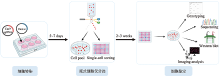
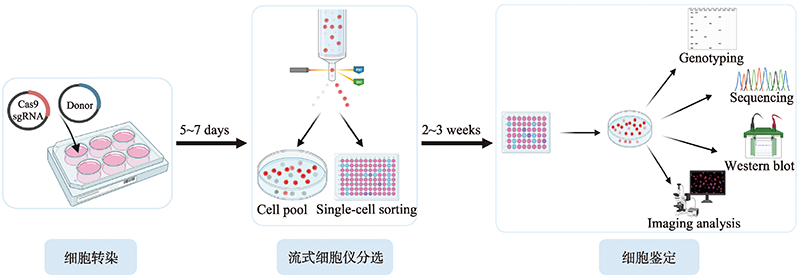
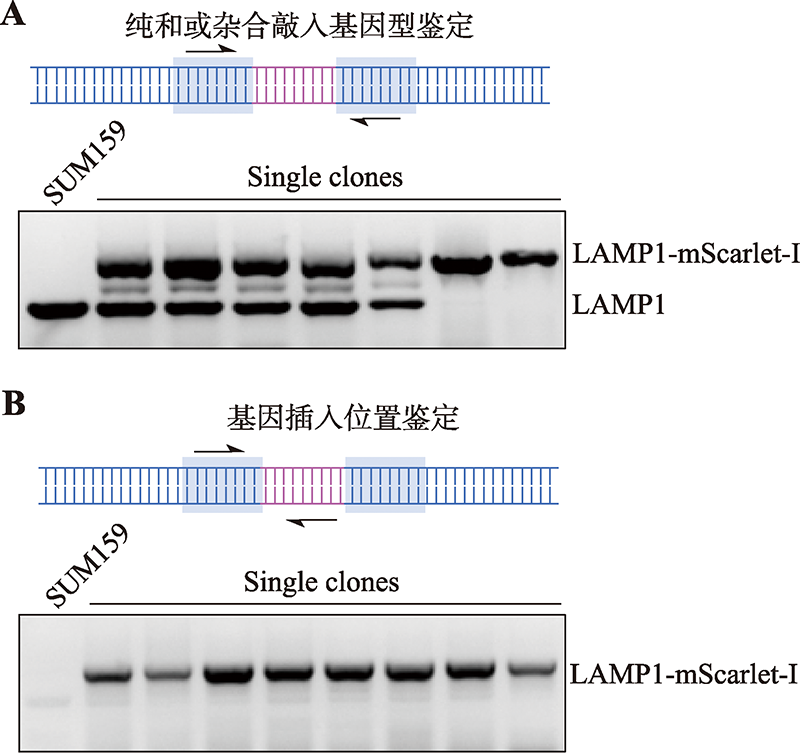
Hereditas(Beijing) ›› 2023, Vol. 45 ›› Issue (2): 165-175.doi: 10.16288/j.yczz.22-395
Previous Articles Next Articles
The protocol of tagging endogenous proteins with fluorescent tags using CRISPR-Cas9 genome editing
Zhongsheng Wu1,2( ), Yu Gao1,2(
), Yu Gao1,2( ), Yongtao Du1,2(
), Yongtao Du1,2( ), Song Dang1, Kangmin He1,2(
), Song Dang1, Kangmin He1,2( )
)
- 1. State Key Laboratory of Molecular Developmental Biology, Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, Beijing 100101, China
2. University of Chinese Academy of Sciences, Beijing 100049, China
-
Received:2022-12-02Revised:2023-01-06Online:2023-02-20Published:2023-01-09 -
Contact:He Kangmin E-mail:zswu@genetics.ac.cn;gaoyu@genetics.ac.cn;duyongtao@genetics.ac.cn;kmhe@genetics.ac.cn -
Supported by:the National Natural Science Foundation of China(91957106);the National Natural Science Foundation of China(31970659);the Ministry of Science and Technology of the People’s Republic of China(2021YFA0804802);the Ministry of Science and Technology of the People’s Republic of China(2022YFA1304500)
Cite this article
Zhongsheng Wu, Yu Gao, Yongtao Du, Song Dang, Kangmin He. The protocol of tagging endogenous proteins with fluorescent tags using CRISPR-Cas9 genome editing[J]. Hereditas(Beijing), 2023, 45(2): 165-175.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
Table 1
Reagents and materials"
| 试剂与耗材名称 | 品牌 | 货号 |
|---|---|---|
| KAPA HiFi HotStart ReadyMix Kit | KAPA Biosystems | KK2631 |
| FastPure Gel DNA Extraction Mini Kit | Vazyme | DC301-01 |
| pEASY-Uni Seamless Cloning and Assembly Kit | TransGen | CU101-03 |
| Quick Ligation? Kit | NEB | M2200L |
| T4 Polynucleotide Kinase | NEB | M0201S |
| CutSmart? Buffer | NEB | B7204S |
| Sma I | NEB | R0141S |
| FastDigest? Bpi I | ThermoFisher Scientific | FD1014 |
| FastAP Thermosensitive Alkaline Phosphatase | ThermoFisher Scientific | EF0652 |
| FastDigest Buffer | ThermoFisher Scientific | B64 |
| GoTaq? DNA Polymerase | Promega | M300A |
| TIANamp Genomic DNA Kit | TIANGEN | DP304-02 |
| NucleoBond Xtra Midi Plus | Macherey-Nagel | 740412.50 |
| Lipofectamine? 3000 | Invitrogen | L3000015 |
| Trypsin-EDTA (0.25%), phenol red | Gibco | 25200114 |
| MEM α, Nucleosides, no Phenol Red | Gibco | 41061029 |
| Janelia Fluor? 549 HaloTag? Ligand | Promega | GA1111 |
| QuickExtract? DNA Extraction Solution | Lucigen | QE09050 |
| LAMP-1 Antibody (H4A3) | Santa Cruz | sc-20011 |
| GAPDH Antibody | Proteintech | 10494-1-AP |
| Cell Strainers | Falcon | 352340 |
| Round-Bottom Polystyrene Test Tubes | Falcon | 352054 |
| 4 Chamber glass bottom dishes | Cellvis | D35C4-20-1.5-N |
| [1] |
Schneider AFL, Hackenberger CPR. Fluorescent labelling in living cells. Curr Opin Biotechnol, 2017, 48: 61-68.
doi: 10.1016/j.copbio.2017.03.012 |
| [2] |
Crivat G, Taraska JW. Imaging proteins inside cells with fluorescent tags. Trends Biotechnol, 2012, 30(1): 8-16.
doi: 10.1016/j.tibtech.2011.08.002 pmid: 21924508 |
| [3] |
Cranfill PJ, Sell BR, Baird MA, Allen JR, Lavagnino Z, de Gruiter HM, Kremers GJ, Davidson MW, Ustione A, Piston DW. Quantitative assessment of fluorescent proteins. Nat Methods, 2016, 13(7): 557-562.
doi: 10.1038/nmeth.3891 pmid: 27240257 |
| [4] |
Hirano M, Ando R, Shimozono S, Sugiyama M, Takeda N, Kurokawa H, Deguchi R, Endo K, Haga K, Takai-Todaka R, Inaura S, Matsumura Y, Hama H, Okada Y, Fujiwara T, Morimoto T, Katayama K, Miyawaki A. A highly photostable and bright green fluorescent protein. Nat Biotechnol, 2022, 40(7): 1132-1142.
doi: 10.1038/s41587-022-01278-2 |
| [5] |
Los GV, Encell LP, McDougall MG, Hartzell DD, Karassina N, Zimprich C, Wood MG, Learish R, Ohana RF, Urh M, Simpson D, Mendez J, Zimmerman K, Otto P, Vidugiris G, Zhu J, Darzins A, Klaubert DH, Bulleit RF, Wood KV. HaloTag: a novel protein labeling technology for cell imaging and protein analysis. ACS Chem Biol, 2008, 3(6): 373-382.
doi: 10.1021/cb800025k pmid: 18533659 |
| [6] |
Hoelzel CA, Zhang X. Visualizing and manipulating biological processes by using HaloTag and SNAP-Tag technologies. Chembiochem, 2020, 21(14): 1935-1946.
doi: 10.1002/cbic.202000037 pmid: 32180315 |
| [7] |
Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc, 2013, 8(11): 2281-2308.
doi: 10.1038/nprot.2013.143 pmid: 24157548 |
| [8] |
Tamura R, Kamiyama D. CRISPR-Cas9-mediated knock- in approach to insert the GFP11 tag into the genome of a human cell line. Methods Mol Biol, 2023, 2564: 185-201.
doi: 10.1007/978-1-0716-2667-2_8 pmid: 36107342 |
| [9] |
Surve S, Sorkin A. CRISPR/Cas9 gene editing of HeLa cells to tag proteins with mNeonGreen. Bio Protoc, 2022, 12(10): e4415.
doi: 10.21769/BioProtoc.4415 pmid: 35813028 |
| [10] |
Koch B, Nijmeijer B, Kueblbeck M, Cai Y, Walther N, Ellenberg J. Generation and validation of homozygous fluorescent knock-in cells using CRISPR-Cas9 genome editing. Nat Protoc, 2018, 13(6): 1465-1487.
doi: 10.1038/nprot.2018.042 pmid: 29844520 |
| [11] |
Cabantous S, Terwilliger TC, Waldo GS. Protein tagging and detection with engineered self-assembling fragments of green fluorescent protein. Nat Biotechnol, 2005, 23(1): 102-107.
doi: 10.1038/nbt1044 pmid: 15580262 |
| [12] |
Feng SY, Sekine S, Pessino V, Li H, Leonetti MD, Huang B. Improved split fluorescent proteins for endogenous protein labeling. Nat Commun, 2017, 8(1): 370.
doi: 10.1038/s41467-017-00494-8 pmid: 28851864 |
| [13] |
Alford SC, Ding YD, Simmen T, Campbell RE. Dimerization-dependent green and yellow fluorescent proteins. ACS Synth Biol, 2012, 1(12): 569-575.
doi: 10.1021/sb300050j pmid: 23656278 |
| [14] |
Rodriguez EA, Campbell RE, Lin JY, Lin MZ, Miyawaki A, Palmer AE, Shu XK, Zhang J, Tsien RY. The growing and glowing toolbox of fluorescent and photoactive proteins. Trends Biochem Sci, 2017, 42(2): 111-129.
doi: S0968-0004(16)30173-6 pmid: 27814948 |
| [15] |
Cormack BP, Valdivia RH, Falkow S. FACS-optimized mutants of the green fluorescent protein (GFP). Gene, 1996, 173(1 Spec No):33-38.
doi: 10.1016/0378-1119(95)00685-0 pmid: 8707053 |
| [16] |
Zacharias DA, Violin JD, Newton AC, Tsien RY. Partitioning of lipid-modified monomeric GFPs into membrane microdomains of live cells. Science, 2002, 296(5569): 913-916.
doi: 10.1126/science.1068539 pmid: 11988576 |
| [17] |
Shaner NC, Lambert GG, Chammas A, Ni YH, Cranfill PJ, Baird MA, Sell BR, Allen JR, Day RN, Israelsson M, Davidson MW, Wang JW. A bright monomeric green fluorescent protein derived from Branchiostoma lanceolatum. Nat Methods, 2013, 10(5): 407-409.
doi: 10.1038/nmeth.2413 pmid: 23524392 |
| [18] |
Shaner NC, Campbell RE, Steinbach PA, Giepmans BNG, Palmer AE, Tsien RY. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat Biotechnol, 2004, 22(12): 1567-1572.
doi: 10.1038/nbt1037 pmid: 15558047 |
| [19] |
Merzlyak EM, Goedhart J, Shcherbo D, Bulina ME, Shcheglov AS, Fradkov AF, Gaintzeva A, Lukyanov KA, Lukyanov S, Gadella TWJ, Chudakov DM. Bright monomeric red fluorescent protein with an extended fluorescence lifetime. Nat Methods, 2007, 4(7): 555-557.
doi: 10.1038/nmeth1062 pmid: 17572680 |
| [20] |
Bindels DS, Haarbosch L, van Weeren L, Postma M, Wiese KE, Mastop M, Aumonier S, Gotthard G, Royant A, Hink MA, Gadella TWJ. mScarlet: a bright monomeric red fluorescent protein for cellular imaging. Nat Methods, 2017, 14(1): 53-56.
doi: 10.1038/nmeth.4074 pmid: 27869816 |
| [21] |
Mukherjee S, Hung ST, Douglas N, Manna P, Thomas C, Ekrem A, Palmer AE, Jimenez R. Engineering of a brighter variant of the fusionRed fluorescent protein using lifetime Flow Cytometry and structure-guided mutations. Biochemistry, 2020, 59(39): 3669-3682.
doi: 10.1021/acs.biochem.0c00484 |
| [22] | Leonetti MD, Sekine S, Kamiyama D, Weissman JS, Huang B. A scalable strategy for high-throughput GFP tagging of endogenous human proteins. Proc Natl Acad Sci USA, 2016, 113(25): E3501-E3508. |
| [23] |
Los GV, Wood K. The HaloTag: a novel technology for cell imaging and protein analysis. Methods Mol Biol, 2007, 356: 195-208.
pmid: 16988404 |
| [24] |
Keppler A, Gendreizig S, Gronemeyer T, Pick H, Vogel H, Johnsson K. A general method for the covalent labeling of fusion proteins with small molecules in vivo. Nat Biotechnol, 2003, 21(1): 86-89.
pmid: 12469133 |
| [25] |
Juillerat A, Gronemeyer T, Keppler A, Gendreizig S, Pick H, Vogel H, Johnsson K. Directed evolution of O6- alkylguanine-DNA alkyltransferase for efficient labeling of fusion proteins with small molecules in vivo. Chem Biol, 2003, 10(4): 313-317.
doi: 10.1016/S1074-5521(03)00068-1 |
| [26] |
Gautier A, Juillerat A, Heinis C, Corrêa IR, Kindermann M, Beaufils F, Johnsson K. An engineered protein tag for multiprotein labeling in living cells. Chem Biol, 2008, 15(2): 128-136.
doi: 10.1016/j.chembiol.2008.01.007 |
| [27] |
Grimm JB, Brown TA, English BP, Lionnet T, Lavis LD. Synthesis of Janelia Fluor HaloTag and SNAP-Tag ligands and their use in cellular imaging experiments. Methods Mol Biol, 2017, 1663: 179-188.
doi: 10.1007/978-1-4939-7265-4_15 pmid: 28924668 |
| [28] |
Trinh R, Gurbaxani B, Morrison SL, Seyfzadeh M. Optimization of codon pair use within the (GGGGS)3 linker sequence results in enhanced protein expression. Mol Immunol, 2004, 40(10): 717-722.
pmid: 14644097 |
| [29] |
van Rosmalen M, Krom M, Merkx M. Tuning the flexibility of glycine-serine linkers to allow rational design of multidomain proteins. Biochemistry, 2017, 56(50): 6565-6574.
doi: 10.1021/acs.biochem.7b00902 pmid: 29168376 |
| [30] | Gasiunas G, Barrangou R, Horvath P, Siksnys V. Cas9- crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc Natl Acad Sci USA, 2012, 109(39): E2579-E2586. |
| [31] |
Bukhari H, Müller T. Endogenous fluorescence tagging by CRISPR. Trends Cell Biol, 2019, 29(11): 912-928.
doi: S0962-8924(19)30140-0 pmid: 31522960 |
| [32] |
Ratz M, Testa I, Hell SW, Jakobs S. CRISPR/Cas9- mediated endogenous protein tagging for RESOLFT super-resolution microscopy of living human cells. Sci Rep, 2015, 5: 9592.
doi: 10.1038/srep09592 |
| [33] |
Hendel A, Kildebeck EJ, Fine EJ, Clark J, Punjya N, Sebastiano V, Bao G, Porteus MH. Quantifying genome- editing outcomes at endogenous loci with SMRT sequencing. Cell Rep, 2014, 7(1): 293-305.
doi: 10.1016/j.celrep.2014.02.040 pmid: 24685129 |
| [34] |
Paquet D, Kwart D, Chen A, Sproul A, Jacob S, Teo S, Olsen KM, Gregg A, Noggle S, Tessier-Lavigne M.Efficient introduction of specific homozygous and heterozygous mutations using CRISPR/Cas9. Nature, 2016, 533(7601): 125-129.
doi: 10.1038/nature17664 |
| [35] |
Concordet JP, Haeussler M. CRISPOR: intuitive guide selection for CRISPR/Cas9 genome editing experiments and screens. Nucleic Acids Res, 2018, 46(W1): W242-W245.
doi: 10.1093/nar/gky354 |
| [36] |
Doench JG, Fusi N, Sullender M, Hegde M, Vaimberg EW, Donovan KF, Smith I, Tothova Z, Wilen C, Orchard R, Virgin HW, Listgarten J, Root DE.Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat Biotechnol, 2016, 34(2): 184-191.
doi: 10.1038/nbt.3437 pmid: 26780180 |
| [37] |
He KM, Marsland R, Upadhyayula S, Song E, Dang S, Capraro BR, Wang WM, Skillern W, Gaudin R, Ma MH, Kirchhausen T. Dynamics of phosphoinositide conversion in clathrin-mediated endocytic traffic. Nature, 2017, 552(7685): 410-414.
doi: 10.1038/nature25146 |
| [38] |
He KM, Song E, Upadhyayula S, Dang S, Gaudin R, Skillern W, Bu K, Capraro BR, Rapoport I, Kusters I, Ma MH, Kirchhausen T. Dynamics of Auxilin 1 and GAK in clathrin-mediated traffic. J Cell Biol, 2020, 219(3): e201908142.
doi: 10.1083/jcb.201908142 |
| [39] | Li GL, Yang SX, Wu ZF, Zhang XW. Recent developments in enhancing the efficiency of CRISPR/Cas9-mediated knock-in in animals. Hereditas(Beijing), 2020, 42(7): 641-656. |
| 李国玲, 杨善欣, 吴珍芳, 张献伟. 提高CRISPR/Cas9介导的动物基因组精确插入效率研究进展. 遗传, 2020, 42(7): 641-656. |
| [1] | Tingting Yan, Lei Zhang, Yudong Li, Xinle Liang. Research on the blended learning mode of “Microbial Breeding Experiments” based on WeChat [J]. Hereditas(Beijing), 2018, 40(7): 601-606. |
| [2] | Taiming Li, Wenjun Lan, Can Huang, Chun Zhang, Xiaomei Liu. Establishment and identification of the near-infrared fluorescence labeled exosomes in breast cancer cell lines [J]. HEREDITAS(Beijing), 2016, 38(5): 427-435. |
| [3] | LI Pan-Pan, YONG Jing-Ru, JI Yun-Ling, ZHANG Ying, DIAO Liang, JIA Shi-Lin, LI Dong, WANG Hui-Li, BAO Ji-Yu, LI Pei-Zhen. Functional analysis of promoter fragments of salt-tolerance related genes in Spirulina [J]. HEREDITAS, 2011, 33(10): 1134-1140. |
| [4] | LIU Chang-Qing, Guo Yu, LIU Shuai, BAO A-Dong, LU Tao-Feng, LIU Hong-Kun, GUAN Wei-Jun, MA Yue-Hui. Characterization of expression of α1-acid glycoprotein gene in Beijing fatty chicken (Gallus gallus) [J]. HEREDITAS, 2009, 31(6): 620-628. |
| [5] | SHEN Wei-Feng, WENG Hong-Biao, NIU Bao-Long, HE Li-Hua, LIU Yan, QI Xiao-Peng, MENG Zhi-Qi . Agrobacterium tumefaciens-mediated transformation of the pathogen of Botrytis cinerea [J]. HEREDITAS, 2008, 30(4): 515-520. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||