mu1-related transposable elements of maize preferentially insert into low copy number DNA. Genetics, 1995,140(1):315-324.
pmid: 7635296 |
| [105] |
Hanley S, Edwards D, Stevenson D, Haines S, Hegarty M, Schuch W, Edwards KJ . Identification of transposon- tagged genes by the random sequencing of Mutator- tagged DNA fragments from Zea mays. Plant J, 2000,23(4):557-566.
doi: 10.1046/j.1365-313x.2000.00830.x pmid: 10972882 |
| [106] |
Wong GK, Wang J, Tao L, Tan J, Zhang J, Passey DA, Yu J . Compositional gradients in gramineae genes. Genome Res, 2002,12(6):851-856.
doi: 10.1101/gr.189102 pmid: 12045139 |
| [107] |
Liu S, Yeh CT, Ji T, Ying K, Wu H, Tang HM, Fu Y, Nettleton D, Schnable PS . Mu transposon insertion sites and meiotic recombination events co-localize with epigenetic marks for open chromatin across the maize genome. PLoS Genet, 2009,5(11):e1000733.
doi: 10.1371/journal.pgen.1000733 pmid: 19936291 |
| [108] |
Sultana T, Zamborlini A, Cristofari G, Lesage P . Integration site selection by retroviruses and transposable elements in eukaryotes. Nat Rev Genet, 2017,18(5):292-308.
doi: 10.1038/nrg.2017.7 pmid: 28286338 |
| [109] |
Springer NM, Anderson SN, Andorf CM, Ahern KR, Bai F, Barad O, Barbazuk WB, Bass HW, Baruch K, Ben-Zvi G, Buckler ES, Bukowski R, Campbell MS, Cannon E, Chomet P, Dawe RK, Davenport R, Dooner HK, Du LH, Du C, Easterling KA, Gault C, Guan JC, Hunter CT, Jander G, Jiao Y, Koch KE, Kol G, Köllner TG, Kudo T, Li Q, Lu F, Mayfield-Jones D, Mei W, Mccarty DR, Noshay JM, Portwood JN, Ronen G, Settles AM, Shem-Tov D, Shi J, Soifer I, Stein JC, Stitzer MC, Suzuki M, Vera DL, Vollbrecht E, Vrebalov JT, Ware D, Wei S, Wimalanathan K, Woodhouse MR, Xiong W, Brutnell TP . The maize W22 genome provides a foundation for functional genomics and transposon biology. Nat Genet, 2018,50(9):1282-1288.
doi: 10.1038/s41588-018-0158-0 pmid: 30061736 |
| [110] |
Shepherd NS, Rhoades MM, Dempsey E . Genetic and molecular characterization of a-mrh-Mrh, a new mutable system of Zea mays. Dev Genet, 1989,10(6):507-519.
doi: 10.1002/dvg.1020100610 pmid: 2557991 |
| [1] | . Advances in studies of Sleeping Beauty transposon [J]. HEREDITAS, 2007, 29(7): 785-792. |
| [2] | FU Guang-Ming, SU Qiao, AN Li-Jia. The Progresses of Tol2 Transposable Element of Medaka Fish [J]. HEREDITAS, 2006, 28(7): 899-905. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||