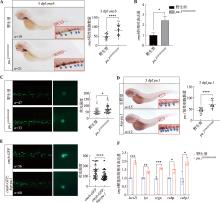
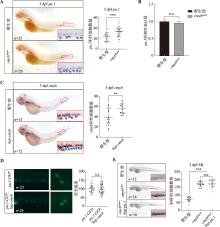
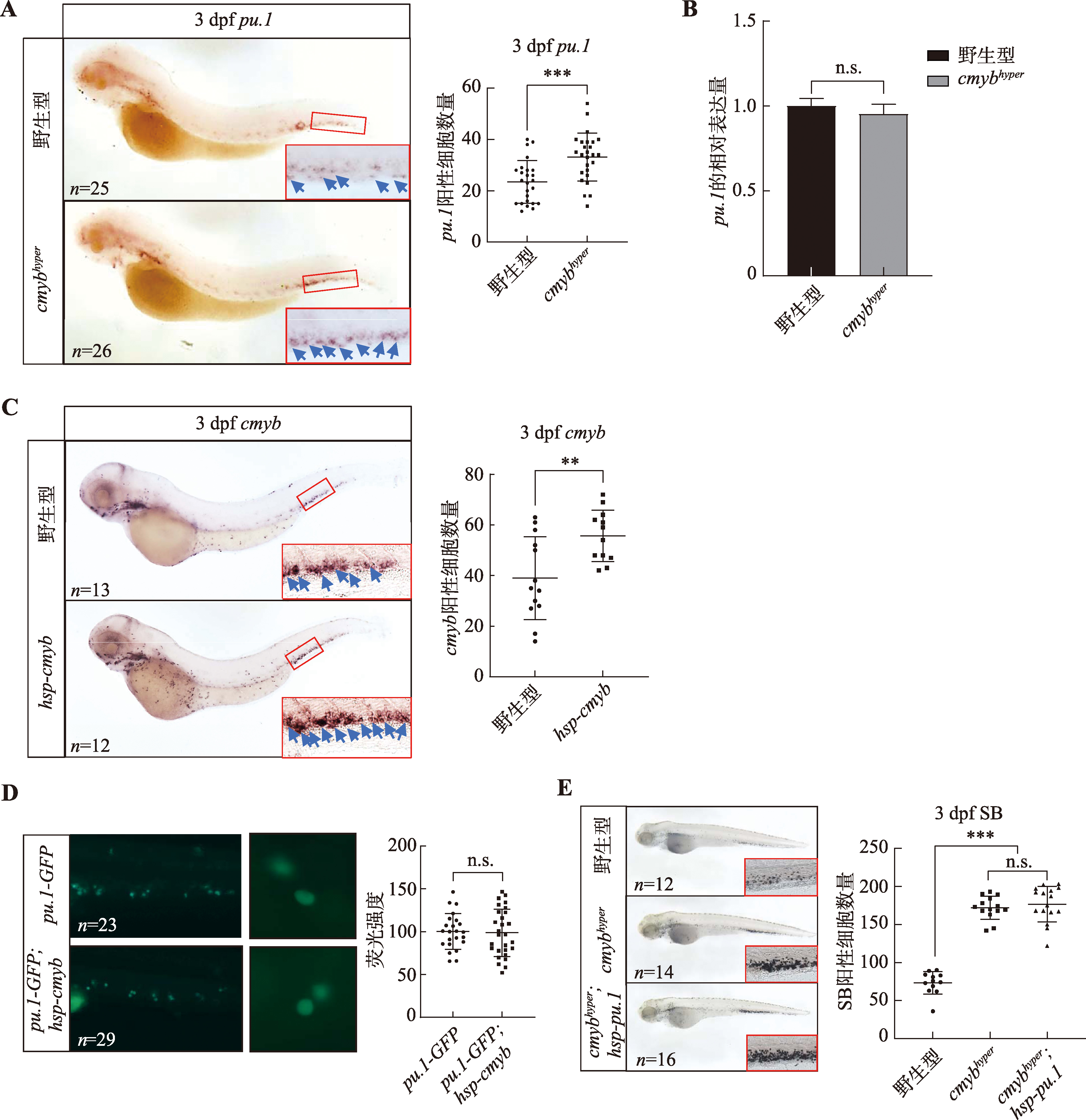
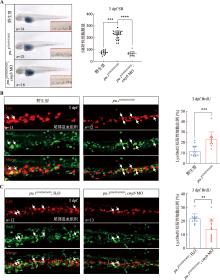
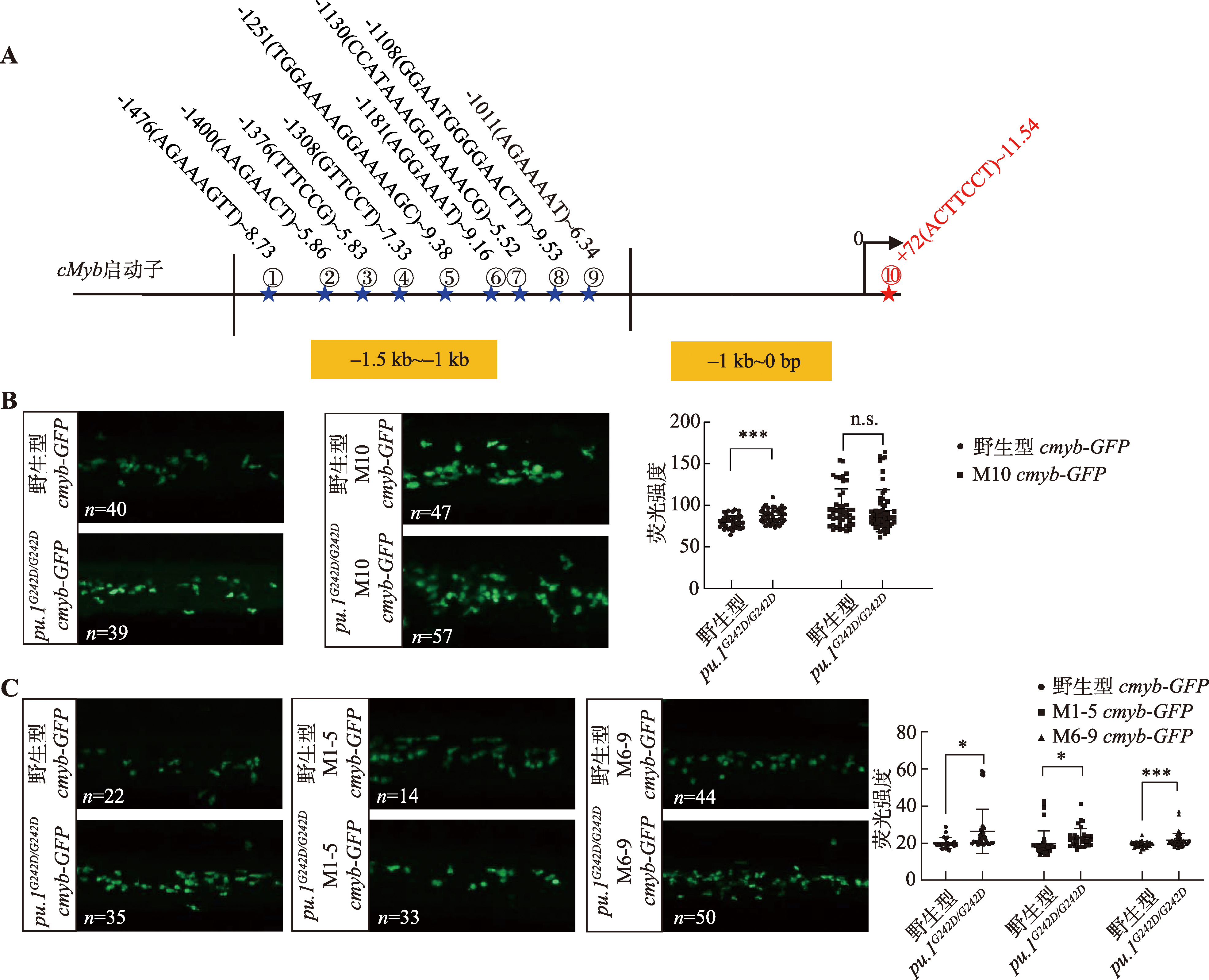
Hereditas(Beijing) ›› 2024, Vol. 46 ›› Issue (4): 319-332.doi: 10.16288/j.yczz.23-312
• Research Article • Previous Articles Next Articles
The interaction of Pu.1 and cMyb in zebrafish neutrophil development
Jiaxin Hong( ), Song’en Xu, Wenqing Zhang(
), Song’en Xu, Wenqing Zhang( ), Wei Liu(
), Wei Liu( )
)
- Division of Development Biology & Regenerative Medicine, South China University of Technology, Guangzhou 510006, China
-
Received:2023-12-18Revised:2024-01-30Online:2024-04-20Published:2024-03-11 -
Contact:Wenqing Zhang, Wei Liu E-mail:hjx_0925@163.com;mczhangwq@scut.edu.cn;liuwei7@scut.edu.cn -
Supported by:National Natural Science Foundation of China (NSFC)/Research Grants Council (RGC) Joint Research Scheme(32161160326);National Natural Science Foundation of China(82070163)
Cite this article
Jiaxin Hong, Song’en Xu, Wenqing Zhang, Wei Liu. The interaction of Pu.1 and cMyb in zebrafish neutrophil development[J]. Hereditas(Beijing), 2024, 46(4): 319-332.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
Table 3
The primer sequences used in cmyb promoter multi-site mutation plasmids construction"
| 突变位点 | 引物序列(5′→3′) |
|---|---|
| 1 AACTTTCT | F:ATAACGTCTGCTGGTTACTTTTGGTTATTCTGCAATTACG R:CCAAAAGTAACCAGCAGACGTTATCTGTCACAACACACCC |
| 2 AGTTCTT | F:ACAAACTTAAACAAGCTGCTCTACTTTTCCGTCCAGACCTGA R:AAAGTAGAGCAGCTTGTTTAAGTTTGTCTCCGACATGTCCAA |
| 3 CGGAAA | F:TTGACGTCCAGACCTGACCGTGAAATTCAGTAACAAAATGT R:ATTTCACGGTCAGGTCTGGACGTCAAAGTAGAGCAGCTTGT |
| 4 AGGAAC | F:ATGAATTTCACTTTGCGTCGAGTCACAGTGTTACACAATAC R:GTGACTCGACGCAAAGTGAAATTCATAAGCAACCAAGTGGA |
| 5 GCTTTTCCTTTTCCA | F:TGGAAAATGAGGAGCGAAACTGTTGCAACCCAATGAATGC R:GGTTGCAACAGTTTCGCTCCTCATTTTCCAGAAATGTTCC |
| 6 ATTTCCT | F:TTATTAAAATGATTCACAGTCAATTAACGATGACACAGGG R:CCCTGTGTCATCGTTAATTGACTGTGAATCATTTTAATAA |
| 7 CGTTTTCCTTTATGG | F:CCATAAGTGAAGACGTGCATTTAGGAATGGGGAACTTTAA R:CCATTCCTAAATGCACGTCTTCACTTATGGAGAGACAGTT |
| 8 AAGTTCCCCATTCC | F:GGACTGGGACACTTTAAAAACTGCCCCAATCGAAATAATG R:TTGGGGCAGTTTTTAAAGTGTCCCAGTCCTAAATGCACGT |
| 9 ATTTTCT | F:AGAAGCTTCTGCACAGATATTTAGGCGCAACACTGCTGGA R:CGCCTAAATATCTGTGCAGAAGCTTCTTTCTTCTGCGAAG |
| 10 AGGAAGT | F:GAGAGCTTAATATCAGCGCTTCTGCCTCGAAGAGGAGCTG R:CAGCTCCTCTTCGAGGCAGAAGCGCTGATATTAAGCTCTC |
| [1] |
Amulic B, Cazalet C, Hayes GL, Metzler KD, Zychlinsky A. Neutrophil function: From mechanisms to disease. Annu Rev Immunol, 2012, 30: 459-489.
doi: 10.1146/annurev-immunol-020711-074942 pmid: 22224774 |
| [2] |
Borregaard N. Neutrophils, from marrow to microbes. Immunity, 2010, 33(5): 657-670.
doi: 10.1016/j.immuni.2010.11.011 pmid: 21094463 |
| [3] |
Galli SJ, Borregaard N, Wynn TA. Phenotypic and functional plasticity of cells of innate immunity: Macrophages, mast cells and neutrophils. Nat Immunol, 2011, 12(11): 1035-1044.
doi: 10.1038/ni.2109 pmid: 22012443 |
| [4] |
Mantovani A, Cassatella MA, Costantini C, Jaillon S. Neutrophils in the activation and regulation of innate and adaptive immunity. Nat Rev Immunol, 2011, 11(8): 519-531.
doi: 10.1038/nri3024 pmid: 21785456 |
| [5] |
Carnevale S, Ceglie ID, Grieco G, Rigatelli A, Bonavita E, Jaillon S. Neutrophil diversity in inflammation and cancer. Front Immunol, 2023, 14: 1180810.
doi: 10.3389/fimmu.2023.1180810 |
| [6] | Lawrence SM, Corriden R, Nizet V. The ontogeny of a neutrophil: Mechanisms of granulopoiesis and homeostasis. Microbiol Mol Biol Rev, 2018, 82(1): e00057-17. |
| [7] |
Antony-Debré I, Paul A, Leite J, Mitchell K, Kim HM, Carvajal LA, Todorova TI, Huang K, Kumar A, Farahat AA, Bartholdy B, Narayanagari SR, Chen JH, Ambesi- Impiombato A, Ferrando AA, Mantzaris I, Gavathiotis E, Verma A, Will B, Boykin DW, Wilson WD, Poon GM, Steidl U. Pharmacological inhibition of the transcription factor pu.1 in leukemia. J Clin Invest, 2017, 127(12): 4297-4313.
doi: 10.1172/JCI92504 pmid: 29083320 |
| [8] |
Anderson KL, Smith KA, Pio F, Torbett BE, Maki RA. Neutrophils deficient in pu.1 do not terminally differentiate or become functionally competent. Blood, 1998, 92(5): 1576-1585.
pmid: 9716585 |
| [9] |
Iwasaki H, Somoza C, Shigematsu H, Duprez EA, Iwasaki-Arai J, Mizuno SI, Arinobu Y, Geary K, Zhang P, Dayaram T, Fenyus ML, Elf S, Chan SS, Kastner P, Huettner CS, Murray R, Tenen DG, Akashi K. Distinctive and indispensable roles of pu.1 in maintenance of hematopoietic stem cells and their differentiation. Blood, 2005, 106(5): 1590-1600.
doi: 10.1182/blood-2005-03-0860 pmid: 15914556 |
| [10] |
Rosenbauer F, Wagner K, Kutok JL, Iwasaki H, Le Beau MM, Okuno Y, Akashi K, Fiering S, Tenen DG. Acute myeloid leukemia induced by graded reduction of a lineage-specific transcription factor, pu.1. Nat Genet, 2004, 36(6): 624-630.
pmid: 15146183 |
| [11] |
Sun J, Liu W, Li L, Chen J, Wu M, Zhang Y, Leung AYH, Zhang W, Wen Z, Liao W. Suppression of pu.1 function results in expanded myelopoiesis in zebrafish. Leukemia, 2013, 27(9): 1913-1917.
doi: 10.1038/leu.2013.67 pmid: 23455395 |
| [12] |
Burda P, Curik N, Kokavec J, Basova P, Mikulenkova D, Skoultchi AI, Zavadil J, Stopka T. Pu.1 activation relieves gata-1-mediated repression of cebpa and cbfb during leukemia differentiation. Mol Cancer Res, 2009, 7(10): 1693-1703.
doi: 10.1158/1541-7786.MCR-09-0031 pmid: 19825991 |
| [13] |
Walsh JC, DeKoter RP, Lee HJ, Smith ED, Lancki DW, Gurish MF, Friend DS, Stevens RL, Anastasi J, Singh H. Cooperative and antagonistic interplay between pu.1 and gata-2 in the specification of myeloid cell fates. Immunity, 2002, 17(5): 665-676.
pmid: 12433372 |
| [14] |
Chang HC, Han L, Jabeen R, Carotta S, Nutt SL, Kaplan MH. Pu.1 regulates tcr expression by modulating gata-3 activity. J Immunol, 2009, 183(8): 4887-4894.
doi: 10.4049/jimmunol.0900363 |
| [15] |
Yang Z, Wara-Aswapati N, Chen C, Tsukada J, Auron PE. Nf-il6 (c/ebpβ) vigorously activates il1b gene expression via a spi-1 (pu.1) protein-protein tether. J Biol Chem, 2000, 275(28): 21272-21277.
doi: 10.1074/jbc.M000145200 pmid: 10801783 |
| [16] |
Rekhtman N, Choe KS, Matushansky I, Murray S, Stopka T, Skoultchi AI. Pu.1 and prb interact and cooperate to repress gata-1 and block erythroid differentiation. Mol Cell Biol, 2003, 23(21): 7460-7474.
doi: 10.1128/MCB.23.21.7460-7474.2003 pmid: 14559995 |
| [17] |
Wei F, Zaprazna K, Wang JW, Atchison ML. Pu.1 can recruit bcl6 to DNA to repress gene expression in germinal center b cells. Mol Cell Biol, 2009, 29(17): 4612-4622.
doi: 10.1128/MCB.00234-09 pmid: 19564417 |
| [18] |
Meraro D, Hashmueli S, Koren B, Azriel A, Oumard A, Kirchhoff S, Hauser H, Nagulapalli S, Atchison ML, Levi BZ. Protein-protein and DNA-protein interactions affect the activity of lymphoid-specific ifn regulatory factors. J Immunol, 1999, 163(12): 6468-6478.
pmid: 10586038 |
| [19] |
Zhao XH, Bartholdy B, Yamamoto Y, Evans EK, Alberich- Jordà M, Staber PB, Benoukraf T, Zhang P, Zhang JY, Trinh BQ, Crispino JD, Hoang T, Bassal MA, Tenen DG. Pu.1-c-jun interaction is crucial for pu.1 function in myeloid development. Commun Biol, 2022, 5(1): 961.
doi: 10.1038/s42003-022-03888-7 pmid: 36104445 |
| [20] | Yeamans C, Wang DH, Paz-Priel I, Torbett BE, Tenen DG, Friedman AD. C/ebpα binds and activates the pu.1 distal enhancer to induce monocyte lineage commitment. Blood, 2007, 110(9): 3136-3142. |
| [21] |
Lipsick JS, Wang DM. Transformation by v-myb. Oncogene, 1999, 18(19): 3047-3055.
doi: 10.1038/sj.onc.1202745 pmid: 10378700 |
| [22] |
Clarke ML, Lemma RB, Walton DS, Volpe G, Noyvert B, Gabrielsen OS, Frampton J. Myb insufficiency disrupts proteostasis in hematopoietic stem cells, leading to age-related neoplasia. Blood, 2023, 141(15): 1858-1870.
doi: 10.1182/blood.2022019138 pmid: 36603185 |
| [23] |
Takao S, Forbes L, Uni M, Cheng SY, Pineda JMB, Tarumoto Y, Cifani P, Minuesa G, Chen C, Kharas MG, Bradley RK, Vakoc CR, Koche RP, Kentsis A. Convergent organization of aberrant myb complex controls oncogenic gene expression in acute myeloid leukemia. eLife, 2021, 10: e65905.
doi: 10.7554/eLife.65905 |
| [24] |
Mucenski ML, McLain K, Kier AB, Swerdlow SH, Schreiner CM, Miller TA, Pietryga DW, Scott WJ, Potter SS. A functional c-myb gene is required for normal murine fetal hepatic hematopoiesis. Cell, 1991, 65(4): 677-689.
doi: 10.1016/0092-8674(91)90099-k pmid: 1709592 |
| [25] |
Sandberg ML, Sutton SE, Pletcher MT, Wiltshire T, Tarantino LM, Hogenesch JB, Cooke MP. C-myb and p 300 regulate hematopoietic stem cell proliferation and differentiation. Dev Cell, 2005, 8(2): 153-166.
doi: 10.1016/j.devcel.2004.12.015 pmid: 15691758 |
| [26] |
Bjerregaard MD, Jurlander J, Klausen P, Borregaard N, Cowland JB. The in vivo profile of transcription factors during neutrophil differentiation in human bone marrow. Blood, 2003, 101(11): 4322-4332.
doi: 10.1182/blood-2002-03-0835 pmid: 12560239 |
| [27] |
Basova P, Pospisil V, Savvulidi F, Burda P, Vargova K, Stanek L, Dluhosova M, Kuzmova E, Jonasova A, Steidl U, Laslo P, Stopka T. Aggressive acute myeloid leukemia in pu.1/p53 double-mutant mice. Oncogene, 2014, 33(39): 4735-4745.
doi: 10.1038/onc.2013.414 pmid: 24121269 |
| [28] |
Liu W, Wu M, Huang Z, Lian J, Chen J, Wang T, Leung AYH, Liao Y, Zhang Z, Liu Q, Yen K, Lin S, Zon LI, Wen Z, Zhang Y, Zhang W. C-myb hyperactivity leads to myeloid and lymphoid malignancies in zebrafish. Leukemia, 2017, 31(1): 222-233.
doi: 10.1038/leu.2016.170 pmid: 27457538 |
| [29] |
Hall C, Flores MV, Storm T, Crosier K, Crosier P. The zebrafish lysozyme C promoter drives myeloid-specific expression in transgenic fish. Bmc Dev Biol, 2007, 7: 42.
pmid: 17477879 |
| [30] |
Ward AC, McPhee DO, Condron MM, Varma S, Cody SH, Onnebo SMN, Paw BH, Zon LI, Lieschke GJ. The zebrafish spi1 promoter drives myeloid-specific expression in stable transgenic fish. Blood, 2003, 102(9): 3238-3240.
doi: 10.1182/blood-2003-03-0966 pmid: 12869502 |
| [31] |
Jin H, Li L, Xu J, Zhen FH, Zhu L, Liu PP, Zhang MJ, Zhang WQ, Wen ZL.Runx1 regulates embryonic myeloid fate choice in zebrafish through a negative feedback loop inhibiting pu.1 expression. Blood, 2012, 119(22): 5239-5249.
doi: 10.1182/blood-2011-12-398362 pmid: 22493295 |
| [32] | Westerfield M. The zebrafish book: A guide for the laboratory use of zebrafish (Danio rerio). 4th ed.,University of Oregon Press, Eugene, 1995. |
| [33] | Chitramuthu BP, Bennett HPJ. High resolution whole mount in situ hybridization within zebrafish embryos to study gene expression and function. J Vis Exp, 2013, (80): e50644. |
| [34] |
Lu JA, Huang CY, Lin ZY, Tang Z, Ma N, Huang ZB. The role of the cd99l2 gene on leukocyte interstitial migration in zebrafish. Hereditas(Beijing), 2022, 44(9): 798-809.
doi: 10.16288/j.yczz.22-193 pmid: 36384956 |
| 卢荆澳, 黄春燕, 林芷茵, 唐政, 马宁, 黄志斌. cd99l2基因调控斑马鱼白细胞组织间的迁移机制. 遗传, 2022, 44(9): 798-809. | |
| [35] |
Soza-Ried C, Hess I, Netuschil N, Schorpp M, Boehm T. Essential role of c-myb in definitive hematopoiesis is evolutionarily conserved. Proc Natl Acad Sci USA, 2010, 107(40): 17304-17308.
doi: 10.1073/pnas.1004640107 pmid: 20823231 |
| [36] |
Bellon T, Perrotti D, Calabretta B. Granulocytic differentiation of normal hematopoietic precursor cells induced by transcription factor pu.1 correlates with negative regulation of the c-myb promoter. Blood, 1997, 90(5): 1828-1839.
pmid: 9292515 |
| [37] |
Bies J, Mukhopadhyaya R, Pierce J, Wolff L. Only late, nonmitotic stages of granulocyte differentiation in 32dcl3 cells are blocked by ectopic expression of murine c-myb and its truncated forms. Cell Growth Differ, 1995, 6(1): 59-68.
pmid: 7536440 |
| [38] |
Gotea V, Visel A, Westlund JM, Nobrega MA, Pennacchio LA, Ovcharenko I. Homotypic clusters of transcription factor binding sites are a key component of human promoters and enhancers. Genome Res, 2010, 20(5): 565-577.
doi: 10.1101/gr.104471.109 pmid: 20363979 |
| [39] |
Ezer D, Zabet NR, Adryan B. Homotypic clusters of transcription factor binding sites: a model system for understanding the physical mechanics of gene expression. Comput Struct Biotechnol J, 2014, 10(17): 63-69.
doi: 10.1016/j.csbj.2014.07.005 |
| [40] | Dudek H, Tantravahi RV, Rao VN, Reddy ES, Reddy EP. Myb and ets proteins cooperate in transcriptional activation of the mim-1 promoter. Proc Natl Acad Sci USA, 1992, 89(4): 1291-1295. |
| [41] |
Oelgeschläger M, Nuchprayoon I, Lüscher B, Friedman AD. C/ebp, c-myb, and pu.1 cooperate to regulate the neutrophil elastase promoter. Mol Cell Biol, 1996, 16(9): 4717-4725.
doi: 10.1128/MCB.16.9.4717 pmid: 8756629 |
| [42] |
Ward AC, Loeb DM, Soede-Bobok AA, Touw IP, Friedman AD. Regulation of granulopoiesis by transcription factors and cytokine signals. Leukemia, 2000, 14(6): 973-990.
pmid: 10865962 |
| [1] | Piao Sun, Ying Li, Fan Liu, Lu Wang. Generation and analysis of TPI deficiency zebrafish model [J]. Hereditas(Beijing), 2024, 46(3): 232-241. |
| [2] | Kailun Li, Jingao Lu, Xiaohui Chen, Wenqing Zhang, Wei Liu. The role of the allantoin in promoting fracture healing in osteoclast-deficient zebrafish [J]. Hereditas(Beijing), 2023, 45(4): 341-353. |
| [3] | Jing’ao Lu, Chunyan Huang, Zhiyin Lin, Zheng Tang, Ning Ma, Zhibin Huang. The role of the cd99l2 gene on leukocyte interstitial migration in zebrafish [J]. Hereditas(Beijing), 2022, 44(9): 798-809. |
| [4] | Pengfei Zheng, Haibo Xie, Panpan Zhu, Chengtian Zhao. Distribution pattern of floor plate neurons in zebrafish [J]. Hereditas(Beijing), 2022, 44(6): 510-520. |
| [5] | Tingting Zhang, Feng Liu. Study on a detection method of protein tyrosine sulfation modification in zebrafish [J]. Hereditas(Beijing), 2022, 44(2): 178-186. |
| [6] | Tingting Jia, Lei Lei, Xinyuan Wu, Shunyou Cai, Yixuan Chen, Yu Xue. Study on the mechanism of metformin on zebrafish skeletal development and damage repair [J]. Hereditas(Beijing), 2022, 44(1): 68-79. |
| [7] | Jiani Guo, Fan Liu, Lu Wang. Zebrafish blood disease models and applications [J]. Hereditas(Beijing), 2020, 42(8): 725-738. |
| [8] | Feng Xiong,Xunwei Xie,Luyuan Pan,Kuoyu Li,Liyue Liu,Yun Zhang,Linglu Li,Yonghua Sun. Development of resources, technologies and services at the China Zebrafish Resource Center [J]. Hereditas(Beijing), 2018, 40(8): 683-692. |
| [9] | Jingjin Xu, Wenjuan Zhang, Jingyi Wang, Liyun Yao, Yutian Pan, Yixin Ou, Yu Xue. The active component screening of Anoectochilus roxburghii and the functional study on inhibition of melanogenesis in zebrafish [J]. Hereditas(Beijing), 2017, 39(12): 1178-1187. |
| [10] | Shanshan Liu, Cuizhen Zhang, Gang Peng. Effects of starvation on the expression of feeding related neuropeptides in the larval zebrafish hypothalamus [J]. Hereditas(Beijing), 2016, 38(9): 821-830. |
| [11] | Fenghua Zhang, Houpeng Wang, Siyu Huang, Feng Xiong, Zuoyan Zhu, Yonghua Sun. A comparison of the knockout efficiencies of two codon-optimized Cas9 coding sequences in zebrafish embryos [J]. HEREDITAS(Beijing), 2016, 38(2): 144-154. |
| [12] | YAN Li-Feng GU Ai-Hua. Progress and application of zebrafish in regenerative medicine [J]. HEREDITAS, 2013, 35(7): 856-866. |
| [13] | LI Li, LUO Ling-Fei. Zebrafish as the model system to study organogenesis and regeneration [J]. HEREDITAS, 2013, 35(4): 421-432. |
| [14] | XU Ran-Ran ZHANG Cong-Wei CAO Yu WANG Qiang. mir122 deficiency inhibits differentiation of zebrafish hepa-toblast into hepatocyte [J]. HEREDITAS, 2013, 35(4): 488-494. |
| [15] | SHEN Yan, HUANG Peng, ZHANG Bo. A protocol for TALEN construction and gene targeting in zebrafish [J]. HEREDITAS, 2013, 35(4): 533-544. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||