Hereditas(Beijing) ›› 2020, Vol. 42 ›› Issue (9): 870-881.doi: 10.16288/j.yczz.20-189
• Review • Previous Articles Next Articles
New molecular diagnostic technologies for clinical detection of SARS-CoV-2
Chunmei Xie1, Haiping Wu1, Xueping Ma1, Guohua Zhou1,2( )
)
- 1. Department of Clinical Pharmacy, Jinling Hospital, Medical School of Nanjing University, Nanjing 210002, China
2. School of Pharmaceutical Sciences, Southern Medical University, Guangzhou 510515, China
-
Received:2020-06-22Revised:2020-08-19Online:2020-09-20Published:2020-09-04 -
Contact:Zhou Guohua E-mail:ghzhou@nju.edu.cn -
Supported by:Supported by the National Natural Science Foundation of China No(61871403);Jiangsu Provincial Science Fund for Distinguished Young Scholars No(BK20180005)
Cite this article
Chunmei Xie, Haiping Wu, Xueping Ma, Guohua Zhou. New molecular diagnostic technologies for clinical detection of SARS-CoV-2[J]. Hereditas(Beijing), 2020, 42(9): 870-881.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
Table 1
SARS-CoV-2 nucleic acid detection kits approved by The National Medical Products Administration (different from RT-qPCR technology)"
| 试剂盒名称 | 公司 | 分子诊断技术 | 扩增方式 | 检测方式 | 检测靶标 |
|---|---|---|---|---|---|
| 6项呼吸道病毒核酸检测试剂盒(恒温扩增芯片法) | 成都博奥晶芯生物科技有限公司 | 恒温扩增芯片技术 | 环介导等温扩增 | 荧光检测 | S、N |
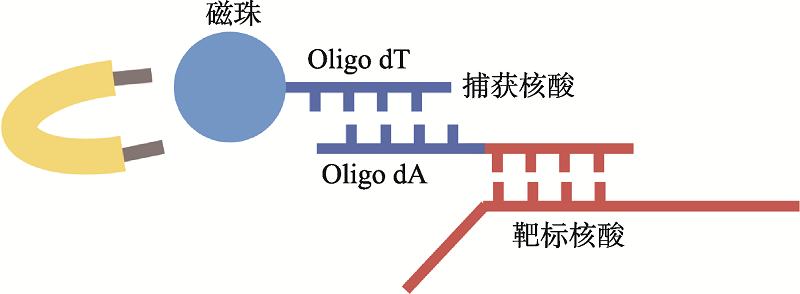
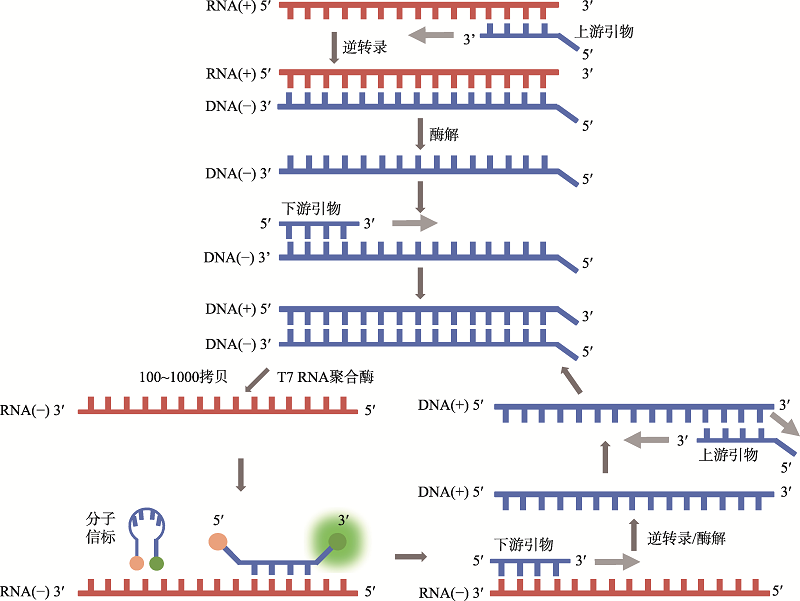
| 新型冠状病毒2019-nCoV核酸检测试剂盒(RNA捕获探针法) | 上海仁度生物科技有限公司 | SAT-RNA实时荧光恒温扩增技术 | T7 RNA聚合酶扩增 | 荧光检测 | ORFlab |
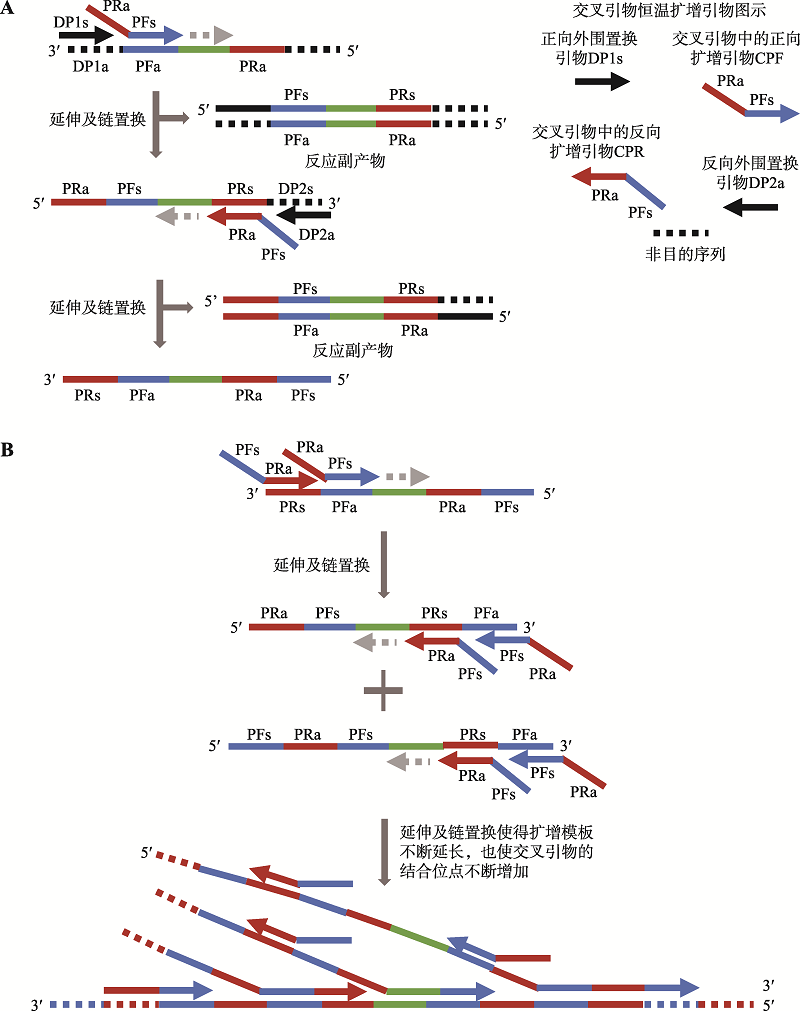
| 新型冠状病毒2019-nCoV核酸检测试剂盒(恒温扩增-实时荧光法) | 杭州优思达生物技术有限公司 | 交叉引物恒温扩增——实时荧光技术 | 交叉引物恒温扩增 | 荧光检测 | ORF1ab、N |
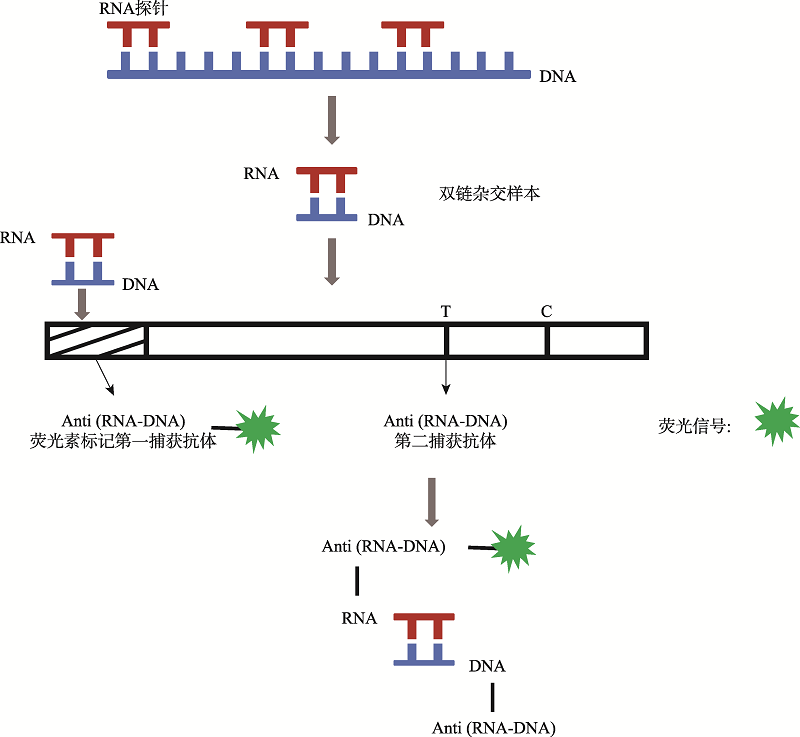
| 新型冠状病毒2019-nCoV核酸检测试剂盒(杂交捕获免疫荧光法) | 安邦(厦门)生物科技有限公司 | 杂交捕获免疫荧光技术 | 无需扩增 | 试纸条可视化判读 | ORF1ab、N、E |
| 新型冠状病毒2019-nCoV核酸检测试剂盒(联合探针锚定聚合测序法) | 华大生物科技(武汉)有限公司 | 联合探针锚定聚合测序技术 | DNA纳米球扩增 | 光信号识别 | 核酸序列 |
Table 2
Summary and comparison of molecular diagnostic technologies for clinical SARS-CoV-2 nucleic acid detection"
| 分子诊断技术 | 反应时间(min) | 检测样本类型 | 检测限 | 临床灵敏度(%) | 临床特异性(%) | 优点 | 缺点 |
|---|---|---|---|---|---|---|---|
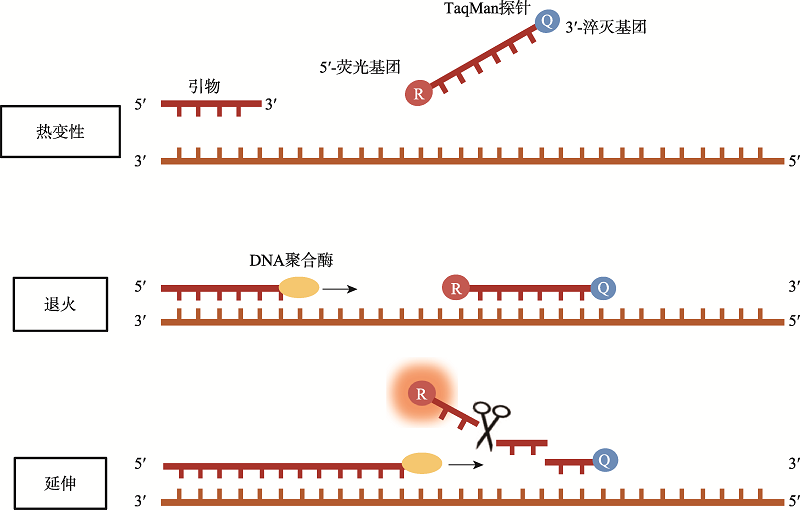
| RT-qPCR技术 | 120~180 | 肺泡灌洗液、咽拭子等样本 | 100~300拷贝/mL | 100 | 99 | 高灵敏、高特异、可定量 | 耗时长、存在假阳性及假阴性问题 |
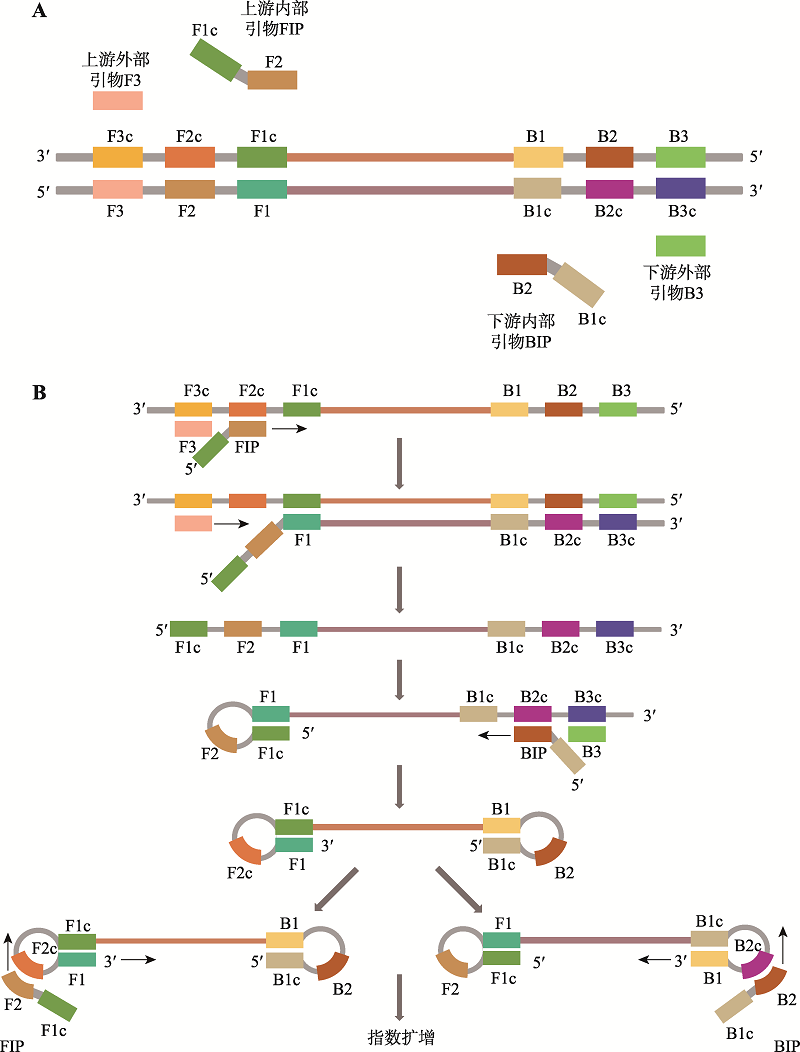
| 恒温扩增芯片技术 | 90 | 鼻拭子、咽拭子样本 | 15拷贝/反应 | - | - | 可同时检测多个靶标 | 未完全实现自动化 |
| SAT-RNA实时荧光恒温扩增技术 | 90 | 咽拭子、痰液样本 | - | - | - | 全流程自动化、高通量 | 价格昂贵、循环反应过程复杂 |
| 交叉引物恒温扩增——实时 荧光技术 | 30 | 咽拭子、痰液、血清、血浆、粪便样本 | 10拷贝/反应 | 100 | 100 | 全流程自动化、操作简便 | 单次只能检测2个 样本 |
| 杂交捕获免疫荧光技术 | 45 | 咽拭子、痰液样本 | 500 拷贝/mL | 88.61 | 94.92 | 无需提取纯化、无需核酸扩增、实现一步现场快速检测 | 无法定量、灵敏度 较低 |
| 联合探针锚定聚合测序技术 | 60 | 鼻拭子、咽拭子样本 | - | - | - | 快速高效、准确度高、通量大、可监测病毒突变 | 结果易受环境温度、检测试剂的影响 |
| [1] |
Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, Si HR, Zhu Y, Li B, Huang CL, Chen HD, Chen J, Luo Y, Guo H, Jiang RD, Liu MQ, Chen Y, Shen XR, Wang X, Zheng XS, Zhao K, Chen QJ, Deng F, Liu LL, Yan B, Zhang FX, Wang YY, Xiao GF, Shi ZL . A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature, 2020,579(7798):270-273.
doi: 10.1038/s41586-020-2012-7 pmid: 32015507 |
| [2] |
Wang C, Horby PW, Hayden FG, Gao GF . A novel coronavirus outbreak of global health concern. Lancet, 2020,395(10223):470-473.
doi: 10.1016/S0140-6736(20)30185-9 pmid: 31986257 |
| [3] |
Chan JFW, Kok KH, Zhu Z, Chu H, To KKW, Yuan SF, Yuen KY . Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg Microbes Infect, 2020,9(1):221-236.
doi: 10.1080/22221751.2020.1719902 pmid: 31987001 |
| [4] |
Lu RJ, Zhao X, Li J, Niu PH, Yang B, Wu HL, Wang WL, Song H, Huang BY, Zhu N, Bi YH, Ma XJ, Zhan FX, Wang L, Hu T, Zhou H, Hu ZH, Zhou WM, Zhao L, Chen J, Meng Y, Wang J, Lin Y, Yuan JY, Xie ZH, Ma JM, Liu WJ, Wang DY, Xu WB, Holmes EC, Gao GF, Wu GZ, Chen WJ, Shi WF, Tan WJ . Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet, 2020,395(10224):565-574.
doi: 10.1016/S0140-6736(20)30251-8 pmid: 32007145 |
| [5] |
Hoffmann M, Kleine-Weber H, Schroeder S, Krüger N, Herrler T, Erichsen S, Schiergens TS, Herrler G, Wu NH, Nitsche A, Müller MA, Drosten C, Pöhlmann S, . SARS- CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell, 2020, 181(2): 271-280.e8.
doi: 10.1016/j.cell.2020.02.052 pmid: 32142651 |
| [6] |
Zhu N, Zhang DY, Wang WL, Li XW, Yang B, Song JD, Zhao X, Huang BY, Shi WF, Lu RJ, Niu PH, Zhan FX, Ma XJ, Wang DY, Xu WB, Wu GZ, Gao GF, Tan WJ . A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med, 2020,382(8):727-733.
doi: 10.1056/NEJMoa2001017 pmid: 31978945 |
| [7] |
Saiki RK, Scharf S, Faloona F, Mullis KB, Horn GT, Erlich HA, Arnheim N . Enzymatic amplification of beta- globin genomic sequences and restriction site analysis for diagnosis of sickle cell anemia. Science, 1985,230(4732):1350-1354.
doi: 10.1126/science.2999980 pmid: 2999980 |
| [8] | General Office of National Health Commission of the People's Republic of China, Office of National Administration of Traditional Chinese Medicine. Diagnosis and treatment of corona virus disease-19(7th trial edition). Chin Med, 2020,15(6):801-805. |
| 中华人民共和国国家卫生健康委员会办公厅, 国家中医药管理局办公室. 新型冠状病毒肺炎诊疗方案(试行第七版). 中国医药, 2020,15(6):801-805. | |
| [9] | Xu JH, Wang SL, Zhang SX, Lai GX, Lan XP, Wu XL, Yu ZY. Methods for nucleic acid detection of 2019 novel coronavirus. Int J Lab Med, 2020, 3:1-19. |
| 许金和, 王水良, 张胜行, 赖国祥, 兰小鹏, 吴小丽, 余宗阳 . 新型冠状病毒核酸检测方法. 国际检验医学杂志, 2020,3:1-19. | |
| [10] |
Craighead H . Future lab-on-a-chip technologies for interrogating individual molecules. Nature, 2006,442(7101):387-393.
doi: 10.1038/nature05061 pmid: 16871206 |
| [11] | Wu JD, Dong ML, Santos S, Rigatto C, Liu Y, Lin F . Lab- on-a-chip platforms for detection of cardiovascular disease and cancer biomarkers. Sensors(Basel), 2017,17(12):2394. |
| [12] | He XP, Zou BJ, Qi XM, Chen S, Lu Y, Huang Q, Zhou GH . Methods of isothermal nucleic acid amplification-based microfluidic chips for pathogen microorganism detection. Hereditas(Beijing), 2019,41(7):611-624. |
| 何祥鹏, 邹秉杰, 齐谢敏, 陈杉, 陆妍, 黄青, 周国华 . 基于核酸等温扩增的病原微生物微流控检测技术. 遗传, 2019,41(7):611-624. | |
| [13] |
Yuan XF, Lv JZ, Lin XM, Zhang CY, Deng JH, Wang CX, Fan XP, Wang YG, Xu H, Wu SQ . Multiplex detection of six swine viruses on an integrated centrifugal disk using loop-mediated isothermal amplification. J Vet Diagn Invest, 2019,31(3):415-425.
doi: 10.1177/1040638719841096 pmid: 30947641 |
| [14] |
Yan H, Zhu YZ, Zhang Y, Wang L, Chen JG, Lu Y, Xu YC, Xing WL . Multiplex detection of bacteria on an integrated centrifugal disk using bead-beating lysis and loop-mediated amplification. Sci Rep, 2017,7(1):1460.
doi: 10.1038/s41598-017-01415-x pmid: 28469259 |
| [15] | 刘伟 . 基于RNA靶标SAT解脲脲原体感染分子生物学检测方法. CN108611403A, 2018-10-02. |
| [16] | 高通量、全流程一体化的新冠核酸检测平台. , 2020-03-29. |
| [17] |
Fang RD, Li X, Hu L, You QM, Li J, Wu J, Xu P, Zhong HY, Luo Y, Mei J, Gao Q . Cross-priming amplification for rapid detection of mycobacterium tuberculosis in sputum specimens. J Clin Microbiol, 2009,47(3):845-847.
doi: 10.1128/JCM.01528-08 pmid: 19116359 |
| [18] | 杭州一企业紧急研制新冠病毒核酸检测试剂盒最快30分钟出结果. , 2020-01-31. |
| [19] | You QM . Cross priming amplification of target nucleic acids. CA2748822, 2016 -09-06-18. |
| [20] | 祁军, 尤其敏, 柴宏森, 左锋, 王馨, 詹曦菁, 刘振宇, 刘寅, 杨春江, 刘智勇, 徐高连, 刘启军, 张霞, 崔景柏, 王宏莹, 高秋萍, 吴汀滢 . 运用交叉引物核酸恒温扩增技术检测肠出血性大肠埃希菌的试剂及其扩增方法和检测方法. CN102363816A, 2012-02-29. |
| [21] | 汪大明, 钟乾兴, 胡啸, 张利伟, 肖江群, 王保丹, 江应玲, 乐宜萃 . 一种核酸杂交捕获免疫荧光检测方法、免疫荧光层析试条及试剂盒. CN109613236A, 2019-04-12. |
| [22] | 新型冠状病毒核酸-安邦(厦门)生物科技有限公司. , 2020-03. |
| [23] | 一文读懂测序技术在新冠病毒检测中的应用. , 2020-03-27. |
| [24] | Guo Z, Wang F, Guo YX, Liu XW, Yang XG, Song YF, Wang DG . Potential value of droplet digital PCR in detection of SARS-CoV-2. J Lanzhou Univ(Med Sci). 2020,46(02):14-18. |
| 郭兆, 王芳, 郭妍譞, 刘向文, 杨旭光, 宋焱峰, 王德贵 . 微滴式数字PCR技术在新型冠状病毒检测中的潜在价值. 兰州大学学报(医学版), 2020,46(2):14-18. | |
| [25] |
Yu FT, Yan LT, Wang N, Yang SY, Wang LH, Tang YX, Gao GJ, Wang S, Ma CJ, Xie RM, Wang F, Tan C, Zhu LX, Guo Y, Zhang FJ . Quantitative detection and viral load analysis of SARS-CoV-2 in infected patients. Clin Infect Dis, 2020,71(15):793-798.
doi: 10.1093/cid/ciaa345 pmid: 32221523 |
| [26] | Wahed AAE, Patel P, Heidenreich D, Hufert FT, Weidmann M . Reverse transcription recombinase polymerase amplification assay for the detection of middle east respiratory syndrome coronavirus. PLoS Curr, 2013,5(8):813-818. |
| [27] |
Gootenberg JS, Abudayyeh OO, Lee JW, Essletzbichler P, Dy AJ, Joung J, Verdine V, Donghia N, Daringer NM, Freije CA, Myhrvold C, Bhattacharyya RP, Livny J, Regev A, Koonin EV, Hung DT, Sabeti PC, Collins JJ, Zhang F . Nucleic acid detection with CRISPR-Cas13a/C2c2. Science, 2017,356(6336):438-442.
doi: 10.1126/science.aam9321 pmid: 28408723 |
| [28] | Wang XJ, Zhong MT, Liu Y, Ma PX, Dang L, Meng QZ, Wan WW, Ma XD, Liu J, Yang G, Yang ZF, Huang XX, Liu M . Rapid and sensitive detection of COVID-19 using CRISPR/Cas12a-based detection with naked eye readout, CRISPR/Cas12a-NER. Sci Bull(Beijing), 2020,65(17):1436-1439. |
| [29] | Enabling coronavirus detection using CRISPR-Cas13: Open-access SHERLOCK research protocols and design resources | Broad Institute. |
| [1] | Wenming Zhao, Shuhui Song, Meili Chen, Dong Zou, Lina Ma, Yingke Ma, Rujiao Li, Lili Hao, Cuiping Li, Dongmei Tian, Bixia Tang, Yanqing Wang, Junwei Zhu, Huanxin Chen, Zhang Zhang, Yongbiao Xue, Yiming Bao. The 2019 novel coronavirus resource [J]. Hereditas(Beijing), 2020, 42(2): 212-221. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||