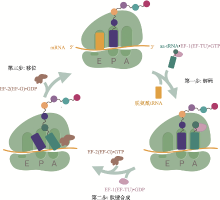
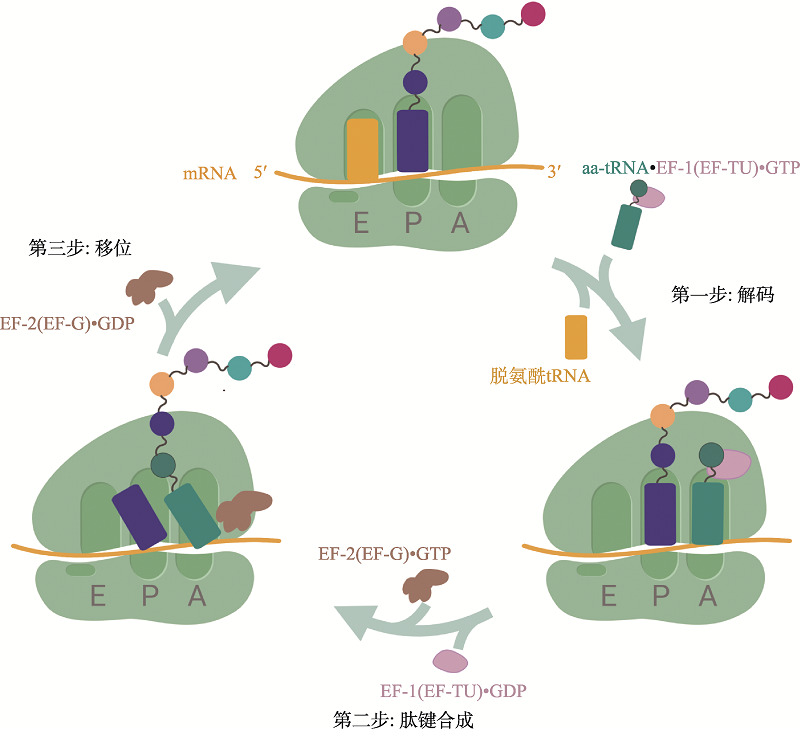
Hereditas(Beijing) ›› 2020, Vol. 42 ›› Issue (7): 613-631.doi: 10.16288/j.yczz.20-074
• Invited Review • Next Articles
Cis-regulatory mechanisms and biological effects of translation elongation
Taolan Zhao1, Shuo Zhang1,2, Wenfeng Qian1,2( )
)
- 1. State Key Laboratory of Plant Genomics, Institute of Genetics and Developmental Biology, Innovation Academy for Seed Design, Chinese Academy of Sciences, Beijing 100101, China
2. University of Chinese Academy of Sciences, Beijing 100049, China
-
Received:2020-03-18Revised:2020-06-28Online:2020-07-20Published:2020-07-07 -
Contact:Qian Wenfeng E-mail:wfqian@genetics.ac.cn -
Supported by:Supported by the National Key R&D Program of China No(2019YFA0508700);the National Natural Science Foundation of China Nos(31900455);the National Natural Science Foundation of China Nos(91331112)
Cite this article
Taolan Zhao, Shuo Zhang, Wenfeng Qian. Cis-regulatory mechanisms and biological effects of translation elongation[J]. Hereditas(Beijing), 2020, 42(7): 613-631.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
| [1] | Zheng CX, Ma XF, Zhang YH, Li HJ, Zhang GF . Research progress in the mechanism of translation initiation of eukaryotic mRNAs. Hereditas(Beijing), 2018,40(8):607-619. |
| 郑超星, 马小凤, 张永华, 李洪杰, 张根发 . 真核生物mRNA翻译起始机制研究进展. 遗传, 2018,40(8):607-619. | |
| [2] | Stein KC, Frydman J . The stop-and-go traffic regulating protein biogenesis: how translation kinetics controls proteostasis. J Biol Chem, 2019,294(6):2076-2084. |
| [3] | Richter JD, Coller J . Pausing on polyribosomes: make way for elongation in translational control. Cell, 2015,163(2):292-300. |
| [4] | Choi J, Grosely R, Prabhakar A, Lapointe CP, Wang Jf, Puglisi JD . How messenger RNA and nascent chain sequences regulate translation elongation. Annu Rev Biochem, 2018,87:421-449. |
| [5] | Buskirk AR, Green R . Ribosome pausing, arrest and rescue in bacteria and eukaryotes. Philos Trans R Soc Lond B Biol Sci, 2017,372(1716):20160183. |
| [6] | Voorhees RM, Ramakrishnan V . Structural basis of the translational elongation cycle. Annu Rev Biochem, 2013,82:203-236. |
| [7] | Lin JZ, Gagnon MG, Bulkley D, Steitz TA . Conformational changes of elongation factor G on the ribosome during tRNA translocation. Cell, 2015,160(0):219-227. |
| [8] | Gagnon MG, Lin JZ, Bulkley D, Steitz TA . Crystal structure of elongation factor 4 bound to a clockwise ratcheted ribosome. Science, 2014,345(6197):684-687. |
| [9] | Mustoe AM, Brooks CL, Al-Hashimi HM . Hierarchy of RNA functional dynamics. Annu Rev Biochem, 2014,83:441-466. |
| [10] | Javed A, Christodoulou J, Cabrita LD, Orlova EV . The ribosome and its role in protein folding: looking through a magnifying glass. Acta Crystallogr D Struct Biol, 2017,73(pt 6):509-521. |
| [11] | Nygaard R, Romaniuk JAH, Rice DM, Cegelski L . Whole ribosome NMR: dipolar couplings and contributions to whole cells. J Phys Chem B, 2017,121(40):9331-9335. |
| [12] | Bai XC, Fernandez IS, McMullan G, Scheres SH. Ribosome structures to near-atomic resolution from thirty thousand cryo-EM particles. eLife, 2013,2:e00461. |
| [13] | Amunts A, Brown A, Bai XC, Llacer JL, Hussain T, Emsley P, Long F, Murshudov G, Scheres SHW, Ramakrishnan V . Structure of the yeast mitochondrial large ribosomal subunit. Science, 2014,343(6178):1485-1489. |
| [14] | Liu Z, Gutierrez-Vargas C, Wei J, Grassucci RA, Sun M, Espina N, Madison-Antenucci S, Tong L, Frank J . Determination of the ribosome structure to a resolution of 2.5 Å by single-particle cryo-EM. Protein Sci, 2017,26(1):82-92. |
| [15] | Loveland AB, Demo G, Grigorieff N, Korostelev AA . Ensemble cryo-EM elucidates the mechanism of translation fidelity. Nature, 2017,546(7656):113-117. |
| [16] | Fischer N, Neumann P, Konevega AL, Bock LV, Ficner R, Rodnina MV, Stark H . Structure of the E. coli ribosome-EF-Tu complex at <3 Å resolution by Cs-corrected cryo-EM. Nature, 2015,520(7548):567-570. |
| [17] | Zhang YQ, Mandava CS, Cao W, Li XJ, Zhang DJ, Li NN, Zhang YX, Zhang XX, Qin Y, Mi KX, Lei JL, Sanya S, Gao N . HflX is a ribosome-splitting factor rescuing stalled ribosomes under stress conditions. Nat Struct Mol Biol, 2015,22(11):906-913. |
| [18] | Johansson M, Bouakaz E, Lovmar M, Ehrenberg M . The kinetics of ribosomal peptidyl transfer revisited. Mol Cell, 2008,30(5):589-598. |
| [19] | Wang JF, Kwiatkowski M, Forster AC . Kinetics of ribosome-catalyzed polymerization using artificial aminoacyl-tRNA substrates clarifies inefficiencies and improvements. ACS Chem Biol, 2015,10(10):2187-2192. |
| [20] | Wohlgemuth I, Brenner S, Beringer M, Rodnina MV . Modulation of the rate of peptidyl transfer on the ribosome by the nature of substrates. J Biol Chem, 2008,283(47):32229-32235. |
| [21] | Chu D, Kazana E, Bellanger N, Singh T , Tuite MF, von der Haar T. Translation elongation can control translation initiation on eukaryotic mRNAs. EMBO J, 2014,33(1):21-34. |
| [22] | Morisaki T, Stasevich TJ . Quantifying single mRNA translation kinetics in living cells. Cold Spring Harb Perspect Biol, 2018,10(11):a032078. |
| [23] | Yan XW, Hoek TA, Vale RD, Tanenbaum ME . Dynamics of translation of single mRNA molecules in vivo. Cell, 2016,165(4):976-989. |
| [24] | Wu B, Eliscovich C, Yoon YJ, Singer RH . Translation dynamics of single mRNAs in live cells and neurons. Science, 2016,352(6292):1430-1435. |
| [25] | Morisaki T, Lyon K, DeLuca KF, DeLuca JG, English BP, Zhang ZJ, Lavis LD, Grimm JB, Viswanathan S, Looger LL, Lionnet T, Stasevich TJ. Real-time quantification of single RNA translation dynamics in living cells. Science, 2016,352(6292):1425-1429. |
| [26] | Steitz JA . Polypeptide chain initiation: nucleotide sequences of the three ribosomal binding sites in bacteriophage R17 RNA. Nature, 1969,224(5223):957-964. |
| [27] | Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS . Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science, 2009,324(5924):218-223. |
| [28] | Pelechano V, Wei W, Steinmetz LM . Widespread co-translational RNA decay reveals ribosome dynamics. Cell, 2015,161(6):1400-1412. |
| [29] | Michel AM, Baranov PV . Ribosome profiling: a Hi-Def monitor for protein synthesis at the genome-wide scale. Wiley Interdiscip Rev RNA, 2013,4(5):473-490. |
| [30] | Zhao J, Zhang G . Translatomics: methods and applications. Chem Life, 2017,37(1):70-79. |
| 赵晶, 张弓 . 翻译组学: 方法及应用. 生命的化学, 2017,37(1):70-79. | |
| [31] | Ingolia NT, Lareau LF, Weissman JS . Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell, 2011,147(4):789-802. |
| [32] | Qian WF, Yang JR, Pearson NM, Maclean C, Zhang JZ . Balanced codon usage optimizes eukaryotic translational efficiency. PLoS Genet, 2012,8(3):e1002603. |
| [33] | Weinberg DE, Shah P, Eichhorn SW, Hussmann JA, Plotkin JB, Bartel DP . Improved ribosome-footprint and mRNA measurements provide insights into dynamics and regulation of yeast translation. Cell Rep, 2016,14(7):1787-1799. |
| [34] | Hussmann JA, Patchett S, Johnson A, Sawyer S, Press WH. Understanding biases in ribosome profiling experiments reveals signatures of translation dynamics in yeast. PLoS Genet, 2015,11(12):e1005732. |
| [35] | Subramaniam AR, Zid BM, O'Shea EK. An integrated approach reveals regulatory controls on bacterial translation elongation. Cell, 2014,159(5):1200-1211. |
| [36] | Guydosh NR, Green R . Dom34 rescues ribosomes in 3' untranslated regions. Cell, 2014,156(5):950-962. |
| [37] | Yang JR, Chen XS, Zhang JZ . Codon-by-codon modulation of translational speed and accuracy via mRNA folding. PLoS Biol, 2014,12(7):e1001910. |
| [38] | Dever TE, Dinman JD, Green R . Translation elongation and recoding in eukaryotes. Cold Spring Harb Perspect Biol, 2018,10(8):a032649. |
| [39] | Chen PJ, Huang YS . CPEB2-eEF2 interaction impedes HIF-1α RNA translation. EMBO J, 2012,31(4):959-971. |
| [40] | Grelet S, Howe PH,. hnRNP E1 at the crossroads of translational regulation of epithelial-mesenchymal transition. J Cancer Metastasis Treat, 2019,5:16. |
| [41] | Desnoyers G, Bouchard MP, Masse E . New insights into small RNA-dependent translational regulation in prokaryotes. Trends Genet, 2013,29(2):92-98. |
| [42] | Ambros V, Chen XM . The regulation of genes and genomes by small RNAs. Development, 2007,134(9):1635-1641. |
| [43] | Grantham R, Gautier C, Gouy M, Mercier R, Pavé A . Codon catalog usage and the genome hypothesis. Nucleic Acids Res, 1980,8(1):r49-r62. |
| [44] | Ikemura T . Correlation between the abundance of Escherichia coli transfer RNAs and the occurrence of the respective codons in its protein genes: A proposal for a synonymous codon choice that is optimal for the E. coli translational system. J Mol Biol, 1981,151(3):389-409. |
| [45] | Hershberg R, Petrov DA . Selection on codon bias. Annu Rev Genet, 2008,42:287-299. |
| [46] | Plotkin JB, Kudla G . Synonymous but not the same: the causes and consequences of codon bias. Nat Rev Genet, 2011,12(1):32-42. |
| [47] | Gingold H, Pilpel Y . Determinants of translation efficiency and accuracy. Mol Syst Biol, 2011,7:481. |
| [48] | Akashi H . Synonymous codon usage in Drosophila melanogaster: natural selection and translational accuracy. Genetics, 1994,136(3):927-935. |
| [49] | Stoletzki N, Eyre-Walker A . Synonymous codon usage in Escherichia coli: selection for translational accuracy. Mol Biol Evol, 2007,24(2):374-381. |
| [50] | Ikemura T . Correlation between the abundance of yeast transfer RNAs and the occurrence of the respective codons in protein genes. Differences in synonymous codon choice patterns of yeast and Escherichia coli with reference to the abundance of isoaccepting transfer RNAs. J Mol Biol, 1982,158(4):573-597. |
| [51] | Ikemura T . Codon usage and tRNA content in unicellular and multicellular organisms. Mol Biol Evol, 1985,2(1):13-34. |
| [52] | Novoa EM, Ribas de Pouplana L. Speeding with control: codon usage, tRNAs, and ribosomes. Trends Genet, 2012,28(11):574-581. |
| [53] | Buhr F, Jha S, Thommen M, Mittelstaet J, Kutz F, Schwalbe H, Rodnina MV, Komar AA . Synonymous codons direct cotranslational folding toward different protein conformations. Mol Cell, 2016,61(3):341-351. |
| [54] | Curran JF, Yarus M . Rates of aminoacyl-tRNA selection at 29 sense codons in vivo. J Mol Biol, 1989,209(1):65-77. |
| [55] | Sørensen MA, Kurland CG, Pedersen S . Codon usage determines translation rate in Escherichia coli. J Mol Biol, 1989,207(2):365-377. |
| [56] | Gardin J, Yeasmin R, Yurovsky A, Cai Y, Skiena S, Futcher B . Measurement of average decoding rates of the 61 sense codons in vivo. eLife, 2014,3:e03735. |
| [57] | Kertesz M, Iovino N, Unnerstall U, Gaul U, Segal E . The role of site accessibility in microRNA target recognition. Nat Genet, 2007,39(10):1278-1284. |
| [58] | Beaudoin JD, Novoa EM, Vejnar CE, Yartseva V, Takacs CM, Kellis M, Giraldez AJ . Analyses of mRNA structure dynamics identify embryonic gene regulatory programs. Nat Struct Mol Biol, 2018,25(8):677-686. |
| [59] | Rouskin S, Zubradt M, Washietl S, Kellis M, Weissman JS . Genome-wide probing of RNA structure reveals active unfolding of mRNA structures in vivo. Nature, 2014,505(7485):701-705. |
| [60] | Chartrand P, Meng XH, Huttelmaier S, Donato D, Singer RH . Asymmetric sorting of ash1p in yeast results from inhibition of translation by localization elements in the mRNA. Mol Cell, 2002,10(6):1319-1330. |
| [61] | Qu XH, Wen JD, Lancaster L, Noller HF, Bustamante C, Tinoco I . The ribosome uses two active mechanisms to unwind messenger RNA during translation. Nature, 2011,475(7354):118-121. |
| [62] | Kertesz M, Wan Y, Mazor E, Rinn JL, Nutter RC, Chang HY, Segal E . Genome-wide measurement of RNA secondary structure in yeast. Nature, 2010,467(7311):103-107. |
| [63] | Tang Y, Assmann SM, Bevilacqua PC . Protein structure is related to RNA structural reactivity in vivo. J Mol Biol, 2016,428(5):758-766. |
| [64] | Watts JM, Dang KK, Gorelick RJ, Leonard CW, Bess JW, Swanstrom R, Burch CL, Weeks KM . Architecture and secondary structure of an entire HIV-1 RNA genome. Nature, 2009,460(7256):711-716. |
| [65] | Dinman JD . Mechanisms and implications of programmed translational frameshifting. Wiley Interdiscip Rev RNA, 2012,3(5):661-673. |
| [66] | Biswas P, Jiang X, Pacchia AL, Dougherty JP, Peltz SW . The human immunodeficiency virus type 1 ribosomal frameshifting site is an invariant sequence determinant and an important target for antiviral therapy. J Virol, 2004,78(4):2082-2087. |
| [67] | Wu F, Zhao S, Yu B, Chen YM, Wang W, Song ZG, Hu Y, Tao ZW, Tian JH, Pei YY, Yuan ML, Zhang YL, Dai FH, Liu Y, Wang QM, Zheng JJ, Xu L, Holmes EC, Zhang YZ . A new coronavirus associated with human respiratory disease in China. Nature, 2020,579(7798):265-269. |
| [68] | Caliskan N, Peske F, Rodnina MV . Changed in translation: mRNA recoding by -1 programmed ribosomal frameshifting. Trends Biochem Sci, 2015,40(5):265-274. |
| [69] | Mustoe AM, Busan S, Rice GM, Hajdin CE, Peterson BK, Ruda VM, Kubica N, Nutiu R, Baryza JL, Weeks KM. Pervasive regulatory functions of mRNA structure revealed by high-resolution SHAPE probing. Cell, 2018, 173(1):181-195.e118. |
| [70] | Su Z, Tang Y, Ritchey LE, Tack DC, Zhu M, Bevilacqua PC, Assmann SM . Genome-wide RNA structurome reprogramming by acute heat shock globally regulates mRNA abundance. Proc Natl Acad Sci USA, 2018,115(48):12170-12175. |
| [71] | Pavlov MY, Watts RE, Tan Z, Cornish VW, Ehrenberg M, Forster AC . Slow peptide bond formation by proline and other N-alkylamino acids in translation. Proc Natl Acad Sci USA, 2009,106(1):50-54. |
| [72] | Muto H, Ito K . Peptidyl-prolyl-tRNA at the ribosomal P-site reacts poorly with puromycin. Biochem Biophys Res Commun, 2008,366(4):1043-1047. |
| [73] | Melnikov S, Mailliot J, Rigger L, Neuner S, Shin BS, Yusupova G, Dever TE, Micura R, Yusupov M . Molecular insights into protein synthesis with proline residues. EMBO Rep, 2016,17(12):1776-1784. |
| [74] | Doerfel LK, Wohlgemuth I, Kothe C, Peske F, Urlaub H, Rodnina MV . EF-P is essential for rapid synthesis of proteins containing consecutive proline residues. Science, 2013,339(6115):85-88. |
| [75] | Ude S, Lassak J, Starosta AL, Kraxenberger T, Wilson DN, Jung K . Translation elongation factor EF-P alleviates ribosome stalling at polyproline stretches. Science, 2013,339(6115):82-85. |
| [76] | Gutierrez E, Shin BS, Woolstenhulme CJ, Kim JR, Saini P, Buskirk AR, Dever TE. eIF5A promotes translation of polyproline motifs. Mol Cell, 2013,51(1):35-45. |
| [77] | Woolstenhulme CJ, Parajuli S, Healey DW, Valverde DP, Petersen EN, Starosta AL, Guydosh NR, Johnson WE, Wilson DN, Buskirk AR . Nascent peptides that block protein synthesis in bacteria. Proc Natl Acad Sci USA, 2013,110(10):E878-E887. |
| [78] | Starosta AL, Lassak J, Peil L, Atkinson GC, Virumäe K, Tenson T, Remme J, Jung K, Wilson DN . Translational stalling at polyproline stretches is modulated by the sequence context upstream of the stall site. Nucleic Acids Res, 2014,42(16):10711-10719. |
| [79] | Peil L, Starosta AL, Lassak J, Atkinson GC, Virumäe K, Spitzer M, Tenson T, Jung K, Remme J, Wilson DN . Distinct XPPX sequence motifs induce ribosome stalling, which is rescued by the translation elongation factor EF-P. Proc Natl Acad Sci USA, 2013,110(38):15265-15270. |
| [80] | Rodnina MV . The ribosome in action: tuning of translational efficiency and protein folding. Protein Sci, 2016,25(8):1390-1406. |
| [81] | Ito K, Chiba S . Arrest peptides: cis-acting modulators of translation. Annu Rev Biochem, 2013,82:171-202. |
| [82] | Tenson T, Ehrenberg M . Regulatory nascent peptides in the ribosomal tunnel. Cell, 2002,108(5):591-594. |
| [83] | Nakatogawa H, Ito K . The ribosomal exit tunnel functions as a discriminating gate. Cell, 2002,108(5):629-636. |
| [84] | Muto H, Nakatogawa H, Ito K . Genetically encoded but nonpolypeptide prolyl-tRNA functions in the A site for SecM-mediated ribosomal stall. Mol Cell, 2006,22(4):545-552. |
| [85] | Tanner DR, Cariello DA, Woolstenhulme CJ, Broadbent MA, Buskirk AR . Genetic identification of nascent peptides that induce ribosome stalling. J Biol Chem, 2009,284(50):34809-34818. |
| [86] | Navon SP, Kornberg G, Chen J, Schwartzman T, Tsai A, Puglisi EV, Puglisi JD, Adir N . Amino acid sequence repertoire of the bacterial proteome and the occurrence of untranslatable sequences. Proc Natl Acad Sci USA, 2016,113(26):7166-7170. |
| [87] | Lu JL, Kobertz WR, Deutsch C . Mapping the electrostatic potential within the ribosomal exit tunnel. J Mol Biol, 2007,371(5):1378-1391. |
| [88] | Lu JL, Deutsch C . Electrostatics in the ribosomal tunnel modulate chain elongation rates. J Mol Biol, 2008,384(1):73-86. |
| [89] | Koutmou KS, Schuller AP, Brunelle JL, Radhakrishnan A, Djuranovic S, Green R . Ribosomes slide on lysine-encoding homopolymeric A stretches. eLife, 2015,4:e05534. |
| [90] | Charneski CA, Hurst LD . Positively charged residues are the major determinants of ribosomal velocity. PLoS Biol, 2013,11(3):e1001508. |
| [91] | Artieri CG, Fraser HB . Accounting for biases in riboprofiling data indicates a major role for proline in stalling translation. Genome Res, 2014,24(12):2011-2021. |
| [92] | Sabi R, Tuller T . A comparative genomics study on the effect of individual amino acids on ribosome stalling. BMC Genomics, 2015,16(Suppl.10):S5. |
| [93] | Ikeuchi K, Izawa T, Inada T . Recent progress on the molecular mechanism of quality controls induced by ribosome stalling. Front Genet, 2018,9:743. |
| [94] | Sharma AK, O'Brien EP. Non-equilibrium coupling of protein structure and function to translation-elongation kinetics. Curr Opin Struct Biol, 2018,49:94-103. |
| [95] | Matsuo Y, Ikeuchi K, Saeki Y, Iwasaki S, Schmidt C, Udagawa T, Sato F, Tsuchiya H, Becker T, Tanaka K, Ingolia NT, Beckmann R, Inada T . Ubiquitination of stalled ribosome triggers ribosome-associated quality control. Nat Commun, 2017,8(1):159. |
| [96] | Ikeuchi K, Tesina P, Matsuo Y, Sugiyama T, Cheng J, Saeki Y, Tanaka K, Becker T, Beckmann R, Inada T . Collided ribosomes form a unique structural interface to induce Hel2-driven quality control pathways. EMBO J, 2019,38(5):e100276. |
| [97] | Brandman O, Stewart-Ornstein J, Wong D, Larson A, Williams CC, Li GW, Zhou S, King D, Shen PS, Weibezahn J, Dunn JG, Rouskin S, Inada T, Frost A, Weissman JS . A ribosome-bound quality control complex triggers degradation of nascent peptides and signals translation stress. Cell, 2012,151(5):1042-1054. |
| [98] | Shen PS, Park J, Qin YD, Li XM, Parsawar K, Larson MH, Cox J, Cheng YF, Lambowitz AM, Weissman JS, Brandman O, Frost A . Protein synthesis. Rqc2p and 60S ribosomal subunits mediate mRNA-independent elongation of nascent chains. Science, 2015,347(6217):75-78. |
| [99] | Verma R, Reichermeier KM, Burroughs AM, Oania RS, Reitsma JM, Aravind L, Deshaies RJ . Vms1 and ANKZF1 peptidyl-tRNA hydrolases release nascent chains from stalled ribosomes. Nature, 2018,557(7705):446-451. |
| [100] | Zurita Rendon O, Fredrickson EK, Howard CJ, Van Vranken J, Fogarty S, Tolley ND, Kalia R, Osuna BA, Shen PS, Hill CP, Frost A, Rutter J . Vms1p is a release factor for the ribosome-associated quality control complex. Nat Commun, 2018,9(1):2197. |
| [101] | Defenouillère Q, Yao YH, Mouaikel J, Namane A, Galopier A, Decourty L, Doyen A, Malabat C, Saveanu C, Jacquier A, Fromont-Racine M . Cdc48-associated complex bound to 60S particles is required for the clearance of aberrant translation products. Proc Natl Acad Sci USA, 2013,110(13):5046-5051. |
| [102] | Verma R, Oania RS, Kolawa NJ, Deshaies RJ . Cdc48/p97 promotes degradation of aberrant nascent polypeptides bound to the ribosome. eLife, 2013,2:e00308. |
| [103] | Doma MK, Parker R . Endonucleolytic cleavage of eukaryotic mRNAs with stalls in translation elongation. Nature, 2006,440(7083):561-564. |
| [104] | Shoemaker CJ, Eyler DE, Green R . Dom34:Hbs1 promotes subunit dissociation and peptidyl-tRNA drop-off to initiate no-go decay. Science, 2010,330(6002):369-372. |
| [105] | Shoemaker CJ, Green R . Kinetic analysis reveals the ordered coupling of translation termination and ribosome recycling in yeast. Proc Natl Acad Sci USA, 2011,108(51):E1392-1398. |
| [106] | Pisareva VP, Skabkin MA, Hellen CU, Pestova TV, Pisarev AV . Dissociation by Pelota, Hbs1 and ABCE1 of mammalian vacant 80S ribosomes and stalled elongation complexes. EMBO J, 2011,30(9):1804-1817. |
| [107] | Dimitrova LN, Kuroha K, Tatematsu T, Inada T . Nascent peptide-dependent translation arrest leads to Not4p-mediated protein degradation by the proteasome. J Biol Chem, 2009,284(16):10343-10352. |
| [108] | Juszkiewicz S, Hegde RS. Initiation of quality control during poly(A)translation requires site-specific ribosome ubiquitination. Mol Cell, 2017, 65(4): 743-750.e744. |
| [109] | Bengtson MH, Joazeiro CAP . Role of a ribosome- associated E3 ubiquitin ligase in protein quality control. Nature, 2010,467(7314):470-473. |
| [110] | Letzring DP, Wolf AS, Brule CE, Grayhack EJ . Translation of CGA codon repeats in yeast involves quality control components and ribosomal protein L1. RNA, 2013,19(9):1208-1217. |
| [111] | Simms CL, Hudson BH, Mosior JW, Rangwala AS, Zaher HS . An active role for the ribosome in determining the fate of oxidized Mrna. Cell Rep, 2014,9(4):1256-1264. |
| [112] | Ishimura R, Nagy G, Dotu I, Zhou HH, Yang XL, Schimmel P, Senju S, Nishimura Y, Chuang JH, Ackerman SL . RNA function. Ribosome stalling induced by mutation of a CNS-specific tRNA causes neurodegeneration. Science, 2014,345(6195):455-459. |
| [113] | Saini P, Eyler DE, Green R, Dever TE . Hypusine- containing protein eIF5A promotes translation elongation. Nature, 2009,459(7243):118-121. |
| [114] | Shah P, Ding Y, Niemczyk M, Kudla G, Plotkin JB . Rate-limiting steps in yeast protein translation. Cell, 2013,153(7):1589-1601. |
| [115] | Carlini DB, Stephan W . In vivo introduction of unpreferred synonymous codons into the Drosophila Adh gene results in reduced levels of ADH protein. Genetics, 2003,163(1):239-243. |
| [116] | Lampson BL, Pershing NL, Prinz JA, Lacsina JR, Marzluff WF, Nicchitta CV, MacAlpine DM, Counter CM,. Rare codons regulate KRas oncogenesis. Curr Biol, 2013,23(1):70-75. |
| [117] | Torrent M, Chalancon G, de Groot NS, Wuster A, Madan Babu M,. Cells alter their tRNA abundance to selectively regulate protein synthesis during stress conditions. Sci Signal, 2018,11(546): eaat6409. |
| [118] | Zhao TL, Huan Q, Sun J, Liu CY, Hou XL, Yu X, Silverman IM, Zhang Y, Gregory BD, Liu CM, Qian WF, Cao XF . Impact of poly(A)-tail G-content on Arabidopsis PAB binding and their role in enhancing translational efficiency. Genome Biol, 2019,20:189. |
| [119] | Chen SY, Li K, Cao WQ, Wang J, Zhao T, Huan Q, Yang YF, Wu SH, Qian WF . Codon-resolution analysis reveals a direct and context-dependent impact of individual synonymous mutations on mRNA level. Mol Biol Evol, 2017,34(11):2944-2958. |
| [120] | Schikora-Tamarit MA, Carey LB . Poor codon optimality as a signal to degrade transcripts with frameshifts. Transcription, 2018,9(5):327-333. |
| [121] | Espinar L, Schikora Tamarit MÀ, Domingo J, Carey LB . Promoter architecture determines cotranslational regulation of mRNA. Genome Res, 2018,28(4):509-518. |
| [122] | Bazzini AA, Del Viso F, Moreno-Mateos MA, Johnstone TG, Vejnar CE, Qin YD, Yao J, Khokha MK, Giraldez AJ . Codon identity regulates mRNA stability and translation efficiency during the maternal-to-zygotic transition. EMBO J, 2016,35(19):2087-2103. |
| [123] | Presnyak V, Alhusaini N, Chen YH, Martin S, Morris N, Kline N, Olson S, Weinberg D, Baker KE, Graveley BR, Coller J . Codon optimality is a major determinant of mRNA stability. Cell, 2015,160(6):1111-1124. |
| [124] | Radhakrishnan A, Chen YH, Martin S, Alhusain N, Green R, Coller J. The DEAD-box protein Dhh1p couples mRNA decay and translation by monitoring codon optimality. Cell, 2016, 167(1): 122-132. e129. |
| [125] | Mishima Y, Tomari Y . Codon usage and 3' UTR length determine maternal mRNA stability in zebrafish. Mol Cell, 2016,61(6):874-885. |
| [126] | Siegel V, Walter P . Each of the activities of signal recognition particle (SRP) is contained within a distinct domain: analysis of biochemical mutants of SRP. Cell, 1988,52(1):39-49. |
| [127] | Halic M, Becke T, Pool MR, Spahn CM, Grassucci RA, Frank J, Beckmann R . Structure of the signal recognition particle interacting with the elongation-arrested ribosome. Nature, 2004,427(6977):808-814. |
| [128] | Crick SL, Ruff KM, Garai K, Frieden C, Pappu RV . Unmasking the roles of N- and C-terminal flanking sequences from exon 1 of huntingtin as modulators of polyglutamine aggregation. Proc Natl Acad Sci USA, 2013,110(50):20075-20080. |
| [129] | Brinkman RR, Mezei MM, Theilmann J, Almqvist E, Hayden MR . The likelihood of being affected with Huntington disease by a particular age, for a specific CAG size. Am J Hum Genet, 1997,60(5):1202-1210. |
| [130] | Li SH, Li XJ . Aggregation of N-terminal huntingtin is dependent on the length of its glutamine repeats. Hum Mol Genet, 1998,7(5):777-782. |
| [131] | Lee JM, Ramos EM, Lee JH, Gillis T, Mysore JS, Hayden MR, Warby SC, Morrison P, Nance M, Ross CA, Margolis RL, Squitieri F, Orobello S, Di Donato S, Gomez-Tortosa E, Ayuso C, Suchowersky O, Trent RJ, McCusker E, Novelletto A, Frontali M, Jones R, Ashizawa T, Frank S, Saint-Hilaire MH, Hersch SM, Rosas HD, Lucente D, Harrison MB, Zanko A, Abramson RK, Marder K, Sequeiros J, Paulsen JS, Landwehrmeyer GB, Myers RH, MacDonald ME, Gusella JF. CAG repeat expansion in Huntington disease determines age at onset in a fully dominant fashion. Neurology, 2012,78(9):690-695. |
| [132] | Rockabrand E, Slepko N, Pantalone A, Nukala VN, Kazantsev A, Marsh JL, Sullivan PG, Steffan JS, Sensi SL, Thompson LM . The first 17 amino acids of Huntingtin modulate its sub-cellular localization, aggregation and effects on calcium homeostasis. Hum Mol Genet, 2007,16(1):61-77. |
| [133] | Nissley DA, O'Brien EP. Altered co-translational processing plays a role in Huntington's pathogenesis—a hypothesis. Front Mol Neurosci, 2016,9:54. |
| [134] | Hartl FU . Protein misfolding diseases. Annu Rev Biochem, 2017,86(1):21-26. |
| [135] | Chiti F, Dobson CM . Protein misfolding, amyloid formation, and human disease: a summary of progress over the last decade. Annu Rev Biochem, 2017,86:27-68. |
| [136] | Soto C, Pritzkow S . Protein misfolding, aggregation, and conformational strains in neurodegenerative diseases. Nat Neurosci, 2018,21(10):1332-1340. |
| [137] | Ciryam P, Morimoto RI, Vendruscolo M, Dobson CM, O'Brien EP. In vivo translation rates can substantially delay the cotranslational folding of the Escherichia coli cytosolic proteome. Proc Natl Acad Sci USA, 2013,110(2):E132-140. |
| [138] | Komar AA . Synonymous codon usage-a guide for co-translational protein folding in the cell. Mol Biol (Mosk), 2019,53(6):883-898. |
| [139] | Clarke TF, Clark PL . Rare codons cluster. PLoS One, 2008,3(10):e3412. |
| [140] | Komar AA, Lesnik T, Reiss C . Synonymous codon substitutions affect ribosome traffic and protein folding during in vitro translation. FEBS Lett, 1999,462(3):387-391. |
| [141] | Zhang G, Ignatova Z . Folding at the birth of the nascent chain: coordinating translation with co-translational folding. Curr Opin Struct Biol, 2011,21(1):25-31. |
| [142] | Rodnina MV, Wintermeyer W . Protein elongation, co-translational folding and targeting. J Mol Biol, 2016,428(10):2165-2185. |
| [143] | Cortazzo P, Cerveñansky C, Marín M, Reiss C, Ehrlich R, Deana A . Silent mutations affect in vivo protein folding in Escherichia coli. Biochem Biophys Res Commun, 2002,293(1):537-541. |
| [144] | Kimchi-Sarfaty C, Oh JM, Kim IW, Sauna ZE, Calcagno AM, Ambudkar SV, Gottesman MM . A “silent” polymorphism in the MDR1 gene changes substrate specificity. Science, 2007,315(5811):525-528. |
| [145] | Walsh IM, Bowman MA, Soto Santarriaga IF, Rodriguez A, Clark PL . Synonymous codon substitutions perturb cotranslational protein folding in vivo and impair cell fitness. Proc Natl Acad Sci USA, 2020,117(7):3528-3534. |
| [146] | Agashe D, Sane M, Phalnikar K, Diwan GD, Habibullah A, Martinez-Gomez NC, Sahasrabuddhe V, Polachek W, Wang J, Chubiz LM, Marx CJ . Large-effect beneficial synonymous mutations mediate rapid and parallel adaptation in a bacterium. Mol Biol Evol, 2016,33(6):1542-1553. |
| [147] | Yang JR . Does mRNA structure contain genetic information for regulating co-translational protein folding? Zool Res, 2017,38(1):36-43. |
| [148] | Faure G, Ogurtsov AY, Shabalina SA, Koonin EV . Role of mRNA structure in the control of protein folding. Nucleic Acids Res, 2016,44(22):10898-10911. |
| [149] | Voss NR, Gerstein M, Steitz TA, Moore PB . The geometry of the ribosomal polypeptide exit tunnel. J Mol Biol, 2006,360(4):893-906. |
| [150] | Bhushan S, Gartmann M, Halic M, Armache JP, Jarasch A, Mielke T, Berninghausen O, Wilson DN, Beckmann R . Alpha-Helical nascent polypeptide chains visualized within distinct regions of the ribosomal exit tunnel. Nat Struct Mol Biol, 2010,17(3):313-317. |
| [151] | Nilsson OB, Hedman R, Marino J, Wickles S, Bischoff L, Johansson M, Müller-Lucks A, Trovato F, Puglisi JD, O'Brien EP, Beckmann R, von Heijne G. Cotranslational protein folding inside the ribosome exit tunnel. Cell Rep, 2015,12(10):1533-1540. |
| [152] | Lu JL, Deutsch C . Folding zones inside the ribosomal exit tunnel. Nat Struct Mol Biol, 2005,12(12):1123-1129. |
| [153] | Woolhead CA, McCormick PJ, Johnson AE. Nascent membrane and secretory proteins differ in FRET-detected folding far inside the ribosome and in their exposure to ribosomal proteins. Cell, 2004,116(5):725-736. |
| [154] | Kaiser CM, Goldman DH, Chodera JD, Tinoco I, Bustamante C . The ribosome modulates nascent protein folding. Science, 2011,334(6063):1723-1727. |
| [155] | Cabrita LD, Cassaignau AME, Launay HMM, Waudby CA, Wlodarski T, Camilloni C, Karyadi ME, Robertson AL, Wang XL, Wentink AS, Goodsell L, Woolhead CA, Vendruscolo M, Dobson CM, Christodoulou J . A structural ensemble of a ribosome-nascent chain complex during cotranslational protein folding. Nat Struct Mol Biol, 2016,23(4):278-285. |
| [156] | Cabrita LD, Hsu ST, Launay H, Dobson CM, Christodoulou J . Probing ribosome-nascent chain complexes produced in vivo by NMR spectroscopy. Proc Natl Acad Sci USA, 2009,106(2):22239-22244. |
| [157] | Sandikci A, Gloge F, Martinez M, Mayer MP, Wade R, Bukau B, Kramer G . Dynamic enzyme docking to the ribosome coordinates N-terminal processing with polypeptide folding. Nat Struct Mol Biol, 2013,20(7):843-850. |
| [158] | Döring K, Ahmed N, Riemer T, Suresh HG, Vainshtein Y, Habich M, Riemer J, Mayer MP, O'Brien EP, Kramer G, Bukau B. Profiling Ssb-nascent chain interactions reveals principles of Hsp70-assisted folding. Cell, 2017, 170(2): 298-311.e220. |
| [159] | Waudby CA, Dobson CM, Christodoulou J . Nature and regulation of protein folding on the ribosome. Trends Biochem Sci, 2019,44(11):914-926. |
| [160] | Liu KX, Maciuba K, Kaiser CM, . The ribosome cooperates with a chaperone to guide multi-domain protein folding. Mol Cell, 2019, 74(2): 310-319.e7. |
| [1] | YANG Chao WANG Qing-Xiong HUANG Yuan XIAO Hong. Analysis of the complete mitochondrial genome sequence of Larus brunnicephalus (Aves, Laridae) [J]. HEREDITAS, 2012, 34(11): 1434-1446. |
| [2] | KE Yang, HUANG Yuan, LEI Fu-Min. Sequencing and analysis of the complete mitochondrial genome of Podoces hendersoni (Ave, Corvidae) [J]. HEREDITAS, 2010, 32(9): 951-960. |
| [3] | . An iterative method for prediction of RNA secondary structures in-cluding pseudoknots based on minimum of free energy and covari-ance [J]. HEREDITAS, 2007, 29(7): 889-897. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||