Hereditas(Beijing) ›› 2023, Vol. 45 ›› Issue (12): 1158-1168.doi: 10.16288/j.yczz.23-179
• Genetics Teaching • Previous Articles
Epigenetics comprehensive experimental course based on the integration of science and education to cultivate students' ability of cutting-edge innovation
Xiufang Ou( ), Ying Wu, Ning Li, Lili Jiang, Bao Liu, Lei Gong(
), Ying Wu, Ning Li, Lili Jiang, Bao Liu, Lei Gong( )
)
- Key Laboratory of Molecular Epigenetics of Ministry of Education (MOE), Northeast Normal University, Changchun 130024, China
-
Received:2023-07-03Revised:2023-11-15Online:2023-12-20Published:2023-11-16 -
Contact:Lei Gong E-mail:ouxf074@nenu.edu.cn;gongl100@nenu.edu.cn -
Supported by:Higher Education Teaching Reform Research Topic of Jilin Province(JLO4169120190727102937);Higher Education Teaching Reform Research Topic of Jilin Province(JLJY202393805644);Higher Education Teaching Reform Research Topic of Jilin Province(JG2020008)
Cite this article
Xiufang Ou, Ying Wu, Ning Li, Lili Jiang, Bao Liu, Lei Gong. Epigenetics comprehensive experimental course based on the integration of science and education to cultivate students' ability of cutting-edge innovation[J]. Hereditas(Beijing), 2023, 45(12): 1158-1168.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
Table 1
Course arrangement"
| 组织形式 | 具体内容 | 学时 |
|---|---|---|
| 课前理论部分 | 1. 教师介绍本课程相关的研究背景,引导学生提出科学问题(2学时) 2. 教师对相关技术进行介绍,包括引物设计软件使用方法和引物设计原则,转座子激活检测相关知识,基因组重测序原理以及DNA甲基化检测相关知识(2学时) | 4 |
| 实验设计汇报 | 1. 课后学生以小组为单位进行实验设计 2. 课上学生进行实验设计汇报,师生共同探讨提出修改建议,确定小组特异的实验方案(按照10组,每组15~20分钟计算,约4学时) | 4 |
| 实验一: 水稻DNA的提取及浓度检测 | 1. 实验操作过程中注意事项的讲解(1学时) 2. DNA的提取(3.5学时) 3. DNA浓度的检测(1.5学时) | 6 |
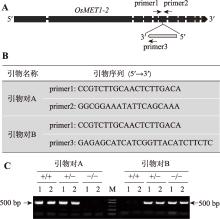
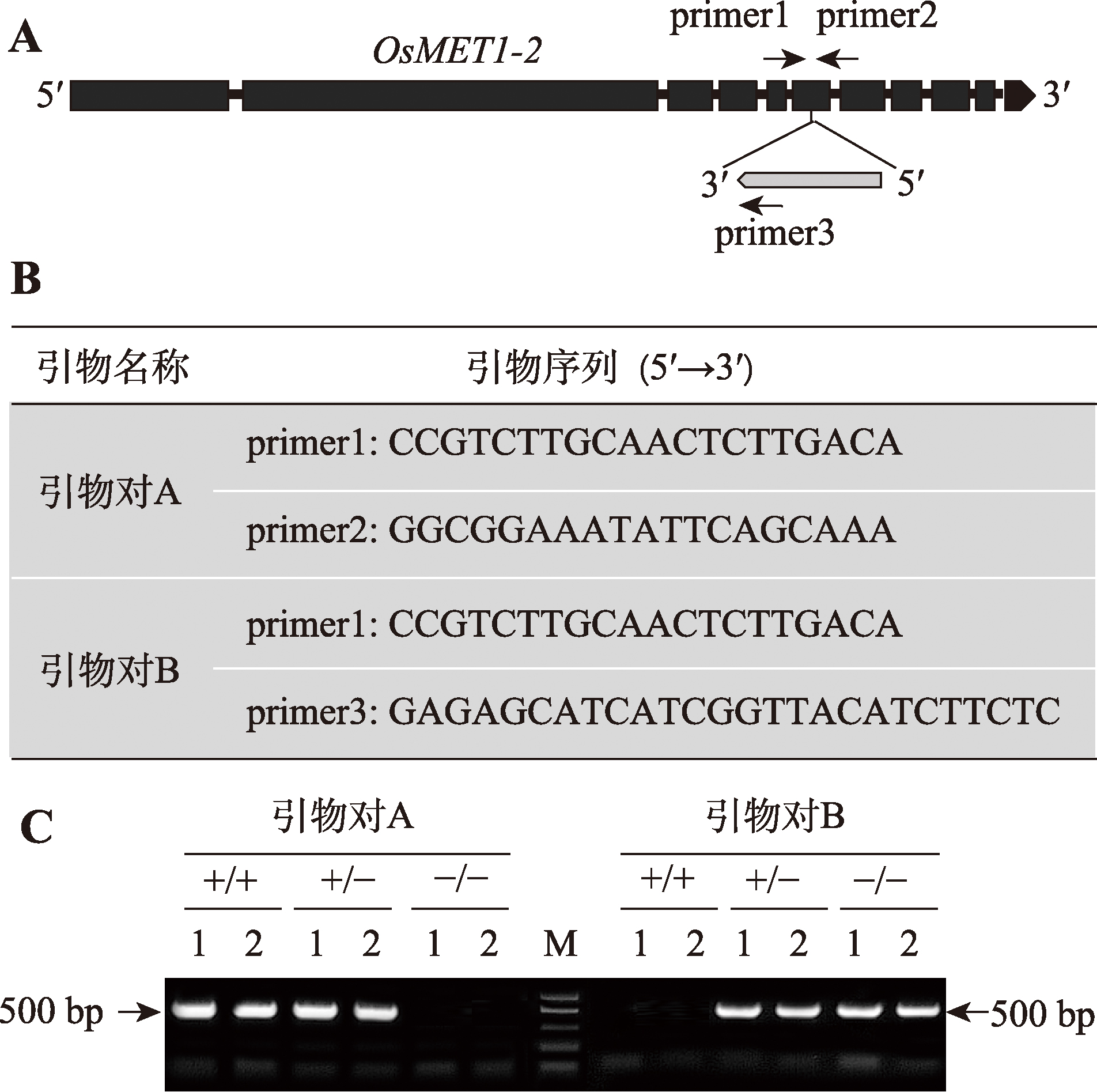
| 实验二: 实验材料基因型的鉴定 | 1. 实验操作过程中注意事项的讲解(0.5学时) 2. PCR扩增(1.5学时) 3. 琼脂糖凝胶电泳(1.5学时) | 3.5 |
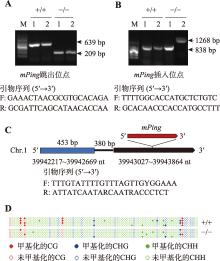
| 实验三: 转座子激活检测 | 1. 实验操作过程中注意事项的讲解(0.5学时) 2. PCR扩增(1.5学时) 3. 琼脂糖凝胶电泳(1.5学时) | 3.5 |
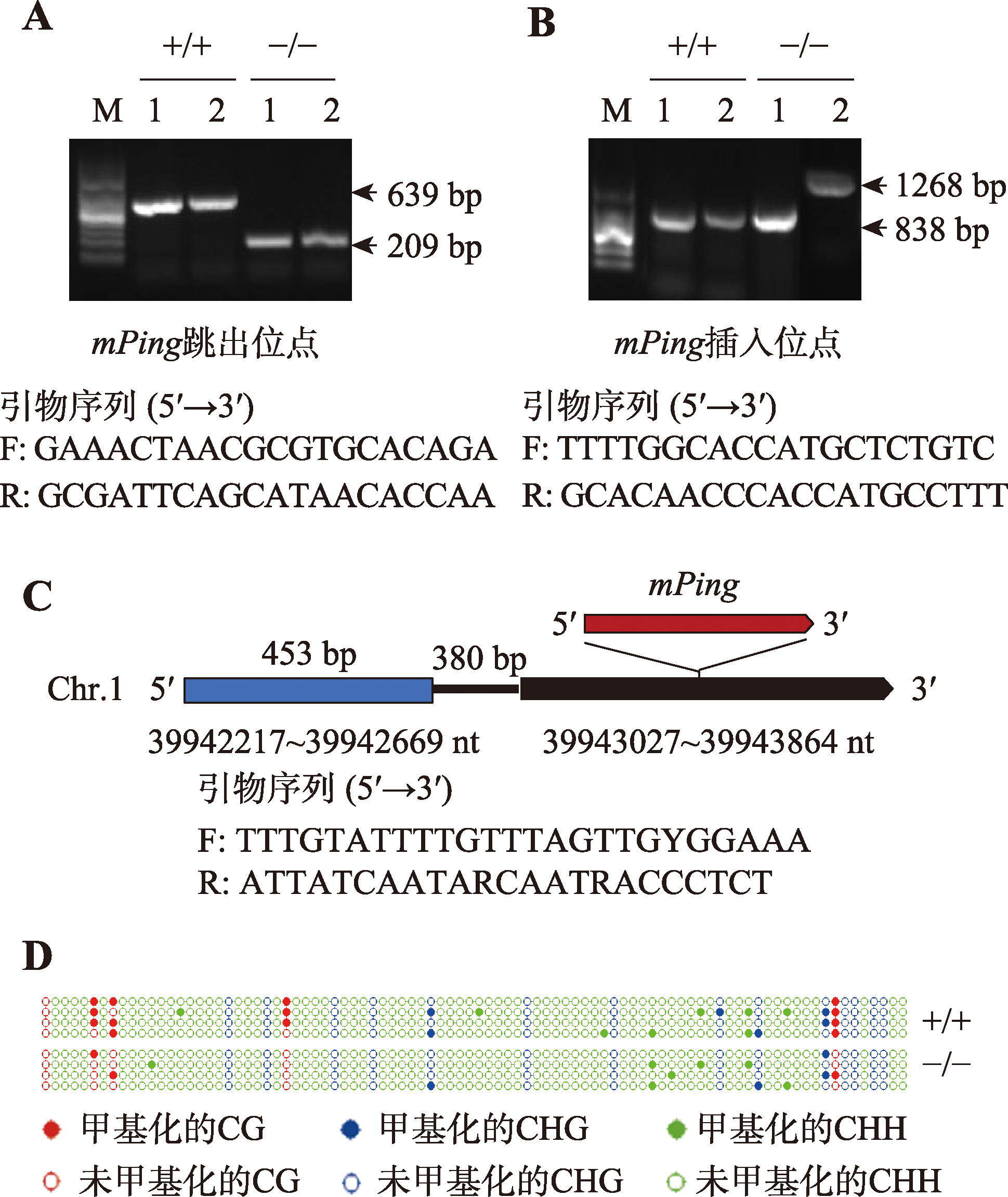
| 实验四: DNA甲基化检测和变异分析 | 1. 实验操作过程中注意事项的讲解(1学时) 2. 亚硫酸盐处理和产物回收、连接、转化(10学时) 3. 阳性克隆筛选、测序(2学时) 4. 测序数据分析计算DNA甲基化水平(2学时) | 15 |
Table 2
Course assessment methods"
| 评价方式 | 具体要求 | 考核目的 | 评价标准 | 比例 |
|---|---|---|---|---|
| 研究综述 | 学生独立查阅文献,撰写植物中关于DNA甲基化研究方法和转座子研究方法的综述 | 考察学生自主学习、查阅文献并进行归纳和总结的能力,提高学生撰写综述规范性和科学性的能力 | 思路是否清晰,逻辑是否严谨,论证是否充分(25分);文献资料是否详实,是否具有代表性(25分);文字表达是否准确、流畅,是否符合学术道德规范(25分);是否运用了本门课程相关理论知识,是否体现了科学研究能力(25分) | 20% |
| 实验设计 | 学生以小组为单位进行实验设计和汇报 | 考察学生提出科学问题、分析问题和解决问题的能力,提高学生团队协作的能力 | 实验设计是否清晰、完整,具有科学性(60分);设计中遇到的问题是否能够自主解决(15分);组内是否具有明确的分工(10分);汇报中是否能够准确回答教师问题(15分) | 20% |
| 实验操作 | 学生独立或与小组成员合作完成实验过程,实验仪器使用规范 | 考察学生实验操作的规范性,以及在实验过程中对遇到的问题进行分析和解决的能力,提高学生的实践能力和高阶科研思维 | 实验前的准备是否充分(10分);实验仪器使用是否规范(60分);实验操作时小组分工是够明确(15分);实验过程中遇到的问题是否能够有效解决(15分) | 20% |
| 实验记录 | 学生要有完整且清晰的手写实验记录 | 考察学生实验记录的科学性和完整性,提高学生的基本科研素养 | 是否及时记录实验过程(30分);实验记录是否清晰、完整(60分);是否体现了实验中的反思(10分) | 10% |
| 研究论文 | 学生按照学术论文要求撰写研究论文 | 考察学生撰写学术论文的规范性和科学性,提高学生针对实验结果和实验中的问题进行分析和讨论的能力 | 研究论文的格式是否规范(25分);逻辑是否清晰(25分);研究方法和结果是否可信(25分);是否对实验结果进行了深入的分析和讨论(25分) | 30% |
Table 3
Student satisfaction survey"
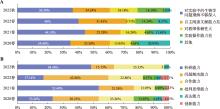
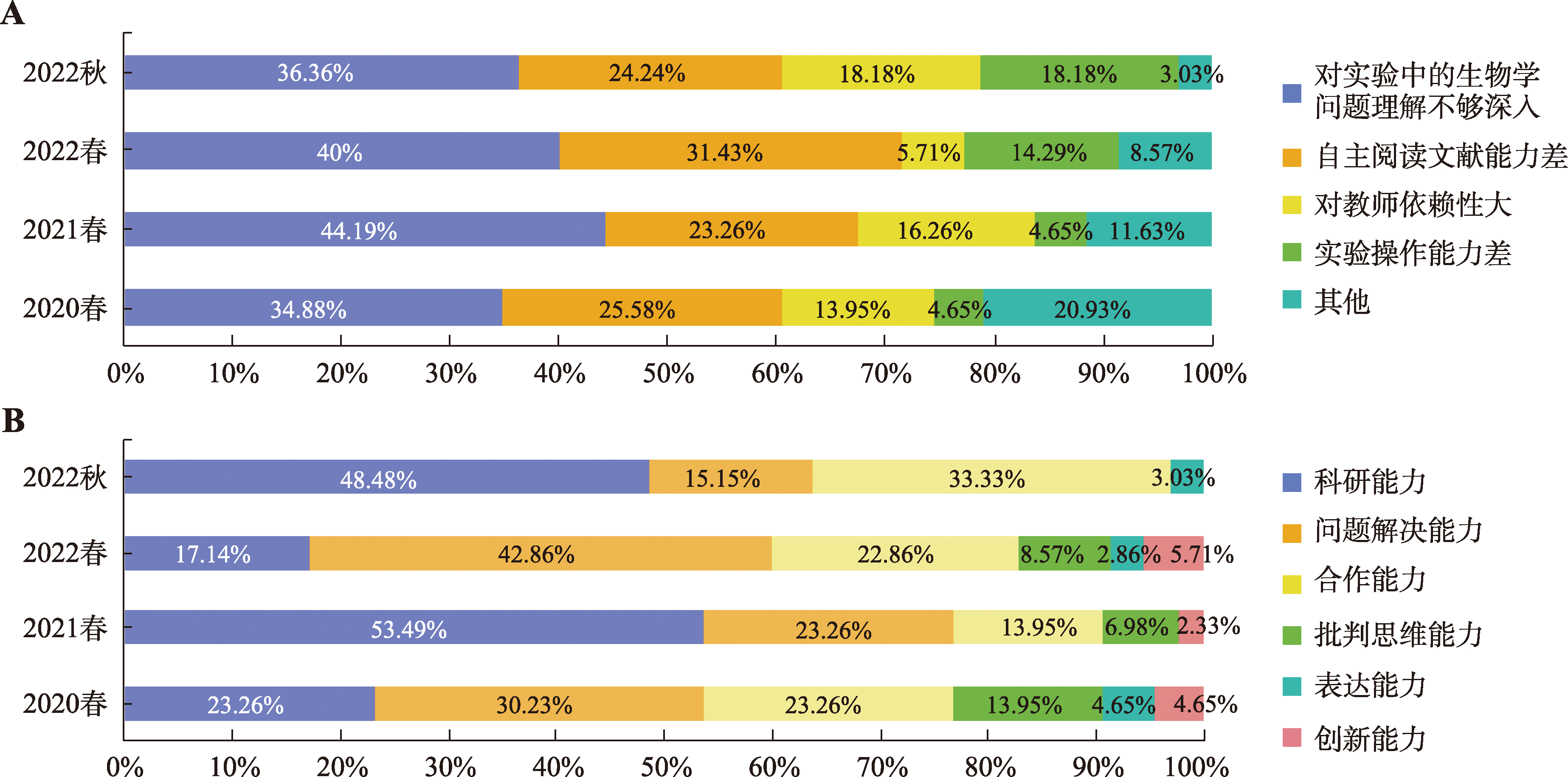
| 主观性问题 | 满意度(%) | |||
|---|---|---|---|---|
| 2020春 (43人) | 2021春 (43人) | 2022春 (35人) | 2022秋 (33人) | |
| 1. 本实验内容的设置突出了研究型的教学内容 | 97.67 | 97.67 | 100 | 93.94 |
| 2. 课前教师对背景的简单介绍有助于您更深刻的了解本实验所要解决的生物学问题 | 95.35 | 100 | 97.14 | 100 |
| 3. 自主查阅文献并完成实验相关研究方法综述能够培养科研思维中的前期调研 | 95.35 | 100 | 97.14 | 100 |
| 4. 以小组为单位进行实验设计有助于培养自主思考能力 | 97.67 | 100 | 94.29 | 100 |
| 5. 小组内平行实验的设计可明确科研中需要的生物学重复和技术重复问题 | 97.67 | 100 | 97.14 | 100 |
| 6. 整体实验的设置体现了以你们为主导的实验过程 | 100 | 97.67 | 97.14 | 96.97 |
| 7. 本实验充分调动了你们主动思考和主动解决问题的能力 | 100 | 97.67 | 97.14 | 96.97 |
| 8. 在整个教学中,您认为评价方式很合理 | 100 | 100 | 97.14 | 93.94 |
| 9. 考核方式的多元性有助于你们更加注重实验过程的严谨和实验思维的培养 | 97.67 | 100 | 94.28 | 100 |
| 10. 您愿意继续采用这种方式进行学习 | 100 | 100 | 85.72 | 93.94 |
| 11. 您觉得这种学习方式能够有效培养你的科研思维和创新能力 | 97.67 | 100 | 91.43 | 100 |
| 12. 实验课的学习过程让您很感兴趣、具有挑战性,并积极完成作业 | 100 | 95.35 | 91.43 | 84.85 |
| 13. 通过实验课这种学习方式,您对自己掌握知识的程度很满意 | 100 | 100 | 85.71 | 90.91 |
| 14. 通过小组合作设计实验方案的方式能够增进您和同学的交流和共同进步 | 97.67 | 100 | 91.43 | 93.94 |
| 15. 您觉得在此学习过程中没有给你增加课业负担 | 88.37 | 79.07 | 65.71 | 66.67 |
| [1] |
Hu LJ, Li N, Xu CM, Zhong SL, Lin XY, Yang JJ, Zhou TQ, Yuliang AZ, Wu Y, Chen YR, Cao XF, Zemach A, Rustgi S, von Wettstein D, Liu B. Mutation of a major CG methylase in rice causes genome-wide hypomethylation, dysregulated genome expression, and seedling lethality. Proc Natl Acad Sci USA, 2014, 111(29): 10642-10647.
doi: 10.1073/pnas.1410761111 pmid: 25002488 |
| [2] |
Hu LJ, Li N, Zhang ZB, Meng XC, Dong QL, Xu CM, Gong L, Liu B. CG hypomethylation leads to complex changes in DNA methylation and transpositional burst of diverse transposable elements in callus cultures of rice. Plant J, 2020, 101(1): 188-203.
doi: 10.1111/tpj.v101.1 |
| [3] |
Berger SL, Kouzarides T, Shiekhattar R, Shilatifard A. An operational definition of epigenetics. Genes Dev, 2009, 23(7): 781-783.
doi: 10.1101/gad.1787609 |
| [4] |
Allis CD, Jenuwein T. The molecular hallmarks of epigenetic control. Nat Rev Genet, 2016, 17(8): 487-500.
doi: 10.1038/nrg.2016.59 pmid: 27346641 |
| [5] |
Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell, 2007, 128(4): 707-719.
doi: 10.1016/j.cell.2007.01.015 pmid: 17320508 |
| [6] |
Feinberg AP. The key role of epigenetics in human disease prevention and mitigation. N Engl J Med, 2018, 378(14): 1323-1334
doi: 10.1056/NEJMra1402513 |
| [7] |
Ashe A, Colot V, Oldroyd BP. How does epigenetics influence the course of evolution? Philos Trans R Soc Lond B Biol Sci, 2021, 376(1826): 20200111.
doi: 10.1098/rstb.2020.0111 |
| [8] |
Lieberman-Lazarovich M, Kaiserli E, Bucher E, Mladenov V. Natural and induced epigenetic variation for crop improvement. Curr Opin Plant Biol, 2022, 70: 102297.
doi: 10.1016/j.pbi.2022.102297 |
| [9] |
Zhang L, Lu QJ, Chang C. Epigenetics in health and disease. Adv Exp Med Biol, 2020, 1253: 3-55.
doi: 10.1007/978-981-15-3449-2_1 pmid: 32445090 |
| [10] | Phillips T. The role of methylation in gene expression. Nature Edu, 2008, 1(1): 116. |
| [11] |
Moore LD, Le T, Fan GP. DNA methylation and its basic function. Neuropsychopharmacology, 2013, 38(1): 23-38.
doi: 10.1038/npp.2012.112 pmid: 22781841 |
| [12] |
Gallusci P, Dai ZW, Génard M, Gauffretau A, Leblanc- Fournier N, Richard-Molard C, Vile D, Brunel-Muguet S. Epigenetics for plant improvement: current knowledge and modeling avenues. Trends Plant Sci, 2017, 22(7): 610-623.
doi: S1360-1385(17)30089-4 pmid: 28587758 |
| [13] |
Law JA, Jacobsen SE. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet, 2010, 11(3): 204-220.
doi: 10.1038/nrg2719 pmid: 20142834 |
| [14] |
Cao XF, Jacobsen SE. Locus-specific control of asymmetric and CpNpG methylation by the DRM and CMT3 methyltransferase genes. Proc Natl Acad Sci USA, 2002, 99(Suppl 4): 16491-16498.
doi: 10.1073/pnas.162371599 |
| [15] |
Stroud H, Greenberg MV, Feng SH, Bernatavichute YV, Jacobsen SE. Comprehensive analysis of silencing mutants reveals complex regulation of the Arabidopsis methylome. Cell, 2013, 152(1-2): 352-364.
doi: 10.1016/j.cell.2012.10.054 pmid: 23313553 |
| [16] |
Zemach A, Yvonne Kim M, Hsieh PH, Coleman-Derr D, Eshed-Williams L, Thao K, Harmer SL, Zilberman D. The Arabidopsis nucleosome remodeler DDM1 allows DNA methyltransferases to access H1-containing heterochromatin. Cell, 2013, 153(1): 193-205.
doi: 10.1016/j.cell.2013.02.033 pmid: 23540698 |
| [17] |
Cokus SJ, Feng SH, Zhang XY, Chen ZG, Merriman B, Haudenschiil CD, Pradhan S, Nelson SF, Pellegrini M, Jacobsen SE. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature, 2008, 452(7184): 215-219.
doi: 10.1038/nature06745 |
| [18] |
Lister R, O’Malley RC, Tonti-Filippini J, Gregory BD, Berry CC, Harvey Millar A, Ecker JR. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell, 2008, 133(3): 523-536.
doi: 10.1016/j.cell.2008.03.029 pmid: 18423832 |
| [19] |
Mathieu O, Reinders J, Caikovski M, Smathajitt C, Paszkowski J.Transgenerational stability of the Arabidopsis epigenome is coordinated by CG methylation. Cell, 2007, 130(5): 851-862.
doi: 10.1016/j.cell.2007.07.007 |
| [20] |
Pavlopoulou A, Kossida S. Plant cytosine-5 DNA methyltransferases: structure, function, and molecular evolution. Genomics, 2007, 90(4): 530-541.
pmid: 17689048 |
| [21] |
Yamauchi T, Johzuka-Hisatomi Y, Fukada-Tanaka S, Terada R, Nakamura I, Iida S. Homologous recombination-mediated knock-in targeting of the MET1a gene for a maintenance DNA methyltransferase reproducibly reveals dosage-dependent spatiotemporal gene expression in rice. Plant J, 2009, 60(2): 386-396.
doi: 10.1111/tpj.2009.60.issue-2 |
| [22] |
Yamauchi T, Johzuka-Hisatomi Y, Terada R, Nakamura I, Iida S. The MET1b gene encoding a maintenance DNA methyltransferase is indispensable for normal development in rice. Plant Mol Biol, 2014, 85(3): 219-232.
doi: 10.1007/s11103-014-0178-9 pmid: 24535433 |
| [23] |
Saze H, Mittelsten Scheid O, Paszkowski J. Maintenance of CpG methylation is essential for epigenetic inheritance during plant gametogenesis. Nat Genet, 2003, 34(1): 65-69.
doi: 10.1038/ng1138 pmid: 12669067 |
| [24] |
Kankel MW, Ramsey DE, Stokes TL, Flowers SK, Haag JR, Jeddeloh JA, Riddle NC, Verbsky ML, Richards EJ. Arabidopsis MET1 cytosine methyltransferase mutants. Genetics, 2003, 163(3): 1109-1122.
doi: 10.1093/genetics/163.3.1109 pmid: 12663548 |
| [25] | Kidwell KK, Osborn TC. Simple plant DNA isolation procedures. In: Beckmann JS, Osborn TC, Springer, eds. Plant Genomes: Methods for Genetic and Physical Mapping. Dordrecht, 1992. |
| [26] |
Jiang N, Bao ZR, Zhang XY, Hirochika H, Eddy SR, McCouch SR, Wessler SR. An active DNA transposon family in rice. Nature, 2003, 421: 163-167.
doi: 10.1038/nature01214 |
| [27] |
Kikuchi K, Terauchi K, Wada M, Hirano HY. The plant MITE mPing is mobilized in anther culture. Nature, 2003, 421: 167-170.
doi: 10.1038/nature01218 |
| [28] | Shan XH, Ou XF, Liu ZL, Dong YZ, Lin XY, Li XW, Liu B. Transpositional activation of mPing in an asymmetric nuclear somatic cell hybrid of rice and Zizania latifolia was accompanied by massive element loss. Theor Appl Genet, 2009, 19: 1325-1333. |
| [29] | Zhang XQ, Li N, Xie XM. Design and exploration of epigenetic comprehensive experiments. Hereditas(Beijing), 2021, 43(12): 1179-1187. |
| 张向前, 李楠, 解新明. 表观遗传学综合性实验设计与探讨. 遗传, 2021, 43(12): 1179-1187. |
| No related articles found! |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||