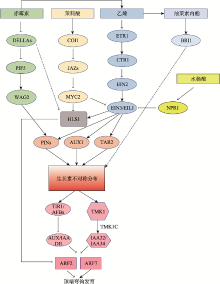
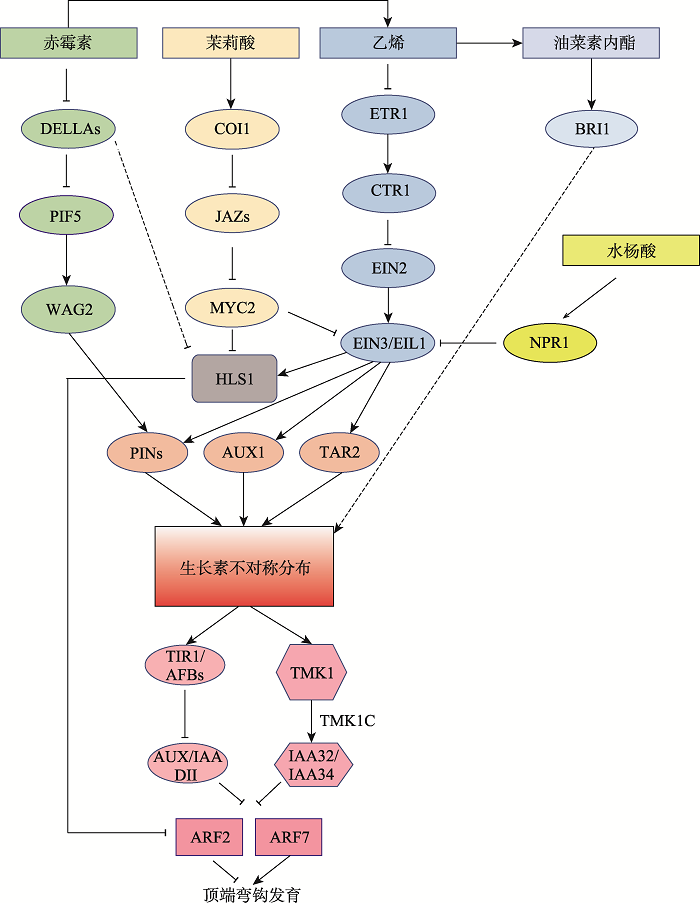
Hereditas(Beijing) ›› 2021, Vol. 43 ›› Issue (8): 723-736.doi: 10.16288/j.yczz.21-105
• Special Section: Excellent Doctoral Thesis • Previous Articles Next Articles
The molecular mechanism of apical hook development in dicot plant
- 1. Shanghai Center for Plant Stress Biology, Center for Excellence in Molecular Plant Sciences, Chinese Academy of Sciences, Shanghai 201602, China
2. FAFU-UCR Joint Center, Horticulture Biology and Metabolomics Center, Haixia Institute of Science and Technology, Fujian Agriculture and Forestry University, Fuzhou 350002, China
-
Received:2021-03-22Revised:2021-06-08Online:2021-08-20Published:2021-07-19 -
Contact:Xu Tongda E-mail:mcao@salk.edu;tdxu@sibs.ac.cn -
Supported by:Supported by the National Natural Science Foundation of China(31870256)
Cite this article
Min Cao, Tongda Xu. The molecular mechanism of apical hook development in dicot plant[J]. Hereditas(Beijing), 2021, 43(8): 723-736.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
| [1] |
Josse EM, Halliday KJ. Skotomorphogenesis: the dark side of light signalling. Curr Biol, 2008, 18(24):R1144-1146.
doi: 10.1016/j.cub.2008.10.034 |
| [2] |
Fankhauser C, Christie JM. Plant phototropic growth. Curr Biol, 2015, 25(9):R384-389.
doi: 10.1016/j.cub.2015.03.020 |
| [3] |
Deng ZP, Wang ZY, Kutschera U. Seedling development in maize cv. B73 and blue light-mediated proteomic changes in the tip vs. stem of the coleoptile. Protoplasma, 2017, 254(3):1317-1322.
doi: 10.1007/s00709-016-1023-6 |
| [4] | Abbas M, Alabadí D, Blázquez MA. Differential growth at the apical hook: all roads lead to auxin. Front Plant Sci, 2013, 4:441. |
| [5] |
Yao LL, Zheng YY, Zhu ZQ. Jasmonate suppresses seedling soil emergence inArabidopsis thaliana. Plant Signal Behav, 2017, 12(6):e1330239.
doi: 10.1080/15592324.2017.1330239 |
| [6] | Shen X, Li YL, Pan Y, Zhong SW. Activation of HLS1 by mechanical stress via ethylene-stabilized EIN3 is crucial for seedling soil emergence. Front Plant Sci, 2016, 7:1571. |
| [7] |
Shi H, Liu RL, Xue C, Shen X, Wei N, Deng XW, Zhong SW. Seedlings transduce the depth and mechanical pressure of covering soil using COP1 and ethylene to regulate EBF1/EBF2 for soil emergence. Curr Biol, 2016, 26(2):139-149.
doi: 10.1016/j.cub.2015.11.053 |
| [8] |
Raz V, Ecker JR. Regulation of differential growth in the apical hook ofArabidopsis. Development, 1999, 126(16):3661-3668.
pmid: 10409511 |
| [9] | Zhu Q, ŽádníkováP, Smet D, Van Der Straeten D, Benková E. Real-time analysis of the apical hook development. Methods Mol Biol, 2017, 1497:1-8. |
| [10] |
Smet D, Žádníková P, Vandenbussche F, Benková E, Van Der Straeten D. Dynamic infrared imaging analysis of apical hook development inArabidopsis: the case of brassinosteroids. New Phytol, 2014, 202(4):1398-1411.
doi: 10.1111/nph.2014.202.issue-4 |
| [11] |
Vandenbussche F, Petrásek J, ZádníkováP, HoyerováK, Pesek B, Raz V, Swarup R, Bennett M, ZazímalováE, BenkováE, Van Der Straeten D. The auxin influx carriers AUX1 and LAX3 are involved in auxin- ethylene interactions during apical hook development inArabidopsis thaliana seedlings. Development, 2010, 137(4):597-606.
doi: 10.1242/dev.040790 pmid: 20110325 |
| [12] |
Zádníková P, Petrásek J, Marhavy P, Raz V, Vandenbussche F, Ding Z, Schwarzerová K, Morita MT, Tasaka M, Hejátko J, Van Der Straeten D, Friml J, Benková E. Role of PIN-mediated auxin efflux in apical hook development ofArabidopsis thaliana. Development, 2010, 137(4):607-617.
doi: 10.1242/dev.041277 pmid: 20110326 |
| [13] | Knee EM, Hangarter RP, Knee M. Interactions of light and ethylene in hypocotyl hook maintenance in Arabidopsis thaliana seedlings. Physiol Plant, 2000, 108(2):208-215. |
| [14] |
Smet D, Žádníková P, Vandenbussche F, Benková E, Van Der Straeten D. Dynamic infrared imaging analysis of apical hook development in Arabidopsis: the case of brassinosteroids. New Phytol, 2014, 202(4):1398-1411.
doi: 10.1111/nph.2014.202.issue-4 |
| [15] |
Wang KLC, Yoshida H, Lurin C, Ecker JR. Regulation of ethylene gas biosynthesis by the Arabidopsis ETO1 protein. Nature, 2004, 428(6986):945-950.
doi: 10.1038/nature02516 |
| [16] |
Woeste KE, Ye C, Kieber JJ. Two Arabidopsis mutants that overproduce ethylene are affected in the posttranscriptional regulation of 1-aminocyclopropane-1- carboxylic acid synthase. Plant Physiol, 1999, 119(2):521-530.
pmid: 9952448 |
| [17] |
Chae HS, Faure F, Kieber JJ. The eto1, eto2, and eto3 mutations and cytokinin treatment increase ethylene biosynthesis in Arabidopsis by increasing the stability of ACS protein. Plant Cell, 2003, 15(2):545-559.
doi: 10.1105/tpc.006882 |
| [18] |
Chang C, Kwok SF, Bleecker AB, Meyerowitz EM. Arabidopsis ethylene-response gene ETR1: similarity of product to two-component regulators. Science, 1993, 262(5133):539-544.
pmid: 8211181 |
| [19] |
Zhao XC, Qu X, Mathews DE, Schaller GE. Effect of ethylene pathway mutations upon expression of the ethylene receptor ETR1 from Arabidopsis. Plant Physiol, 2002, 130(4):1983-1991.
doi: 10.1104/pp.011635 |
| [20] |
Rodríguez FI, Esch JJ, Hall AE, Binder BM, Schaller GE, Bleecker AB. A copper cofactor for the ethylene receptor ETR1 fromArabidopsis. Science, 1999, 283(5404):996-998.
pmid: 9974395 |
| [21] |
Kieber JJ, Rothenberg M, Roman G, Feldmann KA, Ecker JR. CTR1, a negative regulator of the ethylene response pathway in Arabidopsis, encodes a member of the raf family of protein kinases. Cell, 1993, 72(3):427-441.
pmid: 8431946 |
| [22] |
Qiao H, Shen Z, Huang SS, Schmitz RJ, Urich MA, Briggs SP, Ecker JR. Processing and subcellular trafficking of ER-tethered EIN2 control response to ethylene gas. Science, 2012, 338(6105):390-393.
doi: 10.1126/science.1225974 |
| [23] |
Ju CL, Yoon GM, Shemansky JM, Lin DY, Ying ZI, Chang JH, Garrett WM, Kessenbrock M, Groth G, Tucker ML, Cooper B, Kieber JJ, Chang C. CTR1 phosphorylates the central regulator EIN2 to control ethylene hormone signaling from the ER membrane to the nucleus inArabidopsis. Proc Natl Acad Sci USA, 2012, 109(47):19486-19491.
doi: 10.1073/pnas.1214848109 |
| [24] |
Li WY, Ma M, Feng Y, Li HJ, Wang YC, Ma YT, Li MZ, An FY, Guo HW. EIN2-directed translational regulation of ethylene signaling inArabidopsis. Cell, 2015, 163(3):670-683.
doi: 10.1016/j.cell.2015.09.037 |
| [25] |
Alonso JM, Hirayama T, Roman G, Nourizadeh S, Ecker JR. EIN2, a bifunctional transducer of ethylene and stress responses inArabidopsis. Science, 1999, 284(5423):2148-2152.
pmid: 10381874 |
| [26] |
Zhu ZQ, An FY, Feng Y, Li PP, Xue L A M, Jiang ZQ, Kim JM, To TK, Li W, Zhang XY, Yu Q, Dong Z, Chen WQ, Seki M, Zhou JM, Guo HW. Derepression of ethylene-stabilized transcription factors (EIN3/EIL1) mediates jasmonate and ethylene signaling synergy inArabidopsis. Proc Natl Acad Sci USA, 2011, 108(30):12539-12544.
doi: 10.1073/pnas.1103959108 |
| [27] |
Zhang X, Ji YS, Xue C, Ma HH, Xi YL, Huang PX, Wang H, An FY, Li BS, Wang YC, Guo HW. Integrated regulation of apical hook development by transcriptional coupling of EIN3/EIL1 and PIFs inArabidopsis. Plant Cell, 2018, 30(9):1971-1988.
doi: 10.1105/tpc.18.00018 |
| [28] |
An FY, Zhao Q, Ji YS, Li WY, Jiang ZQ, Yu XC, Zhang C, Han Y, He WR, Liu YD, Zhang SQ, Ecker JR, Guo HW. Ethylene-induced stabilization of ETHYLENE INSENSITIVE3 and EIN3-LIKE1 is mediated by proteasomal degradation of EIN3 binding F-box 1 and 2 that requires EIN2 inArabidopsis. Plant Cell, 2010, 22(7):2384-2401.
doi: 10.1105/tpc.110.076588 |
| [29] |
Binder BM, Walker JM, Gagne JM, Emborg TJ, Hemmann G, Bleecker AB, Vierstra RD. The Arabidopsis EIN3 binding F-Box proteins EBF1 and EBF2 have distinct but overlapping roles in ethylene signaling. Plant Cell, 2007, 19(2):509-523.
pmid: 17307926 |
| [30] |
Zheng ZY, Guo YX, Novák O, Dai XH, Zhao YD, Ljung K, Noel JP, Chory J. Coordination of auxin and ethylene biosynthesis by the aminotransferase VAS1. Nat Chem Biol, 2013, 9(4):244-246.
doi: 10.1038/nchembio.1178 pmid: 23377040 |
| [31] |
Stepanova AN, Robertson-Hoyt J, Yun J, Benavente LM, Xie DY, Dolezal K, Schlereth A, Jürgens G, Alonso JM. TAA1-mediated auxin biosynthesis is essential for hormone crosstalk and plant development. Cell, 2008, 133(1):177-191.
doi: 10.1016/j.cell.2008.01.047 pmid: 18394997 |
| [32] |
An FY, Zhang X, Zhu ZQ, Ji YS, He WR, Jiang ZQ, Li MZ, Guo HW. Coordinated regulation of apical hook development by gibberellins and ethylene in etiolatedArabidopsis seedlings. Cell Res, 2012, 22(5):915-927.
doi: 10.1038/cr.2012.29 |
| [33] |
Li H, Johnson P, Stepanova A, Alonso JM, Ecker JR. Convergence of signaling pathways in the control of differential cell growth inArabidopsis. Dev Cell, 2004, 7(2):193-204.
doi: 10.1016/j.devcel.2004.07.002 |
| [34] |
Briggs WR. Plant biology: seedling emergence through soil. Curr Biol, 2016, 26(2):R68-R70.
doi: 10.1016/j.cub.2015.12.003 |
| [35] |
Vriezen WH, Achard P, Harberd NP, Van Der Straeten D. Ethylene-mediated enhancement of apical hook formation in etiolatedArabidopsis thaliana seedlings is gibberellin dependent. Plant J, 2004, 37(4):505-516.
pmid: 14756759 |
| [36] |
AlabadíD, Gil J, Blázquez MA, García-Martínez JL. Gibberellins repress photomorphogenesis in darkness. Plant Physiol, 2004, 134(3):1050-1057.
pmid: 14963246 |
| [37] |
Gallego-BartoloméJ, Arana MV, Vandenbussche F, Zádníková P, Minguet EG, Guardiola V, Van Der Straeten D, Benkova E, Alabadí D, Blázquez MA. Hierarchy of hormone action controlling apical hook development inArabidopsis. Plant J, 2011, 67(4):622-634.
doi: 10.1111/tpj.2011.67.issue-4 |
| [38] |
Willige BC, Ogiso-Tanaka E, Zourelidou M, Schwechheimer C. WAG2 represses apical hook opening downstream from gibberellin and PHYTOCHROME INTERACTING FACTOR 5. Development, 2012, 139(21):4020-4028.
doi: 10.1242/dev.081240 pmid: 22992959 |
| [39] |
Dhonukshe P. PIN polarity regulation by AGC-3 kinases and ARF-GEF: a recurrent theme with context dependent modifications for plant development and response. Plant Signal Behav, 2011, 6(9):1333-1337.
pmid: 21852755 |
| [40] |
Dhonukshe P, Huang F, Galvan-Ampudia CS, Mähönen AP, Kleine-Vehn J, Xu J, Quint A, Prasad K, Friml J, Scheres B, Offringa R. Plasma membrane-bound AGC3 kinases phosphorylate PIN auxin carriers at TPRXS(N/S) motifs to direct apical PIN recycling. Development, 2010, 137(19):3245-3255.
doi: 10.1242/dev.052456 pmid: 20823065 |
| [41] |
Khanna R, Shen Y, Marion CM, Tsuchisaka A, Theologis A, Schäfer E, Quail PH. The basic helix- loop-helix transcription factor PIF5 acts on ethylene biosynthesis and phytochrome signaling by distinct mechanisms. Plant Cell, 2007, 19(12):3915-3929.
doi: 10.1105/tpc.107.051508 |
| [42] |
Song SS, Huang H, Gao H, Wang JJ, Wu DW, Liu XL, Yang SH, Zhai QZ, Li CY, Qi TC, Xie DX. Interaction between MYC2 and ETHYLENE INSENSITIVE3 modulates antagonism between jasmonate and ethylene signaling inArabidopsis. Plant Cell, 2014, 26(1):263-279.
doi: 10.1105/tpc.113.120394 |
| [43] |
Zhang X, Zhu ZQ, An FY, Hao DD, Li PP, Song JH, Yi CQ, Guo HW. Jasmonate-activated MYC2 represses ETHYLENE INSENSITIVE3 activity to antagonize ethylene-promoted apical hook formation inArabidopsis. Plant Cell, 2014, 26(3):1105-1117.
doi: 10.1105/tpc.113.122002 |
| [44] |
Zhang X, Ji YS, Xue C, Ma HH, Xi YL, Huang PX, Wang H, An FY, Li BS, Wang YC, Guo HW. Integrated regulation of apical hook development by transcriptional coupling of EIN3/EIL1 and PIFs inArabidopsis. Plant Cell, 2018, 30(9):1971-1988.
doi: 10.1105/tpc.18.00018 |
| [45] |
Morris K, MacKerness SA, Page T, John CF, Murphy AM, Carr JP, Buchanan-Wollaston V. Salicylic acid has a role in regulating gene expression during leaf senescence. Plant J, 2000, 23(5):677-685.
pmid: 10972893 |
| [46] |
Métraux JP, Signer H, Ryals J, Ward E, Wyss-Benz M, Gaudin J, Raschdorf K, Schmid E, Blum W, Inverardi B. Increase in salicylic acid at the onset of systemic acquired resistance in cucumber. Science, 1990, 250(4983):1004-1006.
pmid: 17746926 |
| [47] |
Guo PR, Li ZH, Huang PX, Li BS, Fang S, Chu JF, Guo HW. A tripartite amplification loop involving the transcription factor WRKY75, salicylic acid, and reactive oxygen species accelerates leaf senescence. Plant Cell, 2017, 29(11):2854-2870.
doi: 10.1105/tpc.17.00438 |
| [48] |
Huang PX, Dong Z, Guo PR, Zhang X, Qiu YP, Li BS, Wang YC, Guo HW. Salicylic acid suppresses apical hook formation via NPR1-mediated repression of EIN3 and EIL1 inArabidopsis. Plant Cell, 2020, 32(3):612-629.
doi: 10.1105/tpc.19.00658 |
| [49] |
Chory J, Nagpal P, Peto CA. Phenotypic and genetic analysis of det2, a new mutant that affects light- regulated seedling development inArabidopsis. Plant Cell, 1991, 3(5):445-459.
doi: 10.2307/3869351 |
| [50] |
Szekeres M, Németh K, Koncz-Kálmán Z, Mathur J, Kauschmann A, Altmann T, Rédei GP, Nagy F, Schell J, Koncz C. Brassinosteroids rescue the deficiency of CYP90, a cytochrome P450, controlling cell elongation and de-etiolation inArabidopsis. Cell, 1996, 85(2):171-182.
pmid: 8612270 |
| [51] |
De Grauwe L, Vandenbussche F, Tietz O, Palme K, Van Der Straeten D. Auxin, ethylene and brassinosteroids: tripartite control of growth in theArabidopsis hypocotyl. Plant Cell Physiol, 2005, 46(6):827-836.
doi: 10.1093/pcp/pci111 |
| [52] |
Gendron JM, Haque A, Gendron N, Chang T, Asami T, Wang ZY. Chemical genetic dissection of brassinosteroid- ethylene interaction. Mol Plant, 2008, 1(2):368-379.
doi: 10.1093/mp/ssn005 pmid: 19825546 |
| [53] |
Weijers D, Nemhauser J, Yang ZB. Auxin: small molecule, big impact. J Exp Bot, 2018, 69(2):133-136.
doi: 10.1093/jxb/erx463 pmid: 29309681 |
| [54] |
Esmon CA, Pedmale UV, Liscum E. Plant tropisms: providing the power of movement to a sessile organism. Int J Dev Biol, 2005, 49(5-6):665-674.
pmid: 16096973 |
| [55] |
Fiorucci AS, Fankhauser C. Plant strategies for enhancing access to sunlight. Curr Biol, 2017, 27(17):R931-R940.
doi: 10.1016/j.cub.2017.05.085 |
| [56] |
Wyatt SE, Kiss JZ. Plant tropisms: from Darwin to the International Space Station. Am J Bot, 2013, 100(1):1-3.
doi: 10.3732/ajb.1200591 |
| [57] |
Li H, Johnson P, Stepanova A, Alonso JM, Ecker JR. Convergence of signaling pathways in the control of differential cell growth inArabidopsis. Dev Cell, 2004, 7(2):193-204.
doi: 10.1016/j.devcel.2004.07.002 |
| [58] |
Žádníková P, Wabnik K, Abuzeineh A, Gallemi M, Van Der Straeten D, Smith RS, Inzé D, Friml J, Prusinkiewicz P, Benková E. A model of differential growth-guided apical hook formation in plants. Plant Cell, 2016, 28(10):2464-2477.
doi: 10.1105/tpc.15.00569 |
| [59] |
Korasick DA, Enders TA, Strader LC. Auxin biosynthesis and storage forms. J Exp Bot, 2013, 64(9):2541-2555.
doi: 10.1093/jxb/ert080 pmid: 23580748 |
| [60] |
Mashiguchi K, Tanaka K, Sakai T, Sugawara S, Kawaide H, Natsume M, Hanada A, Yaeno T, Shirasu K, Yao H, McSteen P, Zhao YD, Hayashi K, Kamiya Y, Kasahara H. The main auxin biosynthesis pathway inArabidopsis. Proc Natl Acad Sci USA, 2011, 108(45):18512-18517.
doi: 10.1073/pnas.1108434108 |
| [61] |
Zhao Y. Auxin biosynthesis: a simple two-step pathway converts tryptophan to indole-3-acetic acid in plants. Mol Plant, 2012, 5(2):334-338.
doi: 10.1093/mp/ssr104 |
| [62] |
Tao Y, Ferrer JL, Ljung K, Pojer F, Hong FX, Long JA, Li L, Moreno JE, Bowman ME, Ivans LJ, Cheng YF, Lim J, Zhao YD, Ballaré CL, Sandberg G, Noel JP, Chory J. Rapid synthesis of auxin via a new tryptophan- dependent pathway is required for shade avoidance in plants. Cell, 2008, 133(1):164-176.
doi: 10.1016/j.cell.2008.01.049 pmid: 18394996 |
| [63] |
Hofmann NR. YUC and TAA1/TAR proteins function in the same pathway for auxin biosynthesis. Plant Cell, 2011, 23(11):3869.
doi: 10.1105/tpc.111.231112 |
| [64] |
Yang ZB, Geng XY, He CM, Zhang F, Wang R, Horst WJ, Ding ZJ. TAA1-regulated local auxin biosynthesis in the root-apex transition zone mediates the aluminum- induced inhibition of root growth inArabidopsis. Plant Cell, 2014, 26(7):2889-2904.
doi: 10.1105/tpc.114.127993 |
| [65] |
Cheng YF, Dai XH, Zhao YD. Auxin biosynthesis by the YUCCA flavin monooxygenases controls the formation of floral organs and vascular tissues inArabidopsis. Genes Dev, 2006, 20(13):1790-1799.
doi: 10.1101/gad.1415106 |
| [66] |
Cheng YF, Dai XH, Zhao YD. Auxin synthesized by the YUCCA flavin monooxygenases is essential for embryogenesis and leaf formation inArabidopsis. Plant Cell, 2007, 19(8):2430-2439.
doi: 10.1105/tpc.107.053009 |
| [67] |
Zhao Y, Christensen SK, Fankhauser C, Cashman JR, Cohen JD, Weigel D, Chory J. A role for flavin monooxygenase-like enzymes in auxin biosynthesis. Science, 2001, 291(5502):306-309.
pmid: 11209081 |
| [68] |
Stepanova AN, Yun J, Robles LM, Novak O, He WR, Guo HW, Ljung K, Alonso JM. The Arabidopsis YUCCA1 flavin monooxygenase functions in the indole-3-pyruvic acid branch of auxin biosynthesis. Plant Cell, 2011, 23(11):3961-3973.
doi: 10.1105/tpc.111.088047 |
| [69] |
Krecek P, Skupa P, Libus J, Naramoto S, Tejos R, Friml J, Zazímalová E. The PIN-FORMED(PIN) protein family of auxin transporters. Genome Biol, 2009, 10(12):249.
doi: 10.1186/gb-2009-10-12-249 pmid: 20053306 |
| [70] |
Mravec J, Kubes M, Bielach A, Gaykova V, Petrásek J, Skůpa P, Chand S, Benková E, Zazímalová E, Friml J. Interaction of PIN and PGP transport mechanisms in auxin distribution-dependent development. Development, 2008, 135(20):3345-3354.
doi: 10.1242/dev.021071 pmid: 18787070 |
| [71] |
Perét B, Swarup K, Ferguson A, Seth M, Yang YD, Dhondt S, James N, Casimiro I, Perry P, Syed A, Yang HB, Reemmer J, Venison E, Howells C, Perez-Amador MA, Yun J, Alonso J, Beemster GT, Laplaze L, Murphy A, Bennett MJ, Nielsen E, Swarup R. AUX/LAX genes encode a family of auxin influx transporters that perform distinct functions duringArabidopsis development. Plant Cell, 2012, 24(7):2874-2885.
doi: 10.1105/tpc.112.097766 |
| [72] |
Zhou JJ, Luo J. The PIN-FORMED auxin efflux carriers in plants. Int J Mol Sci, 2018, 19(9):2759.
doi: 10.3390/ijms19092759 |
| [73] |
Yang HB, Murphy AS. Functional expression and characterization ofArabidopsis ABCB, AUX 1 and PIN auxin transporters in Schizosaccharomyces pombe. Plant J, 2009, 59(1):179-191.
doi: 10.1111/tpj.2009.59.issue-1 |
| [74] |
Noh B, Murphy AS, Spalding EP. Multidrug resistance- like genes ofArabidopsis required for auxin transport and auxin-mediated development. Plant Cell, 2001, 13(11):2441-2454.
pmid: 11701880 |
| [75] |
Wu GS, Cameron JN, Ljung K, Spalding EP. A role for ABCB19-mediated polar auxin transport in seedling photomorphogenesis mediated by cryptochrome 1 and phytochrome B. Plant J, 2010, 62(2):179-191.
doi: 10.1111/j.1365-313X.2010.04137.x |
| [76] |
Friml J, Wisniewska J, Benkova E, Mendgen K, Palme K. Lateral relocation of auxin efflux regulator PIN3 mediates tropism inArabidopsis. Nature, 2002, 415(6873):806-809.
doi: 10.1038/415806a |
| [77] |
Vieten A, Vanneste S, Wisniewska J, Benková E, Benjamins R, Beeckman T, Luschnig C, Friml J. Functional redundancy of PIN proteins is accompanied by auxin-dependent cross-regulation of PIN expression. Development, 2005, 132(20):4521-4531.
pmid: 16192309 |
| [78] |
Chapman EJ, Estelle M. Mechanism of auxin-regulated gene expression in plants. Annu Rev Genet, 2009, 43:265-285.
doi: 10.1146/annurev-genet-102108-134148 |
| [79] |
Dharmasiri N, Dharmasiri S, Estelle M. The F-box protein TIR1 is an auxin receptor. Nature, 2005, 435(7041):441-445.
doi: 10.1038/nature03543 |
| [80] |
Dharmasiri N, Dharmasiri S, Weijers D, Lechner E, Yamada M, Hobbie L, Ehrismann JS, Jurgens G, Estelle M. Plant development is regulated by a family of auxin receptor F box proteins. Dev Cell, 2005, 9(1):109-119.
pmid: 15992545 |
| [81] |
Kepinski S, Leyser O. TheArabidopsis F-box protein TIR1 is an auxin receptor. Nature, 2005, 435(7041):446-451.
doi: 10.1038/nature03542 |
| [82] |
Calderón Villalobos LI, Lee S, De Oliveira C, Ivetac A, Brandt W, Armitage L, Sheard LB, Tan X, Parry G, Mao H, Zheng N, Napier R, Kepinski S, Estelle M. A combinatorial TIR1/AFB-Aux/IAA co-receptor system for differential sensing of auxin. Nat Chem Biol, 2012, 8(5):477-485.
doi: 10.1038/nchembio.926 pmid: 22466420 |
| [83] |
Maraschin Fdos S, Memelink J, Offringa R. Auxin- induced, SCF(TIR1)-mediated poly-ubiquitination marks AUX/IAA proteins for degradation. Plant J, 2009, 59(1):100-109.
doi: 10.1111/tpj.2009.59.issue-1 |
| [84] |
Tan X, Calderon-Villalobos LI, Sharon M, Zheng CX, Robinson CV, Estelle M, Zheng N. Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature, 2007, 446(7136):640-645.
doi: 10.1038/nature05731 |
| [85] |
Béziat C, Barbez E, Feraru MI, Lucyshyn D, Kleine- Vehn J. Light triggers PILS-dependent reduction in nuclear auxin signalling for growth transition. Nat Plants, 2017, 3:17105.
doi: 10.1038/nplants.2017.105 pmid: 28714973 |
| [86] |
De Grauwe L, Vandenbussche F, Tietz O, Palme K, Van Der Straeten D. Auxin, ethylene and brassinosteroids: tripartite control of growth in the Arabidopsis hypocotyl. Plant Cell Physiol, 2005, 46(6):827-836.
doi: 10.1093/pcp/pci111 |
| [87] |
Kim BC, Soh MC, Kang BJ, Furuya M, Nam HG. Two dominant photomorphogenic mutations of Arabidopsis thaliana identified as suppressor mutations of hy2. Plant J, 1996, 9(4):441-456.
pmid: 8624510 |
| [88] |
Kim BC, Soh MS, Hong SH, Furuya M, Nam HG. Photomorphogenic development of theArabidopsis shy2-1D mutation and its interaction with phytochromes in darkness. Plant J, 1998, 15(1):61-68.
pmid: 9744095 |
| [89] |
Li JS, Dai XH, Zhao YD. A role for auxin response factor 19 in auxin and ethylene signaling inArabidopsis. Plant Physiol, 2006, 140(3):899-908.
doi: 10.1104/pp.105.070987 |
| [90] |
Okushima Y, Mitina I, Quach HL, Theologis A. AUXIN RESPONSE FACTOR 2 (ARF2): a pleiotropic developmental regulator. Plant J, 2005, 43(1):29-46.
pmid: 15960614 |
| [91] |
Okushima Y, Overvoorde PJ, Arima K, Alonso JM, Chan A, Chang C, Ecker JR, Hughes B, Lui A, Nguyen D, Onodera C, Quach H, Smith A, Yu GX, Theologis A. Functional genomic analysis of the AUXIN RESPONSE FACTOR gene family members inArabidopsis thaliana: unique and overlapping functions of ARF7 and ARF19. Plant Cell, 2005, 17(2):444-463.
pmid: 15659631 |
| [92] |
Tatematsu K, Kumagai S, Muto H, Sato A, Watahiki MK, Harper RM, Liscum E, Yamamoto KT. MASSUGU2 encodes Aux/IAA19, an auxin-regulated protein that functions together with the transcriptional activator NPH4/ARF7 to regulate differential growth responses of hypocotyl and formation of lateral roots inArabidopsis thaliana. Plant Cell, 2004, 16(2):379-393.
pmid: 14729917 |
| [93] |
Guilfoyle TJ, Hagen G. Auxin response factors. Curr Opin Plant Biol, 2007, 10(5):453-460.
pmid: 17900969 |
| [94] |
Cao M, Chen R, Li P, Yu YQ, Zheng R, Ge DF, Zheng W, Wang XH, Gu YT, Gelová Z, Friml J, Zhang H, Liu RY, He J, Xu TD. TMK1-mediated auxin signalling regulates differential growth of the apical hook. Nature, 2019, 568(7751):240-243.
doi: 10.1038/s41586-019-1069-7 |
| [95] |
Aloni R, Aloni E, Langhans M, Ullrich CI. Role of cytokinin and auxin in shaping root architecture: regulating vascular differentiation, lateral root initiation, root apical dominance and root gravitropism. Ann Bot, 2006, 97(5):883-893.
doi: 10.1093/aob/mcl027 |
| [96] |
Wang L, Wang B, Jiang L, Liu X, Li XL, Lu ZF, Meng XB, Wang YH, Smith SM, Li JY. Strigolactone signaling inArabidopsis regulates shoot development by targeting D53-Like SMXL repressor proteins for ubiquitination and degradation. Plant Cell, 2015, 27(11):3128-3142.
doi: 10.1105/tpc.15.00605 |
| [97] |
Vishal B, Kumar PP. Regulation of seed germination and abiotic stresses by gibberellins and abscisic acid. Front Plant Sci, 2018, 9:838.
doi: 10.3389/fpls.2018.00838 |
| [98] |
Rovira A, Sentandreu M, Nagatani A, Leivar P, Monte E. The Sequential action of MIDA9/PP2C.D1, PP2C.D2, and PP2C.D5 is necessary to form and maintain the hook after germination in the dark. Front Plant Sci, 2021, 12:636098.
doi: 10.3389/fpls.2021.636098 pmid: 33767720 |
| [99] |
Baral A, Aryal B, Jonsson K, Morris E, Demes E, Takatani S, Verger S, Xu T, Bennett M, Hamant O, Bhalerao RP. External mechanical cues reveal a katanin- independent mechanism behind auxin-mediated tissue bending in plants. Dev Cell, 2021, 56(1): 67-80.e63.
doi: 10.1016/j.devcel.2020.12.008 |
| [100] |
Hillmer RA, Tsuda K, Rallapalli G, Asai S, Truman W, Papke MD, Sakakibara H, Jones JDG, Myers CL, Katagiri F. The highly buffered Arabidopsis immune signaling network conceals the functions of its components. PLoS Genet, 2017, 13(5):e1006639.
doi: 10.1371/journal.pgen.1006639 |
| [1] | Di Wu, Linzhou Huang, Jin Gao, Yonghong Wang. The molecular mechanism of plant gravitropism [J]. HEREDITAS(Beijing), 2016, 38(7): 589-602. |
| [2] | Haiwei Shuai, Yongjie Meng, Xiaofeng Luo, Feng Chen, Ying Qi, Wenyu Yang, Kai Shu. The roles of auxin in seed dormancy and germination [J]. HEREDITAS(Beijing), 2016, 38(4): 314-322. |
| [3] | Huazhao Yuan, Mizhen Zhao, Weimin Wu, HongMei Yu, Yaming Qian, Zhuangwei Wang, Xicheng Wang. Genome-wide identification and expression analysis of auxin-related gene families in grape [J]. HEREDITAS(Beijing), 2015, 37(7): 720-730. |
| [4] | LI Yan-An, QI Lin-Lin, SUN Jia-Jiang, LIU Hong-Yu, LI Chuan-You. Genetic screening and analysis of suppressors of asa1-1 (soa) defective in jasmonate-mediated lateral root formation in Arabidopsis [J]. HEREDITAS, 2011, 33(9): 1003-1010. |
| [5] | LIU Zhen-Hua, YU Yan-Chong, XIANG Feng-Ning. Auxin response factors and plant growth and development [J]. HEREDITAS, 2011, 33(12): 1335-1346. |
| [6] | WANG Wen-Guo, LI Dui, SHU Jia-Yi, WANG Qing-Hua, CHEN Fang. DNA methylation-mediated regulation of OsMAPK2 gene expression in rice callus formation [J]. HEREDITAS, 2010, 32(12): 1275-1280. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||