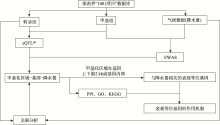
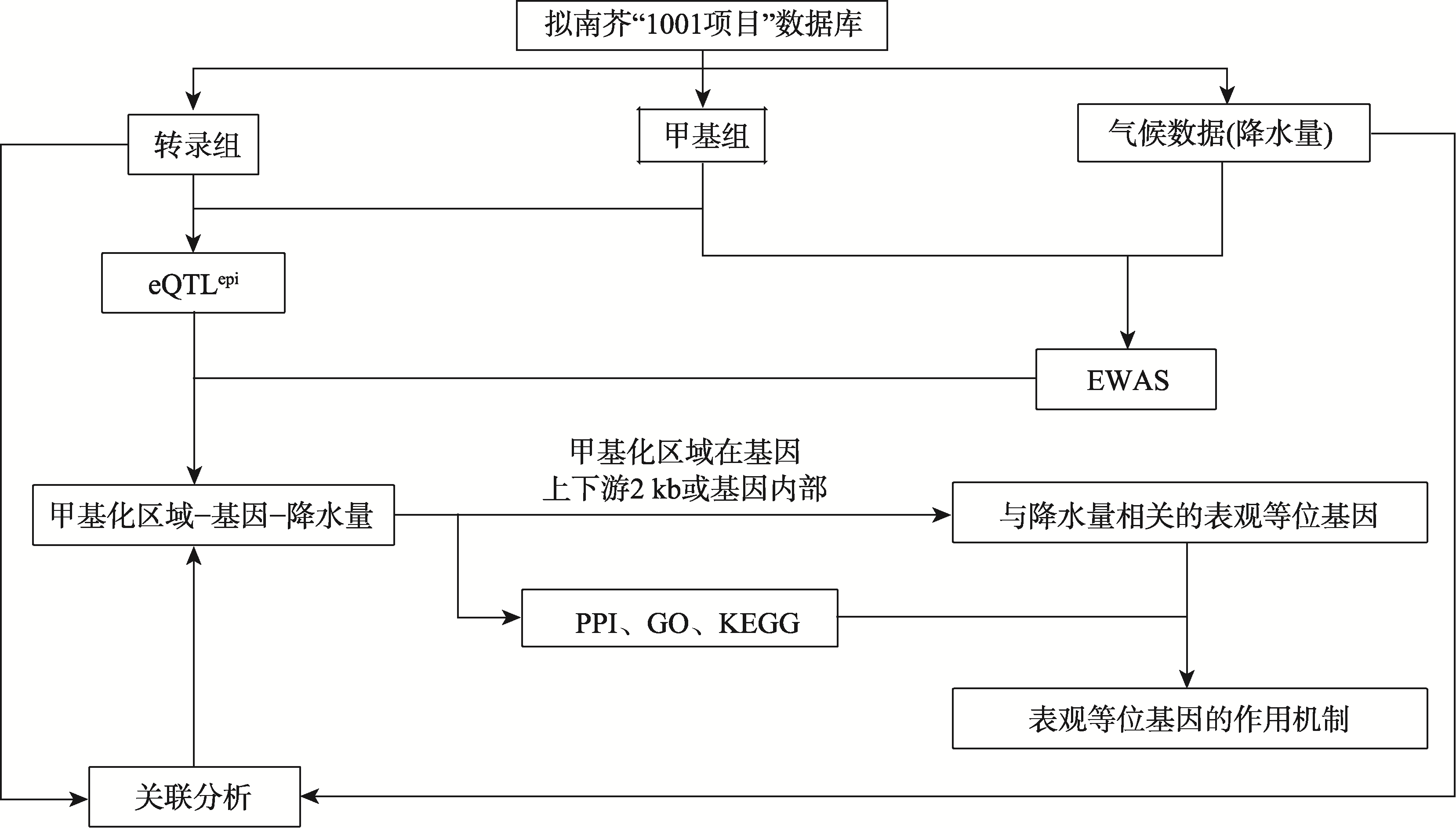
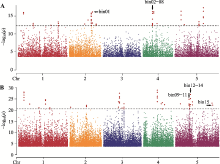
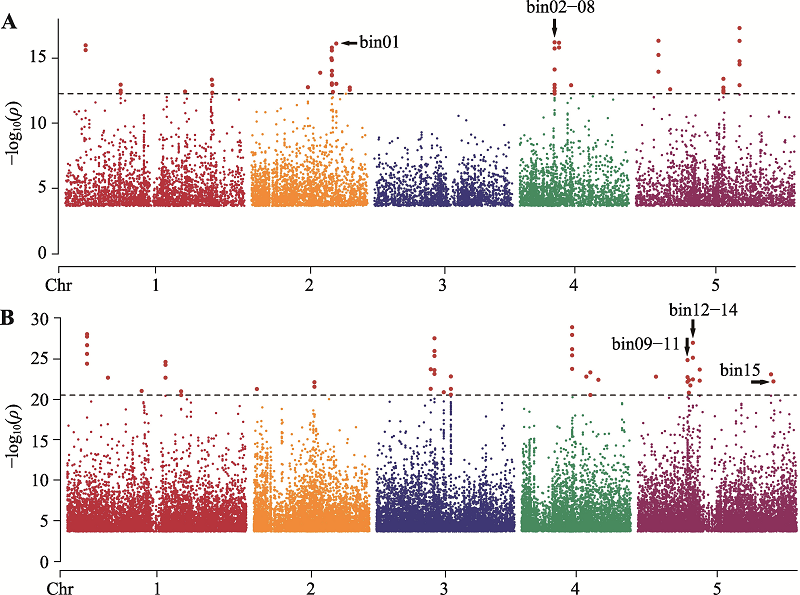
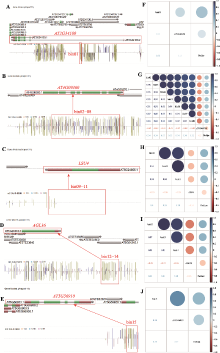
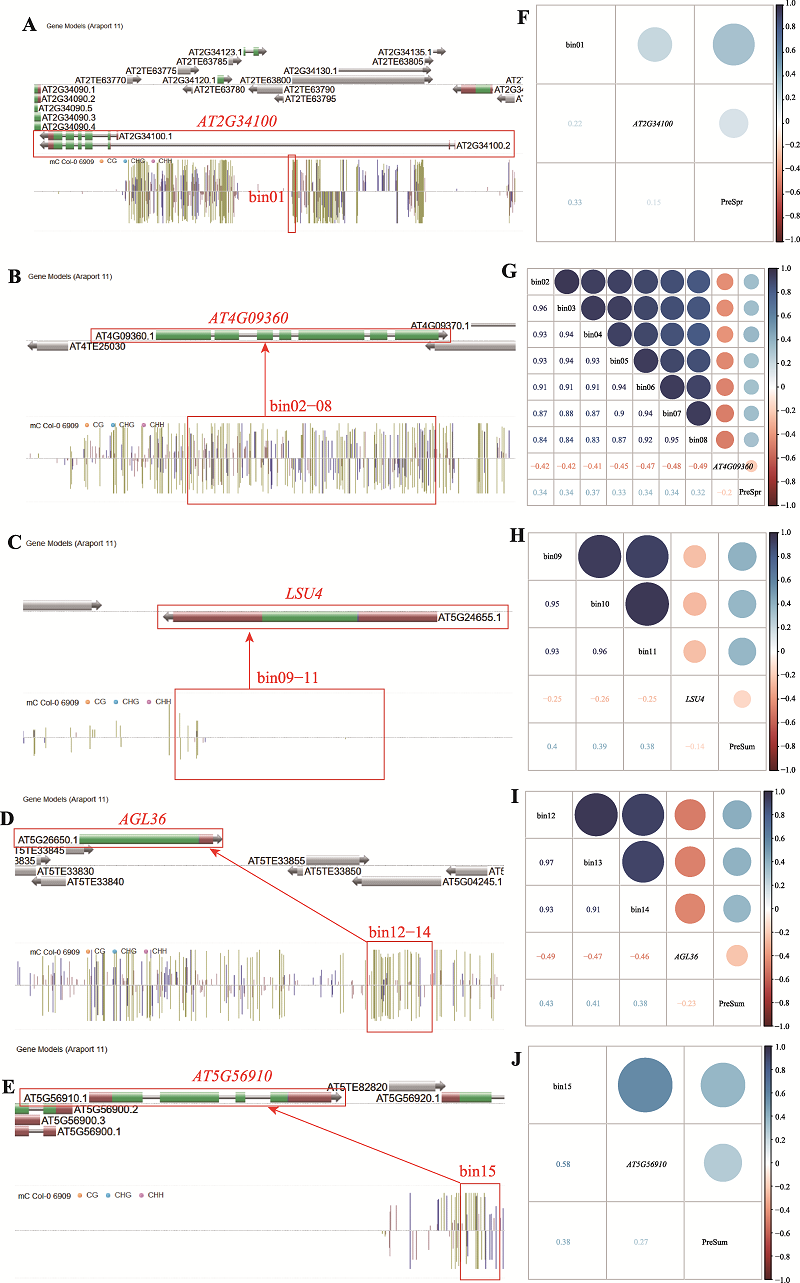
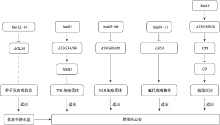
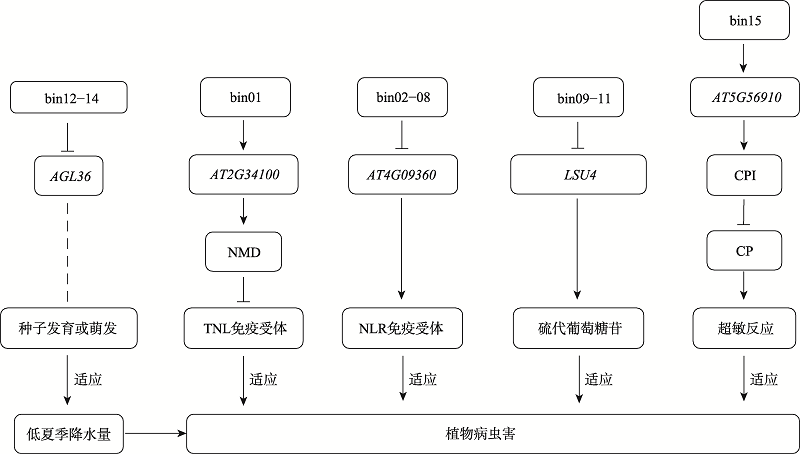
| [1] |
Richards CL, Alonso C, Becker C, Bossdorf O, Bucher E, Colomé-Tatché M, Durka W, Engelhardt J, Gaspar B, Gogol-Döring A, Grosse I, van Gurp TP, Heer K, Kronholm I, Lampei C, Latzel V, Mirouze M, Opgenoorth L, Paun O, Prohaska SJ, Rensing SA, Stadler PF, Trucchi E, Ullrich K, Verhoeven KJF. Ecological plant epigenetics: evidence from model and non-model species, and the way forward. Ecol Lett, 2017,20(12):1576-1590.
|
| [2] |
Zhang HM, Lang ZB, Zhu JK . Dynamics and function of DNA methylation in plants. Nat Rev Mol Cell Biol, 2018,19(8):489-506.
|
| [3] |
Zhao Q, Wang W, Sun YQ . Review on the correlation between DNA methylation and gene expression in plant. Biotechnol Bull, 2016,32(4):16-23.
|
|
赵倩, 王巍, 孙野青 . 植物DNA甲基化与基因表达的关联性研究进展. 生物技术通报, 2016,32(4):16-23.
|
| [4] |
Kakutani T . Epi-alleles in plants: inheritance of epigenetic information over generations. Plant Cell Physiol, 2002,43(10):1106-1111.
|
| [5] |
Luan X, Liu SC, Ke SW, Dai H, Xie XM, Hsieh TF, Zhang XQ . Epigenetic modification of ESP, encoding a putative long noncoding RNA, affects panicle architecture in rice. Rice(NY), 2019,12(1):20.
|
| [6] |
Ma CQ, Jing CJ, Chang B, Yan JY, Liang BW, Liu L, Yang YZ, Zhao ZY . The effect of promoter methylation on MdMYB1 expression determines the level of anthocyanin accumulation in skins of two non-red apple cultivars. BMC Plant Biol, 2018,18(1):108.
|
| [7] |
Song QX, Zhang TZ, Stelly DM, Chen ZJ . Epigenomic and functional analyses reveal roles of epialleles in the loss of photoperiod sensitivity during domestication of allotetraploid cottons. Genome Biol, 2017,18(1):99.
|
| [8] |
Wei XJ, Song XW, Wei LY, Tang SQ, Sun J, Hu PS, Cao XF . An epiallele of rice AK1 affects photosynthetic capacity. J Integr Plant Biol, 2017,59(3):158-163.
|
| [9] |
He L, Wu WW, Zinta G, Yang L, Wang D, Liu RY, Zhang HM, Zheng ZM, Huang H, Zhang QZ, Zhu JK . A naturally occurring epiallele associates with leaf senescence and local climate adaptation in Arabidopsis accessions. Nat Commun, 2018,9(1):460.
|
| [10] |
Kawakatsu T, Huang SSC, Jupe F, Sasaki E, Schmitz RJ, Urich MA, Castanon R, Nery JR, Barragan C, He YP, Chen HM, Dubin M, Lee CR, Wang CM, Bemm F, Becker C, O'Neil R, O'Malley RC, Quarless DX, Schork NJ, Weigel D, Nordborg M, Ecker JR, . Epigenomic diversity in a global collection of Arabidopsis thaliana Accessions. Cell, 2016,166(2):492-505.
|
| [11] |
Shirzadi R, Andersen ED, Bjerkan KN, Gloeckle BM, Heese M, Ungru A, Winge P, Koncz C, Aalen RB, Schnittger A, Grini PE . Genome-wide transcript profiling of endosperm without paternal contribution identifies parent-of-origin-dependent regulation of AGAMOUS- LIKE36. PLoS Genet, 2011,7(2):e1001303.
|
| [12] |
Walia H, Josefsson C, Dilkes B, Kirkbride R, Harada J, Comai L . Dosage-dependent deregulation of an AGAMOUS- LIKE gene cluster contributes to interspecific incompatibility. Curr Biol, 2009,19(13):1128-1132.
|
| [13] |
Ferrero-Serrano Á, Assmann SM . Phenotypic and genome- wide association with the local environment of Arabidopsis. Nat Ecol Evol, 2019,3(2):274-285.
|
| [14] |
Shabalin AA . Matrix eQTL: ultra fast eQTL analysis via large matrix operations. Bioinformatics, 2012,28(10):1353-1358.
|
| [15] |
Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T . Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res, 2003,13(11):2498-2504.
|
| [16] |
Xu FH, Xu BQ . Nonsense-mediated mRNA decay in eukaryotic cells. Chin J Cell Biol, 2008,30(1):55-58.
|
|
徐飞虎, 许宝青 . 真核细胞中无义介导的mRNA降解. 细胞生物学杂志, 2008,30(1):55-58.
|
| [17] |
Wachter A, Hartmann L . NMD: nonsense-mediated defense. Cell Host Microbe, 2014,16(3):273-275.
|
| [18] |
Huo ZG, Li MS, Wang L, Xiao JJ, Huang DP, Wang CY . Impacts of precipitation variations on crop diseases and pests in China. Sci Agric Sin, 2012,45(10):1935-1945.
|
|
霍治国, 李茂松, 王丽, 肖晶晶, 黄大鹏, 王春艳 . 降水变化对中国农作物病虫害的影响. 中国农业科学, 2012,45(10):1935-1945.
|
| [19] |
Zhang L, Huo ZG, Wang L, Jiang YY . Effects of climate change on the occurrence of crop insect pests in China, Chin J Ecol, 2012,31(06):1499-1507.
|
|
张蕾, 霍治国, 王丽, 姜玉英 . 气候变化对中国农作物虫害发生的影响. 生态学杂志, 2012,31(06):1499-1507.
|
| [20] |
Xia ST, Li X . Open a door of defenses: plant resistosome. Bull Bot, 2019,54(03):288-292.
|
|
夏石头, 李昕 . 开启防御之门: 植物抗病小体. 植物学报, 2019,54(03):288-292.
|
| [21] |
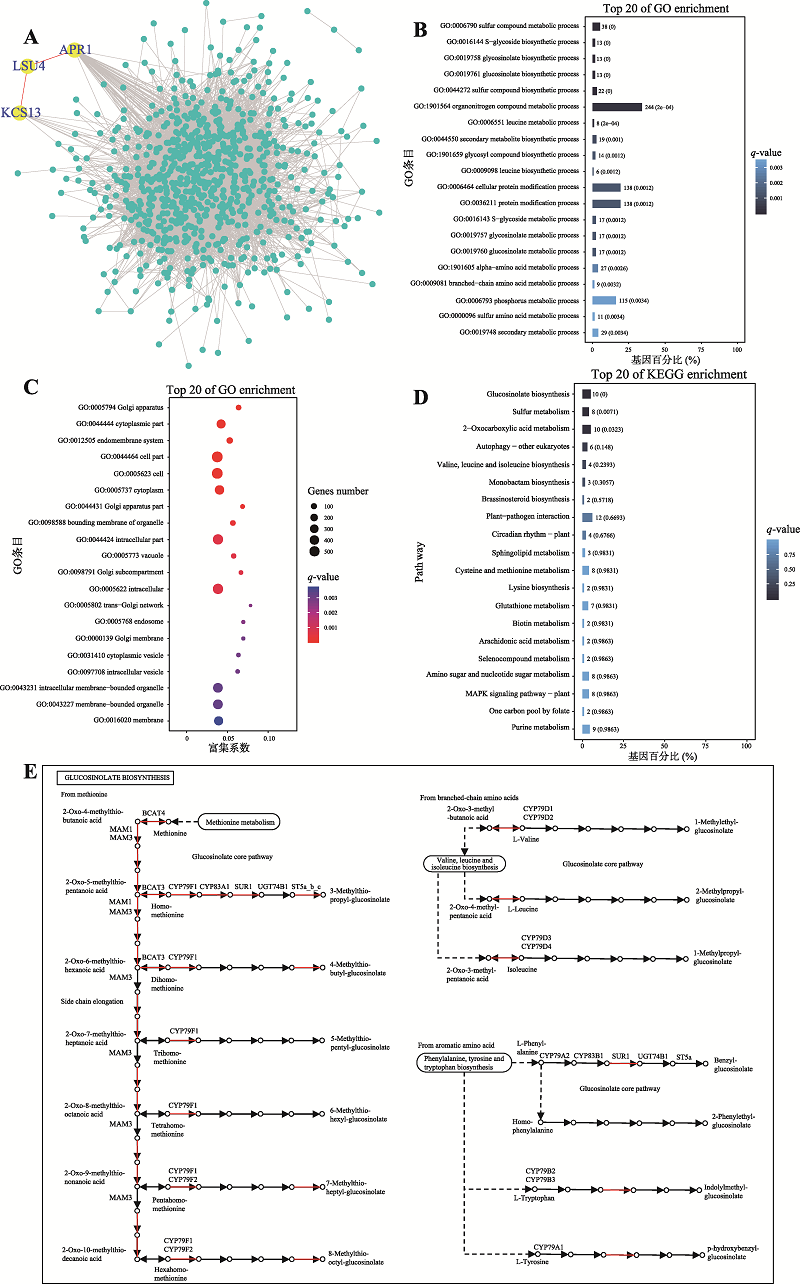
Sirko A, Wawrzyńska A, Rodríguez MC, Sęktas P . The family of LSU-like proteins. Front Plant Sci, 2015,5:774.
|
| [22] |
Zhang JL, Fu C . Response of cysteine protease and cysteine protease inhibitor to adverse stress. Mol Plant Bree, 2018,16(13):4453-4459.
|
|
张洁琳, 付畅 . 半胱氨酸蛋白酶及半胱氨酸蛋白酶抑制剂对逆境胁迫的应答. 分子植物育种, 2018,16(13):4453-4459.
|
| [23] |
Yan MJ, Song PL, Huangfu HY, Hao LF, Guo C, Huangfu JR, Jia XQ, Wu J, Shi ZD, Li ZQ . Research progress on the regulation and synthesis genes of the glucosinolate biosynthesis pathway in Brassica juncea. Inner Mongolia Agric Sci Technol, 2019,47(1):33-41.
|
|
燕孟娇, 宋培玲, 皇甫海燕, 郝丽芬, 郭晨, 皇甫九茹, 贾晓清, 吴晶, 史志丹, 李子钦 . 芥菜型油菜硫代葡萄糖苷基因的研究进展. 北方农业学报, 2019,47(1):33-41.
|
| [24] |
Sánchez-Pujante PJ, Borja-Martínez M, Pedreño MÁ, Almagro L . Biosynthesis and bioactivity of glucosinolates and their production in plant in vitro cultures. Planta, 2017,246(1):19-32.
|
| [25] |
Gallego-Bartolomé J, Gardiner J, Liu W, Papikian A, Ghoshal B, Kuo HY, Zhao JM, Segal DJ, Jacobsen SE . Targeted DNA demethylation of the Arabidopsis genome using the human TET1 catalytic domain. Proc Natl Acad Sci USA, 2018,115(9):E2125-E2134.
|
| [26] |
Johannes F, Schmitz RJ . Spontaneous epimutations in plants. New Phytol, 2019,221(3):1253-1259.
|
| [27] |
Verhoeven KJ, von Holdt BM, Sork VL . Epigenetics in ecology and evolution: what we know and what we need to know. Mol Ecol, 2016,25(8):1631-1638.
|
 )
)