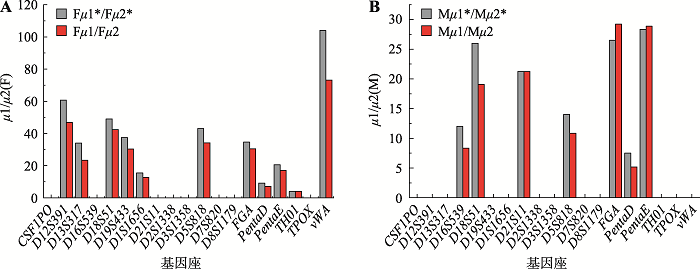
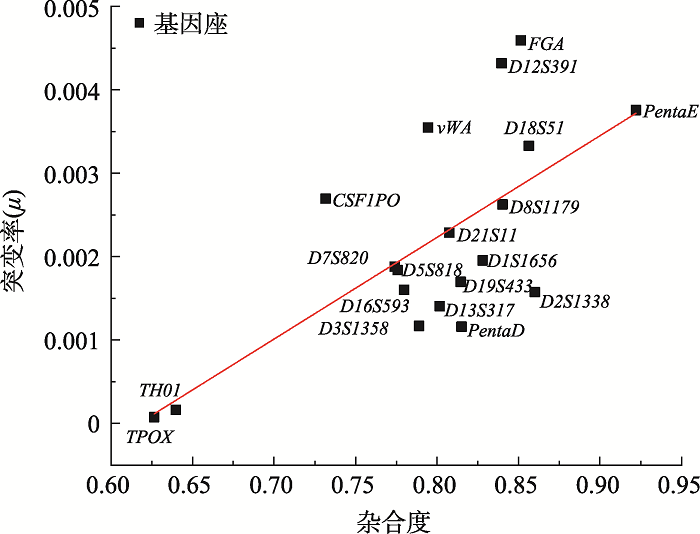
Hereditas(Beijing) ›› 2021, Vol. 43 ›› Issue (10): 949-961.doi: 10.16288/j.yczz.21-197
• Research Article • Previous Articles Next Articles
Actual mutational research of 19 autosomal STRs based on restricted mutation model and big data
Zhiyong Liu2, He Ren3, Chong Chen4, Jingjing Zhang5, Xiaomeng Zhang1, Yan Shi4, Linyu Shi1, Ying Chen4, Feng Cheng1, Li Jia4, Man Chen6, Qingwei Fan7, Jiarong Zhang1, Wanting Li1, Mengchun Wang1, Zilin Ren8, Yacheng Liu4, Ming Ni8, Hongyu Sun2, Jiangwei Yan1( )
)
- 1. School of Forensic Medicine, Shanxi Medical University, Taiyuan 030001, China
2. Faculty of Forensic Medicine, Zhongshan School of Medicine, Sun Yat-sen University, Guangzhou 510080, China
3. Beijing Police College, Beijing 102202, China
4. Beijing Tongda Shoucheng Institute of Forensic Science, Beijing 100192, China
5. Beijing Huayan Judicial Authentication Institute, Beijing 100192, China
6. School of Forensic Medicine, Southern Medical University, Guangzhou 510515, China
7. Faculty of Forensic Medicine, North Sichuan Medical College, Nanchong 637000, China
8. Beijing Institute of Radiation Medicine, Beijing 100850, China
-
Received:2021-06-02Revised:2021-08-18Online:2021-10-20Published:2021-08-31 -
Contact:Yan Jiangwei E-mail:yanjw@sxmu.edu.cn -
Supported by:Supported by the National Natural Science Foundation of China No(82030058)
Cite this article
Zhiyong Liu, He Ren, Chong Chen, Jingjing Zhang, Xiaomeng Zhang, Yan Shi, Linyu Shi, Ying Chen, Feng Cheng, Li Jia, Man Chen, Qingwei Fan, Jiarong Zhang, Wanting Li, Mengchun Wang, Zilin Ren, Yacheng Liu, Ming Ni, Hongyu Sun, Jiangwei Yan. Actual mutational research of 19 autosomal STRs based on restricted mutation model and big data[J]. Hereditas(Beijing), 2021, 43(10): 949-961.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
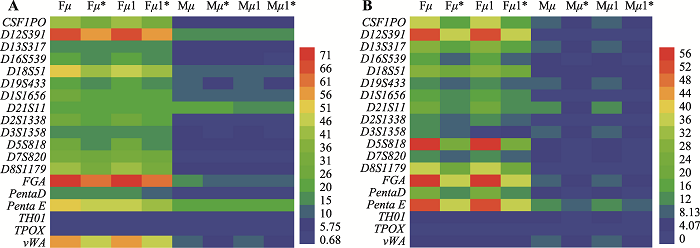
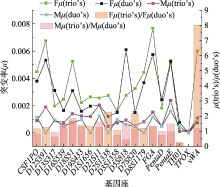
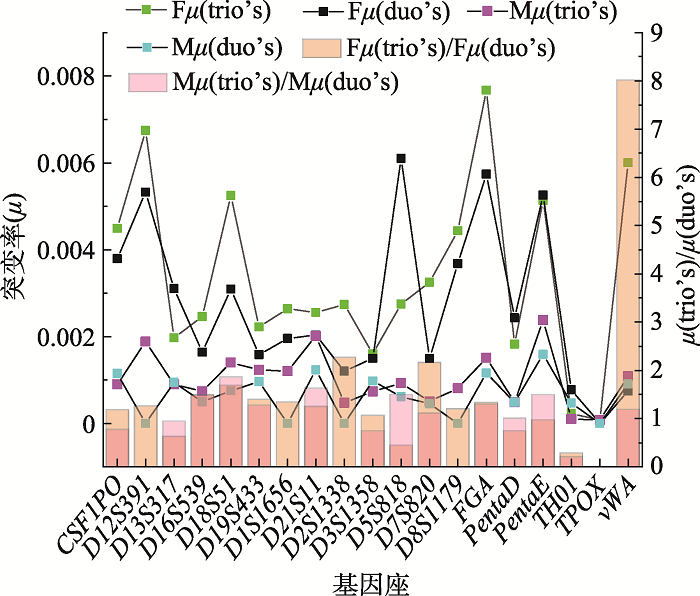
Table 1
The total STR loci-specific actual mutation rates in trio's and duo's"
| 基因座 | 三联体 | 二联体 | ||||
|---|---|---|---|---|---|---|
| Fμ | Mμ | 平均μ | Fμ | Mμ | 平均μ | |
| CSF1PO | 0.00449100 | 0.00089800 | 0.00269450 | 0.00379500 | 0.00115200 | 0.00247350 |
| D12S391 | 0.00674500 | 0.00188500 | 0.00431500 | 0.00532800 | 0.00000000 | 0.00266400 |
| D13S317 | 0.00197200 | 0.00089900 | 0.00143550 | 0.00310500 | 0.00094300 | 0.00202400 |
| D16S539 | 0.00246100 | 0.00073900 | 0.00160000 | 0.00164200 | 0.00049800 | 0.00107000 |
| D18S51 | 0.00524900 | 0.00140400 | 0.00332650 | 0.00309400 | 0.00075100 | 0.00192250 |
| D19S433 | 0.00222300 | 0.00123300 | 0.00172800 | 0.00158100 | 0.00096000 | 0.00127050 |
| D1S1656 | 0.00264300 | 0.00120200 | 0.00192250 | 0.00195900 | 0.00000000 | 0.00097950 |
| D21S11 | 0.00255700 | 0.00201239 | 0.00228470 | 0.00203300 | 0.00123400 | 0.00163350 |
| D2S1338 | 0.00273800 | 0.00047700 | 0.00160750 | 0.00120300 | 0.00000000 | 0.00060150 |
| D3S1358 | 0.00160539 | 0.00073400 | 0.00116970 | 0.00149600 | 0.00097800 | 0.00123700 |
| D5S818 | 0.00274700 | 0.00092600 | 0.00183650 | 0.00610000 | 0.00061700 | 0.00335850 |
| D7S820 | 0.00325000 | 0.00050900 | 0.00187950 | 0.00149800 | 0.00045500 | 0.00097650 |
| D8S1179 | 0.00443800 | 0.00081000 | 0.00262400 | 0.00368100 | 0.00000000 | 0.00184050 |
| FGA | 0.00767100 | 0.00151000 | 0.00459050 | 0.00573600 | 0.00116100 | 0.00344850 |
| Penta D | 0.00182500 | 0.00049400 | 0.00115950 | 0.00243100 | 0.00048900 | 0.00146000 |
| Penta E | 0.00513100 | 0.00237900 | 0.00375500 | 0.00525600 | 0.00158700 | 0.00342150 |
| TH01 | 0.00022611 | 0.00010000 | 0.00016306 | 0.00077600 | 0.00047100 | 0.00062350 |
| TPOX | 0.00007700 | 0.00007700 | 0.00007700 | 0.00000000 | 0.00000000 | 0.00000000 |
| vWA | 0.00600600 | 0.00108800 | 0.00354700 | 0.00074900 | 0.00090900 | 0.00082900 |
| Total | 0.06405550 | 0.01937639 | 0.04171595 | 0.05146300 | 0.01220500 | 0.03183400 |
Table 1 Supplementary
The number of mutations and cases in the 20 STR loci studied in Chinese Han Population"
| 基因座 | 三联体突变数 | 三联体 案例数 | 二联体突变数 | 二联体案例数 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 父源 | 母源 | 父源或母源 | 总计 | 父源 | 母源 | 总计 | 父亲孩子二联体 | 母亲孩子二联体 | ||
| CSF1PO | 65 | 7 | 15 | 87 | 22,113 | 4 | 2 | 6 | 2342 | 3858 |
| D12S391 | 113 | 24 | 21 | 158 | 20,562 | 8 | 0 | 8 | 2057 | 3406 |
| D13S317 | 30 | 11 | 10 | 51 | 22,113 | 4 | 2 | 6 | 2342 | 3858 |
| D16S539 | 38 | 8 | 10 | 56 | 22,113 | 2 | 1 | 3 | 2342 | 3858 |
| D18S51 | 96 | 22 | 10 | 128 | 22,113 | 5 | 2 | 7 | 2342 | 3858 |
| D19S433 | 36 | 19 | 5 | 60 | 22,113 | 2 | 2 | 4 | 2342 | 3858 |
| D1S1656 | 29 | 11 | 8 | 48 | 13,712 | 2 | 0 | 2 | 1361 | 2149 |
| D21S11 | 44 | 41 | 7 | 92 | 22,113 | 3 | 3 | 6 | 2342 | 3858 |
| D2S1338 | 50 | 5 | 9 | 64 | 22,113 | 2 | 0 | 2 | 2342 | 3858 |
| D3S1358 | 29 | 6 | 13 | 48 | 22,113 | 2 | 2 | 4 | 2342 | 3858 |
| D6S1043 | 54 | 8 | 5 | 67 | 20,562 | 0 | 2 | 2 | 2057 | 3406 |
| D5S818 | 40 | 11 | 8 | 59 | 22,113 | 6 | 1 | 7 | 2342 | 3858 |
| D7S820 | 51 | 3 | 13 | 67 | 22,113 | 2 | 1 | 3 | 2342 | 3858 |
| D8S1179 | 75 | 10 | 9 | 94 | 22,113 | 5 | 0 | 5 | 2342 | 3858 |
| FGA | 135 | 18 | 19 | 172 | 22,113 | 9 | 3 | 12 | 2342 | 3858 |
| Penta D | 30 | 7 | 3 | 40 | 20,562 | 3 | 1 | 4 | 2057 | 3406 |
| Penta E | 92 | 40 | 10 | 142 | 20,562 | 8 | 4 | 12 | 2057 | 3406 |
| TH01 | 4 | 1 | 2 | 7 | 22,113 | 1 | 1 | 2 | 2342 | 3858 |
| TPOX | 1 | 1 | 1 | 3 | 22,113 | 0 | 0 | 0 | 2342 | 3858 |
| vWA | 97 | 11 | 16 | 124 | 22,113 | 1 | 2 | 3 | 2342 | 3858 |
| Total | 1109 | 264 | 194 | 1567 | 22,113 | 69 | 29 | 98 | 2342 | 3858 |
Table 2 Supplementary
The detail mutation rate calculation example of the TH01 locus in trio's"
| 基因座 | 孩子 | 父亲 | 母亲 | 突变 来源 | 突变 步数 | F1” | F2” | F3” | M1” | M2” | M3” |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 基因型 | |||||||||||
| TH01 | 8, 9 | 9 | 9 | 父亲 | 1 | 0.5 | 0 | 0 | 0.5 | 0 | 0 |
| 母亲 | |||||||||||
| TH01 | 8,9 | 9 | 9 | 父亲 | 1 | 0.5 | 0 | 0 | 0.5 | 0 | 0 |
| 母亲 | |||||||||||
| TH01 | 9 | 8 | 6, 9 | 父亲 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| TH01 | 6, 11 | 9, 10 | 6, 9 | 父亲 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| TH01 | 9 | 7 | 9 | 父亲 | 2 | 0 | 1 | 0 | 0 | 0 | 0 |
| TH01 | 8 | 6,9 | 8, 9 | 父亲 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| TH01 | 7, 10 | 7 | 9 | 母亲 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| 总计(n) | - | 4 | 1 | 0 | 2 | 0 | 0 | ||||
| 突变” | 0.000181 | 0.000045 | 0 | 0.00009 | 0 | 0 | |||||
Table 3 Supplementary
The STR mutated description of monozygotic twins from 10 cases"
| 案例 | 孩子 | 试剂盒 | 父亲年龄 | 母亲年龄 | 孩子性别 | 突变基因座 | 孩子分型 | 父亲分型 | 母亲分型 | 突变来源 | 突变步数 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 案例1 | MZ1 | AB-Identifiler, AGCU21+1 | 30 | 30 | 男 | FGA | 22,24 | 21,23 | 22,24 | 未知 | ±1 |
| MZ2 | 30 | 30 | 男 | FGA | 22,24 | 21,23 | 22,24 | 未知 | ±1 | ||
| 案例2 | MZ1 | Goldeneye 20A, AGCU21+1 | 33 | 28 | 男 | D6S1043 | 13, 20 | 13, 18 | 18, 21 | 母亲 | -1 |
| MZ2 | 33 | 28 | 男 | D6S1043 | 13, 20 | 13, 18 | 18, 21 | 母亲 | -1 | ||
| 案例3 | MZ1 | Goldeneye 20A | 21 | 23 | 女 | Penta E | 15,25 | 17,24 | 12,15 | 父亲 | +1 |
| MZ2 | 21 | 23 | 女 | Penta E | 15,25 | 17,24 | 12,15 | 父亲 | +1 | ||
| 案例4 | MZ1 | Goldeneye 20A | 26 | 24 | 男 | D18S51 | 12,16 | 14,15 | 12,15 | 父亲 | +1 |
| MZ2 | 26 | 24 | 男 | D18S51 | 12,16 | 14,15 | 12,15 | 父亲 | +1 | ||
| 案例5 | MZ1 | Goldeneye 20A, AGCU21+1 | 40 | 39 | 女 | D21S11 | 30,33.2 | 31,32.2 | 30,31.2 | 父亲 | +1 |
| MZ2 | 40 | 39 | 女 | D21S11 | 30,33.2 | 31,32.2 | 30,31.2 | 父亲 | +1 | ||
| 案例6 | MZ1 | Goldeneye 20A | 44 | 40 | 男 | D6S1043 | 18,21.3 | 12,22.3 | 11,18 | 父亲 | -1 |
| MZ2 | 44 | 40 | 男 | D6S1043 | 18,21.3 | 12,22.3 | 11,18 | 父亲 | -1 | ||
| 案例7 | MZ1 | PowerPlex®21 | 32 | 31 | 女 | Penta E | 16,17 | 16 | 16,18 | 未知 | ±1 |
| MZ2 | 32 | 31 | 女 | Penta E | 16,17 | 16 | 16,18 | 未知 | ±1 | ||
| 案例8 | MZ1 | PowerPlex®21 | 42 | 24 | 男 | D12S391 | 17,22 | 18,22 | 18,19 | 母亲 | -1 |
| MZ2 | 42 | 24 | 男 | D12S391 | 17,22 | 18,22 | 18,19 | 母亲 | -1 | ||
| 案例9 | MZ1 | PowerPlex®21 | 38 | 32 | 男 | CSF1PO | 12,14 | 10,13 | 12 | 父亲 | +1 |
| MZ2 | 38 | 32 | 男 | CSF1PO | 12,14 | 10,13 | 12 | 父亲 | +1 | ||
| 案例10 | MZ1 | PowerPlex®21 | 31 | 31 | 男 | D18S51 | 15,17 | 14,16 | 14,15 | 父亲 | +1 |
| MZ2 | 31 | 31 | 男 | D18S51 | 15,17 | 14,16 | 14,15 | 父亲 | +1 |
Table 4A Supplementary
The 27 cases of 2 mutation events observed in one child"
| 案例 | 试剂盒 | 父亲年龄 | 母亲年龄 | 孩子性别 | 突变基因座 | 孩子分型 | 父亲分型 | 母亲分型 | 突变来源 | 突变步数 |
|---|---|---|---|---|---|---|---|---|---|---|
| 案例1 | AB-Identifiler | 24 | 22 | 女 | D3S1358 | 15 | 15,16 | 16 | 母亲 | -1 |
| D19S433 | 12,14.2 | 13,14 | 14,14.2 | 父亲 | -1 | |||||
| 案例2 | Goldeneye 20A | 25 | 22 | 女 | D18S51 | 16 | 13,15 | 13,16 | 父亲 | +1 |
| D12S391 | 18,21 | 18,20 | 18,20 | 未知 | +1 | |||||
| 案例3 | Goldeneye 20A | 31 | 29 | 男 | D7S820 | 9, 12 | 11 | 9, 10 | 父亲 | +1 |
| CSF1PO | 12, 13 | 10, 13 | 13 | 父亲 | -1 | |||||
| 案例4 | Goldeneye 20A | 26 | 27 | 男 | CSF1PO | 13,14 | 12,14 | 9,14 | 未知 | ±1 |
| FGA | 22,24 | 23,26 | 24,24.2 | 父亲 | -1 | |||||
| 案例5 | Goldeneye 20A | 55 | 25 | 女 | D3S1358 | 16,18 | 16,17 | 15,16 | 父亲 | +1 |
| D12S391 | 19,20 | 18,21 | 19 | 父亲 | -1 | |||||
| 案例6 | Goldeneye 20A | 26 | 22 | 女 | D6S1043 | 12 | 11,13 | 12,13 | 父亲 | ±1 |
| D8S1179 | 14 | 14,15 | 13,15 | 母亲 | ±1 | |||||
| 案例7 | Goldeneye 20A | 37 | 38 | 女 | D12S391 | 20,23 | 21 | 18,23 | 父亲 | -1 |
| FGA | 22,23 | 22,24 | 22,25 | 父亲 | ±1 | |||||
| 案例8 | Goldeneye 20A | 24 | 19 | 男 | D5S818 | 11,14 | 10,14 | 10,12 | 母亲 | ±1 |
| D6S1043 | 14,17 | 13,19 | 17,18 | 父亲 | +1 | |||||
| 案例9 | PowerPlex®21 | 30 | 27 | 女 | D21S11 | 30,31 | 31,31.2 | 31,33 | 未知 | -1 |
| D2S1338 | 23,24 | 18,24 | 19,24 | 未知 | -1 | |||||
| 案例10 | PowerPlex®21 | 35 | 23 | 女 | D21S11 | 30,32.2 | 29 | 31.2,32.2 | 父亲 | +1 |
| D12S391 | 18,22 | 20,21 | 18,19 | 父亲 | +1 | |||||
| 案例11 | PowerPlex®21 | 40 | 34 | 男 | vWA | 17,18 | 19 | 14,17 | 父亲 | -1 |
| FGA | 22,24 | 21,23 | 21,24 | 父亲 | ±1 | |||||
| 案例12 | PowerPlex®21 | 33 | NA | 女 | penta E | 10,22 | 16,23 | NA | 父亲 | -1 |
| D5S818 | 13 | 11,12 | NA | 父亲 | +1 | |||||
| 案例13 | PowerPlex®21 | 29 | 25 | 女 | D8S1179 | 12 | 11,13 | 12,13 | 父亲 | ±1 |
| FGA | 23,24 | 22.2,25 | 20,23 | 父亲 | -1 | |||||
| 案例14 | PowerPlex®21 | 37 | 28 | 女 | D18S51 | 13,21 | 13,15 | 19,22 | 母亲 | -1 |
| vWA | 18 | 16,19 | 18 | 父亲 | -1 | |||||
| 案例15 | PowerPlex®21 | 39 | NA | 女 | Penta E | 16,17 | 10,18 | NA | 父亲 | -1 |
| D18S51 | 14,19 | 18,20 | NA | 父亲 | ±1 | |||||
| 案例16 | PowerPlex®21 | 24 | 25 | 女 | D13S317 | 9,11 | 8,10 | 9,10 | 父亲 | +1 |
| Penta E | 12,15 | 13,16 | 16 | 母亲 | +1 | |||||
| 案例17 | PowerPlex®21 | 30 | 32 | 男 | D18S51 | 14 | 14 | 13,17 | 母亲 | +1 |
| FGA | 23,25 | 21,23 | 18,22 | 父亲 | -1 | |||||
| 案例18 | PowerPlex®21 | 24 | 21 | 女 | TPOX | 8,11 | 8,10 | 8 | 父亲 | +1 |
| D8S1179 | 13,15 | 15,17 | 15,18 | 父亲 | -2 | |||||
| 案例19 | PowerPlex®21 | 28 | 28 | 女 | D6S1043 | 14,17 | 17 | 13,18 | 母亲 | +1 |
| D16S539 | 11,14 | 10,11 | 9,12 | 父亲 | +1 | |||||
| 案例20 | PowerPlex®21 | 37 | 33 | 男 | Penta E | 11,27 | 13,26 | 11,20 | 父亲 | +1 |
| Penta D | 10 | 9 | 10,13 | 父亲 | +1 | |||||
| 案例21 | PowerPlex®21 | 28 | 25 | 女 | D13S317 | 10,12 | 10,11 | 8,10 | 父亲 | +1 |
| D21S11 | 30,31 | 30,31 | 29,32 | 母亲 | +1 | |||||
| 案例22 | PowerPlex®21 | 26 | 23 | 女 | D1S1656 | 10,15 | 15 | 11 | 母亲 | -1 |
| D13S317 | 18,26 | 19,27 | 18,19 | 父亲 | -1 | |||||
| 案例23 | PowerPlex®21 | 39 | 28 | 女 | D16S539 | 10,12 | 9,13 | 12 | 父亲 | +1 |
| D21S11 | 30.2,31.2 | 28,30.2 | 29,32.2 | 母亲 | -1 | |||||
| 案例24 | PowerPlex®21 | 45 | 41 | 女 | D16S539 | 11 | 9,12 | 11,12 | 父亲 | -1 |
| D8S1179 | 13,18 | 13,17 | 12,13 | 父亲 | +1 | |||||
| 案例25 | PowerPlex®21 | 28 | 29 | 女 | Penta D | 11,14 | 8,9 | 12,14 | 父亲 | +2 |
| D21S11 | 30,31.2 | 29,30 | 30 | 未知 | +1.2 | |||||
| 案例26 | PowerPlex®21 | 30 | 28 | 女 | D13S317 | 9 | 9 | 8 | 母亲 | +1 |
| D5S818 | 12,14 | 10,13 | 9,12 | 父亲 | +1 | |||||
| 案例27 | PowerPlex®21 | 40 | 33 | 男 | D1S1656 | 14,16 | 15,17.3 | 12,14 | 父亲 | +1 |
| D13S317 | 8,13 | 9,12 | 8,12 | 父亲 | +1 |
Table 4B Supplementary
The 3 cases of 3 mutation events observed in one child"
| 案例 | 试剂盒 | 父亲年龄 | 母亲年龄 | 孩子性别 | 突变基因座 | 孩子分型 | 父亲分型 | 母亲分型 | 突变来源 | 突变步数 |
|---|---|---|---|---|---|---|---|---|---|---|
| 案例1 | Goldeneye 20A, MicroreaderTM23sp ID Sysrem | 47 | 39 | 男 | D8S1179 | 12,14 | 13 | 14 | 父亲 | -1 |
| D17S1290 | 15,19 | 15,20 | 15,18 | 未知 | ±1 | |||||
| FGA | 22,23 | 24 | 22,24 | 父亲 | -1 | |||||
| 案例2 | PowerPlex®21, MicroreaderTM23sp ID Sysrem | 39 | 26 | 男 | D6S1043 | 12,17 | 17,20 | 13,14 | 母亲 | -1 |
| D8S1132 | 18,21 | 18,19 | 22 | -1 | ||||||
| FGA | 19,24 | 20,25 | 19,25 | 父亲 | -1 | |||||
| 案例3 | PowerPlex®21, MicroreaderTM23sp ID Sysrem | 34 | 33 | 男 | D2S1338 | 19,21 | 19 | 19,20 | 母亲 | +1 |
| D10S1435 | 13 | 13 | 12,14 | ±1 | ||||||
| FGA | 21,25 | 22,24 | 21,23 | 父亲 | +1 |
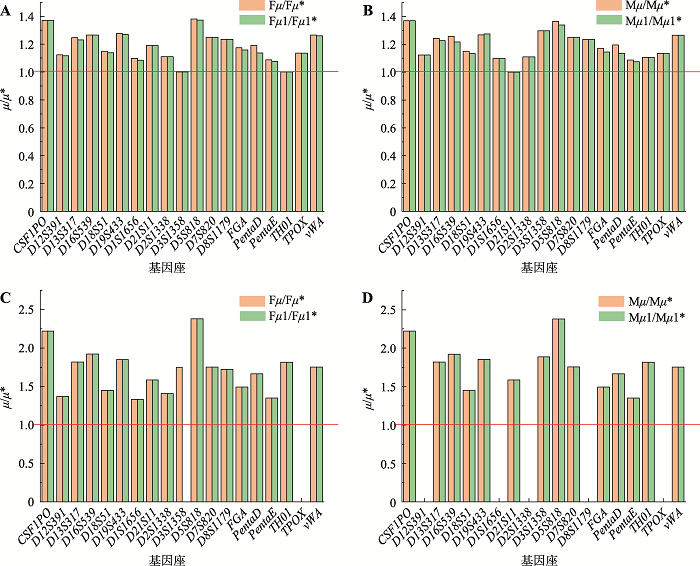
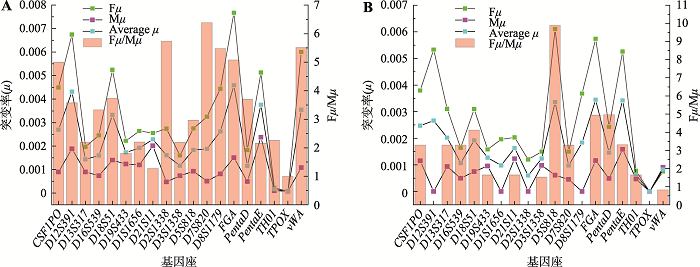
Table 5A Supplementary
The apparent mutation rates in trio's"
| 基因座 | Fμ” | Fμ1” | Fμ2” | Fμ3” | Mμ” | Mμ1” | Mμ2” | Mμ3” |
|---|---|---|---|---|---|---|---|---|
| CSF1PO | 0.00327861 | 0.00327861 | 0.00000000 | 0.00000000 | 0.00065572 | 0.00065572 | 0.00000000 | 0.00000000 |
| D12S391 | 0.00600623 | 0.00590896 | 0.00009727 | 0.00000000 | 0.00167785 | 0.00167785 | 0.00000000 | 0.00000000 |
| D13S317 | 0.00158278 | 0.00153756 | 0.00004522 | 0.00000000 | 0.00072356 | 0.00067833 | 0.00000000 | 0.00004522 |
| D16S539 | 0.00194456 | 0.00194456 | 0.00000000 | 0.00000000 | 0.00058789 | 0.00054267 | 0.00004522 | 0.00000000 |
| D18S51 | 0.00456745 | 0.00443178 | 0.00009044 | 0.00004522 | 0.00122100 | 0.00117578 | 0.00004522 | 0.00000000 |
| D19S433 | 0.00174106 | 0.00169584 | 0.00004522 | 0.00000000 | 0.00097228 | 0.00092706 | 0.00000000 | 0.00004522 |
| D1S1656 | 0.00240665 | 0.00226079 | 0.00014586 | 0.00000000 | 0.00109393 | 0.00109393 | 0.00000000 | 0.00000000 |
| D21S11 | 0.00214806 | 0.00214806 | 0.00000000 | 0.00000000 | 0.00201239 | 0.00192195 | 0.00009044 | 0.00000000 |
| D2S1338 | 0.00246461 | 0.00246461 | 0.00000000 | 0.00000000 | 0.00042961 | 0.00042961 | 0.00000000 | 0.00000000 |
| D3S1358 | 0.00160539 | 0.00160539 | 0.00000000 | 0.00000000 | 0.00056528 | 0.00056528 | 0.00000000 | 0.00000000 |
| D5S818 | 0.00198978 | 0.00194456 | 0.00004522 | 0.00000000 | 0.00067833 | 0.00063311 | 0.00004522 | 0.00000000 |
| D7S820 | 0.00260028 | 0.00260028 | 0.00000000 | 0.00000000 | 0.00040700 | 0.00040700 | 0.00000000 | 0.00000000 |
| D8S1179 | 0.00359517 | 0.00359517 | 0.00000000 | 0.00000000 | 0.00065572 | 0.00065572 | 0.00000000 | 0.00000000 |
| FGA | 0.00653462 | 0.00626328 | 0.00018089 | 0.00009044 | 0.00128883 | 0.00119839 | 0.00004522 | 0.00004522 |
| Penta D | 0.00153195 | 0.00133742 | 0.00014590 | 0.00004863 | 0.00041338 | 0.00036475 | 0.00004863 | 0.00000000 |
| Penta E | 0.00471744 | 0.00449859 | 0.00021885 | 0.00000000 | 0.00218850 | 0.00206692 | 0.00007295 | 0.00004863 |
| TH01 | 0.00022611 | 0.00018089 | 0.00004522 | 0.00000000 | 0.00009044 | 0.00009044 | 0.00000000 | 0.00000000 |
| TPOX | 0.00006783 | 0.00006783 | 0.00000000 | 0.00000000 | 0.00006783 | 0.00006783 | 0.00000000 | 0.00000000 |
| vWA | 0.00474834 | 0.00470312 | 0.00004522 | 0.00000000 | 0.00085922 | 0.00085922 | 0.00000000 | 0.00000000 |
Table 5B Supplementary
The actual mutation rates in trio's"
| 基因座 | Fμ | Fμ1 | Fμ2 | Fμ3 | Mμ | Mμ1 | Mμ2 | Mμ3 |
|---|---|---|---|---|---|---|---|---|
| CSF1PO | 0.00449100 | 0.00449100 | 0.00000000 | 0.00000000 | 0.00089800 | 0.00089800 | 0.00000000 | 0.00000000 |
| D12S391 | 0.00674500 | 0.00660400 | 0.00014100 | 0.00000000 | 0.00188500 | 0.00188500 | 0.00000000 | 0.00000000 |
| D13S317 | 0.00197200 | 0.00189100 | 0.00008100 | 0.00000000 | 0.00089900 | 0.00083200 | -0.00004300 | 0.00011000 |
| D16S539 | 0.00246100 | 0.00246100 | 0.00000000 | 0.00000000 | 0.00073900 | 0.00066000 | 0.00007900 | 0.00000000 |
| D18S51 | 0.00524900 | 0.00504600 | 0.00011900 | 0.00008400 | 0.00140400 | 0.00133400 | 0.00007000 | 0.00000000 |
| D19S433 | 0.00222300 | 0.00215200 | 0.00007100 | 0.00000000 | 0.00123300 | 0.00118200 | -0.00002400 | 0.00007500 |
| D1S1656 | 0.00264300 | 0.00245100 | 0.00019200 | 0.00000000 | 0.00120200 | 0.00120200 | 0.00000000 | 0.00000000 |
| D21S11 | 0.00255700 | 0.00255700 | 0.00000000 | 0.00000000 | 0.00201239 | 0.00192195 | 0.00009044 | 0.00000000 |
| D2S1338 | 0.00273800 | 0.00273800 | 0.00000000 | 0.00000000 | 0.00047700 | 0.00047700 | 0.00000000 | 0.00000000 |
| D3S1358 | 0.00160539 | 0.00160539 | 0.00000000 | 0.00000000 | 0.00073400 | 0.00073400 | 0.00000000 | 0.00000000 |
| D5S818 | 0.00274700 | 0.00266900 | 0.00007800 | 0.00000000 | 0.00092600 | 0.00084800 | 0.00007800 | 0.00000000 |
| D7S820 | 0.00325000 | 0.00325000 | 0.00000000 | 0.00000000 | 0.00050900 | 0.00050900 | 0.00000000 | 0.00000000 |
| D8S1179 | 0.00443800 | 0.00443800 | 0.00000000 | 0.00000000 | 0.00081000 | 0.00081000 | 0.00000000 | 0.00000000 |
| FGA | 0.00767100 | 0.00725200 | 0.00023800 | 0.00018100 | 0.00151000 | 0.00137300 | 0.00004700 | 0.00009000 |
| Penta D | 0.00182500 | 0.00152100 | 0.00021400 | 0.00009000 | 0.00049400 | 0.00041400 | 0.00008000 | 0.00000000 |
| Penta E | 0.00513100 | 0.00484700 | 0.00028400 | 0.00000000 | 0.00237900 | 0.00222200 | 0.00007700 | 0.00008000 |
| TH01 | 0.00022611 | 0.00018089 | 0.00004522 | 0.00000000 | 0.00010000 | 0.00010000 | 0.00000000 | 0.00000000 |
| TPOX | 0.00007700 | 0.00007700 | 0.00000000 | 0.00000000 | 0.00007700 | 0.00007700 | 0.00000000 | 0.00000000 |
| vWA | 0.00600600 | 0.00592500 | 0.00008100 | 0.00000000 | 0.00108800 | 0.00108800 | 0.00000000 | 0.00000000 |
Table 6A Supplementary
The apparent mutation rates in duo's"
| 基因座 | Fμ” | Fμ1” | Fμ2” | Fμ3” | Mμ” | Mμ1” | Mμ2” | Mμ3” |
|---|---|---|---|---|---|---|---|---|
| CSF1PO | 0.00170794 | 0.00170794 | 0.00000000 | 0.00000000 | 0.00051840 | 0.00051840 | 0.00000000 | 0.00000000 |
| D12S391 | 0.00388916 | 0.00388916 | 0.00000000 | 0.00000000 | 0.00000000 | 0.00000000 | 0.00000000 | 0.00000000 |
| D13S317 | 0.00170794 | 0.00170794 | 0.00000000 | 0.00000000 | 0.00051840 | 0.00051840 | 0.00000000 | 0.00000000 |
| D16S539 | 0.00085397 | 0.00085397 | 0.00000000 | 0.00000000 | 0.00025920 | 0.00025920 | 0.00000000 | 0.00000000 |
| D18S51 | 0.00213493 | 0.00213493 | 0.00000000 | 0.00000000 | 0.00051840 | 0.00051840 | 0.00000000 | 0.00000000 |
| D19S433 | 0.00085397 | 0.00085397 | 0.00000000 | 0.00000000 | 0.00051840 | 0.00051840 | 0.00000000 | 0.00000000 |
| D1S1656 | 0.00146951 | 0.00146951 | 0.00000000 | 0.00000000 | 0.00000000 | 0.00000000 | 0.00000000 | 0.00000000 |
| D21S11 | 0.00128096 | 0.00128096 | 0.00000000 | 0.00000000 | 0.00077760 | 0.00077760 | 0.00000000 | 0.00000000 |
| D2S1338 | 0.00085397 | 0.00085397 | 0.00000000 | 0.00000000 | 0.00000000 | 0.00000000 | 0.00000000 | 0.00000000 |
| D3S1358 | 0.00085397 | 0.00042699 | 0.00042699 | 0.00000000 | 0.00051840 | 0.00051840 | 0.00000000 | 0.00000000 |
| D5S818 | 0.00256191 | 0.00256191 | 0.00000000 | 0.00000000 | 0.00025920 | 0.00025920 | 0.00000000 | 0.00000000 |
| D7S820 | 0.00085397 | 0.00085397 | 0.00000000 | 0.00000000 | 0.00025920 | 0.00025920 | 0.00000000 | 0.00000000 |
| D8S1179 | 0.00213493 | 0.00213493 | 0.00000000 | 0.00000000 | 0.00000000 | 0.00000000 | 0.00000000 | 0.00000000 |
| FGA | 0.00384287 | 0.00384287 | 0.00000000 | 0.00000000 | 0.00077760 | 0.00077760 | 0.00000000 | 0.00000000 |
| Penta D | 0.00145843 | 0.00145843 | 0.00000000 | 0.00000000 | 0.00029360 | 0.00029360 | 0.00000000 | 0.00000000 |
| Penta E | 0.00388916 | 0.00388916 | 0.00000000 | 0.00000000 | 0.00117440 | 0.00117440 | 0.00000000 | 0.00000000 |
| TH01 | 0.00042699 | 0.00042699 | 0.00000000 | 0.00000000 | 0.00025920 | 0.00025920 | 0.00000000 | 0.00000000 |
| TPOX | 0.00000000 | 0.00000000 | 0.00000000 | 0.00000000 | 0.00000000 | 0.00000000 | 0.00000000 | 0.00000000 |
| vWA | 0.00042699 | 0.00042699 | 0.00000000 | 0.00000000 | 0.00051840 | 0.00051840 | 0.00000000 | 0.00000000 |
Table 6B Supplementary
The actual mutation rates in duo's"
| 基因座 | Fμ | Fμ1 | Fμ2 | Fμ3 | Mμ | Mμ1 | Mμ2 | Mμ3 |
|---|---|---|---|---|---|---|---|---|
| CSF1PO | 0.00379500 | 0.00379500 | 0.00000000 | 0.00000000 | 0.00115200 | 0.00115200 | 0.00000000 | 0.00000000 |
| D12S391 | 0.00532800 | 0.00532800 | 0.00000000 | 0.00000000 | 0.00000000 | 0.00000000 | 0.00000000 | 0.00000000 |
| D13S317 | 0.00310500 | 0.00310500 | 0.00000000 | 0.00000000 | 0.00094300 | 0.00094300 | 0.00000000 | 0.00000000 |
| D16S539 | 0.00164200 | 0.00164200 | 0.00000000 | 0.00000000 | 0.00049800 | 0.00049800 | 0.00000000 | 0.00000000 |
| D18S51 | 0.00309400 | 0.00309400 | 0.00000000 | 0.00000000 | 0.00075100 | 0.00075100 | 0.00000000 | 0.00000000 |
| D19S433 | 0.00158100 | 0.00158100 | 0.00000000 | 0.00000000 | 0.00096000 | 0.00096000 | 0.00000000 | 0.00000000 |
| D1S1656 | 0.00195900 | 0.00195900 | 0.00000000 | 0.00000000 | 0.00000000 | 0.00000000 | 0.00000000 | 0.00000000 |
| D21S11 | 0.00203300 | 0.00203300 | 0.00000000 | 0.00000000 | 0.00123400 | 0.00123400 | 0.00000000 | 0.00000000 |
| D2S1338 | 0.00120300 | 0.00120300 | 0.00000000 | 0.00000000 | 0.00000000 | 0.00000000 | 0.00000000 | 0.00000000 |
| D3S1358 | 0.00149600 | -0.00155400 | 0.00305000 | 0.00000000 | 0.00097800 | 0.00097800 | 0.00000000 | 0.00000000 |
| D5S818 | 0.00610000 | 0.00610000 | 0.00000000 | 0.00000000 | 0.00061700 | 0.00061700 | 0.00000000 | 0.00000000 |
| D7S820 | 0.00149800 | 0.00149800 | 0.00000000 | 0.00000000 | 0.00045500 | 0.00045500 | 0.00000000 | 0.00000000 |
| D8S1179 | 0.00368100 | 0.00368100 | 0.00000000 | 0.00000000 | 0.00000000 | 0.00000000 | 0.00000000 | 0.00000000 |
| FGA | 0.00573600 | 0.00573600 | 0.00000000 | 0.00000000 | 0.00116100 | 0.00116100 | 0.00000000 | 0.00000000 |
| Penta D | 0.00243100 | 0.00243100 | 0.00000000 | 0.00000000 | 0.00048900 | 0.00048900 | 0.00000000 | 0.00000000 |
| Penta E | 0.00525600 | 0.00525600 | 0.00000000 | 0.00000000 | 0.00158700 | 0.00158700 | 0.00000000 | 0.00000000 |
| TH01 | 0.00077600 | 0.00077600 | 0.00000000 | 0.00000000 | 0.00047100 | 0.00047100 | 0.00000000 | 0.00000000 |
| TPOX | 0.00000000 | 0.00000000 | 0.00000000 | 0.00000000 | 0.00000000 | 0.00000000 | 0.00000000 | 0.00000000 |
| vWA | 0.00074900 | 0.00074900 | 0.00000000 | 0.00000000 | 0.00090900 | 0.00090900 | 0.00000000 | 0.00000000 |
Table 7 Supplementary
The total nonintegral mutations events"
| 基因座 | 基因型 | 突变来源 | 可能的突变步数 | ||
|---|---|---|---|---|---|
| 孩子 | 父亲 | 母亲 | |||
| D19S433 | 13 | 14.2 | 13, 15.2 | 父亲 | -1.2 |
| D19S433 | 13, 16 | 15, 16.2 | 13, 14 | -0.2 | |
| D19S433 | 13 | 15.2 | - | -2.2 | |
| D1S1656 | 16 | 16.3, 17 | 15, 16 | -0.3 | |
| D6S1043 | 17, 23.3 | 18, 24 | 11, 17 | -0.1 | |
| D21S11 | 30, 32 | 29, 30.2 | 32, 32.2 | -0.2 | |
| TH01 | 9 | 6, 9 | 9.3 | 母亲 | -0.3 |
| D21S11 | 30, 31.2 | 29, 30 | 30 | 未知 | +1.2 |
| D12S391 | 19.3, 20 | 19, 20 | 20, 21 | +0.3/-0.1 | |
Table 8 Supplementary
The total mutation events of allele mutation gain and loss"
| 基因座 | 较短组 Short group | 中等组 Medium group | 较长组 Long group | 未确定分组 Undeterminable group | Total | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AR | NG | NL | NU | UM | AR | NG | NL | NU | UM | AR | NG | NL | NU | UM | |||
| CSF1PO | ?7-9.1 | 1 | 0 | 0 | 1 | ?9.2-11.1 | 6 | 4 | 0 | 10 | ?12-15 | 31 | 28 | 3 | 62 | 20 | 93 |
| D12S391 | ?14-18.3 | 8 | 11 | 1 | 20 | ?19-23 | 36 | 64 | 8 | 108 | ?24-28 | 8 | 7 | 0 | 15 | 23 | 166 |
| D13S317 | ?5-7 | 0 | 0 | 0 | 0 | ?8-12 | 22 | 11 | 2 | 35 | ?13-15 | 5 | 12 | 0 | 17 | 5 | 57 |
| D16S539 | ?6-8 | 0 | 0 | 0 | 0 | ?9-12 | 21 | 10 | 4 | 35 | ?13-16 | 6 | 11 | 1 | 18 | 6 | 59 |
| D18S51 | ?10-13 | 9 | 0 | 0 | 9 | ?13.2-20 | 54 | 32 | 5 | 91 | ?21-25 | 15 | 18 | 0 | 33 | 2 | 135 |
| D19S433 | ?9-11.2 | 2 | 0 | 0 | 2 | ?12-16 | 24 | 25 | 2 | 51 | ?16.2-18.2 | 3 | 8 | 0 | 11 | 0 | 64 |
| D1S1656 | ?10-12 | 0 | 1 | 0 | 1 | ?13-17.3 | 20 | 20 | 2 | 42 | ?18-19.3 | 1 | 4 | 0 | 5 | 2 | 50 |
| D21S11 | ?27-29 | 8 | 7 | 0 | 15 | ?29.2-33.1 | 26 | 31 | 3 | 60 | ?33.2-35.2 | 2 | 12 | 0 | 14 | 9 | 98 |
| D2S1338 | ?16-19.2 | 5 | 1 | 0 | 6 | ?19.3-23.3 | 21 | 7 | 0 | 28 | ?24-26 | 9 | 19 | 1 | 29 | 3 | 66 |
| D3S1358 | ?12-13 | 0 | 0 | 0 | 0 | ?14-18 | 21 | 22 | 9 | 52 | ?19-20 | 0 | 0 | 0 | 0 | 0 | 52 |
| D6S1043 | ?9-12 | 2 | 0 | 0 | 2 | ?12.2-18 | 13 | 15 | 4 | 32 | ?18.2-22.3 | 13 | 19 | 2 | 34 | 1 | 69 |
| D5S818 | ?7-9 | 0 | 0 | 0 | 0 | ?9.2-13 | 27 | 16 | 13 | 56 | ?13.2-15 | 2 | 8 | 0 | 10 | 0 | 66 |
| D7S820 | ?7-9 | 2 | 1 | 1 | 4 | ?9.1-11 | 5 | 8 | 2 | 15 | ?11.1-14 | 12 | 19 | 3 | 34 | 17 | 70 |
| D8S1179 | ?8-10 | 4 | 0 | 0 | 4 | ?11-15 | 34 | 19 | 11 | 64 | ?16-18 | 7 | 22 | 1 | 30 | 1 | 99 |
| FGA | 13-18.2 | 1 | 1 | 0 | 2 | 19-24.2 | 51 | 53 | 18 | 122 | 25-28 | 15 | 29 | 3 | 47 | 13 | 184 |
| Penta E | ?5-15 | 16 | 7 | 5 | 28 | ?16-21.3 | 54 | 37 | 6 | 97 | ?22-33.1 | 7 | 14 | 0 | 21 | 8 | 154 |
| PentaD | ?5-9 | 5 | 1 | 0 | 6 | ?10-18 | 18 | 12 | 5 | 35 | ?19-22 | 1 | 0 | 0 | 1 | 2 | 44 |
| TH01 | ?5-7 | 1 | 0 | 0 | 1 | ?8-12 | 2 | 6 | 0 | 8 | ?13-15 | 0 | 0 | 0 | 0 | 0 | 9 |
| TPOX | ?7-8 | 0 | 1 | 0 | 1 | ?8.1-11.2 | 1 | 0 | 0 | 1 | ?12-13 | 0 | 1 | 0 | 1 | 0 | 3 |
| vWA | ?14-15 | 1 | 1 | 1 | 3 | ?16-19 | 41 | 40 | 19 | 100 | ?19.3-21 | 4 | 11 | 0 | 15 | 9 | 127 |
| Total | - | 65 | 32 | 8 | 105 | - | 497 | 432 | 113 | 1042 | - | 141 | 242 | 14 | 397 | 121 | 1665 |
Table 9 Supplementary
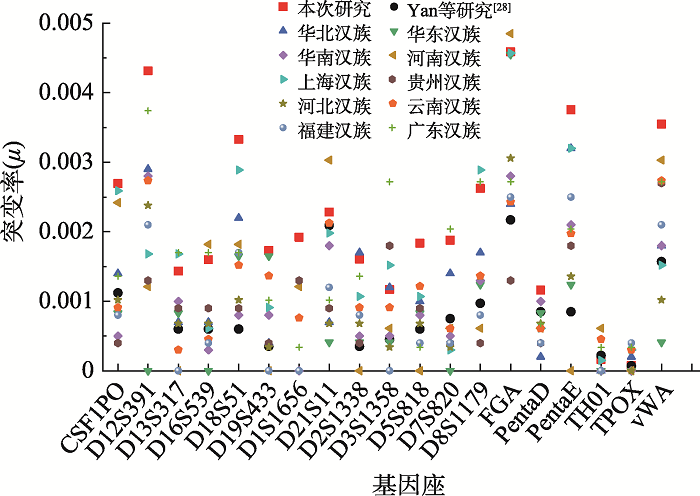
The Han populations mutation rates from other provinces or regions in China"
| 基因座 | 本次 研究 | Yan等 研究[ | 华北 汉族[ | 华东 汉族[ | 华南 汉族[ | 河南 汉族[ | 上海 汉族[ | 贵州 汉族[ | 河北 汉族[ | 云南 汉族[ | 福建 汉族[ | 广东 汉族[ |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n=78739 | n=19754 | n=16612 | n=4494 | n=6303 | n=2475 | n=12413 | n=3879 | n=5006 | n=10941 | n=3279 | n=4094 | |
| CSF1PO | 0.0027 | 0.0011 | 0.0014 | 0.0008 | 0.0005 | 0.0024 | 0.0026 | 0.0004 | 0.0010 | 0.0009 | 0.0008 | 0.0014 |
| D12S391 | 0.0043 | - | 0.0029 | 0.0000 | 0.0028 | 0.0012 | 0.0017 | 0.0013 | 0.0024 | 0.0027 | 0.0021 | 0.0037 |
| D13S317 | 0.0014 | 0.0006 | 0.0007 | 0.0008 | 0.0010 | 0.0000 | 0.0017 | 0.0009 | 0.0007 | 0.0003 | 0.0000 | 0.0017 |
| D16S539 | 0.0016 | 0.0006 | 0.0007 | 0.0000 | 0.0003 | 0.0018 | 0.0006 | 0.0009 | 0.0007 | 0.0005 | 0.0004 | 0.0017 |
| D18S51 | 0.0033 | 0.0006 | 0.0022 | 0.0017 | 0.0008 | 0.0018 | 0.0029 | 0.0009 | 0.0010 | 0.0015 | 0.0017 | 0.0017 |
| D19S433 | 0.0017 | 0.0004 | 0.0004 | 0.0017 | 0.0008 | 0.0000 | 0.0009 | 0.0004 | 0.0003 | 0.0014 | 0.0000 | 0.0010 |
| D1S1656 | 0.0019 | - | - | - | 0.0000 | 0.0012 | - | 0.0013 | - | 0.0008 | 0.0000 | 0.0003 |
| D21S11 | 0.0023 | 0.0021 | 0.0007 | 0.0004 | 0.0018 | 0.0030 | 0.0020 | 0.0009 | 0.0007 | 0.0021 | 0.0012 | 0.0010 |
| D2S1338 | 0.0016 | 0.0004 | 0.0017 | 0.0004 | 0.0005 | 0.0000 | 0.0011 | 0.0004 | 0.0007 | 0.0009 | 0.0008 | 0.0014 |
| D3S1358 | 0.0012 | 0.0005 | 0.0012 | 0.0004 | 0.0005 | 0.0006 | 0.0015 | 0.0018 | 0.0003 | 0.0009 | 0.0000 | 0.0027 |
| D5S818 | 0.0018 | 0.0006 | 0.0010 | 0.0008 | 0.0008 | 0.0000 | 0.0011 | 0.0009 | 0.0007 | 0.0012 | 0.0004 | 0.0003 |
| D7S820 | 0.0019 | 0.0008 | 0.0014 | 0.0000 | 0.0005 | 0.0006 | 0.0003 | 0.0004 | 0.0003 | 0.0006 | 0.0004 | 0.0020 |
| D8S1179 | 0.0026 | 0.0010 | 0.0017 | 0.0012 | 0.0013 | 0.0006 | 0.0029 | 0.0004 | 0.0014 | 0.0014 | 0.0008 | 0.0027 |
| FGA | 0.0046 | 0.0022 | 0.0024 | 0.0046 | 0.0028 | 0.0049 | 0.0046 | 0.0013 | 0.0031 | 0.0024 | 0.0025 | 0.0027 |
| Penta D | 0.0012 | 0.0009 | 0.0002 | 0.0008 | 0.0010 | 0.0006 | 0.0006 | 0.0004 | 0.0007 | 0.0006 | 0.0004 | 0.0007 |
| Penta E | 0.0038 | 0.0009 | 0.0032 | 0.0012 | 0.0021 | 0.0000 | 0.0032 | 0.0018 | 0.0014 | 0.0020 | 0.0025 | 0.0020 |
| TH01 | 0.0002 | 0.0002 | 0.0000 | 0.0000 | 0.0000 | 0.0006 | 0.0002 | - | 0.0000 | 0.0005 | 0.0000 | 0.0003 |
| TPOX | 0.0001 | 0.0001 | 0.0002 | 0.0000 | 0.0000 | 0.0000 | 0.0003 | - | 0.0000 | 0.0003 | 0.0004 | 0.0003 |
| vWA | 0.0035 | 0.0016 | 0.0018 | 0.0004 | 0.0018 | 0.0030 | 0.0015 | 0.0027 | 0.0010 | 0.0027 | 0.0021 | 0.0027 |
| Total | 0.0013 | - | 0.0013 | 0.0008 | - | - | 0.0016 | - | 0.0163 | 0.0012 | 0.0008 | 0.0014 |
| [1] | Subramanian S, Mishra RK, Singh L . Genome-wide analysis of microsatellite repeats in humans: their abundance and density in specific genomic regions. Genome Biol, 2003,4(2):R13. |
| [2] | Bär W, Brinkmann B, Budowle B, Carracedo A, Gill P, Lincoln P, Mayr W, Olaisen B . DNA recommendations. Further report of the DNA Commission of the ISFH regarding the use of short tandem repeat systems. International Society for Forensic Haemogenetics, Int J Legal Med, 1997,110(4):175-176. |
| [3] | Butler JM . Short tandem repeat typing technologies used in human identity testing. Biotechniques, 2007,43(4):2-5. |
| [4] | Kornberg A, Bertsch LL, Jackson JF, Khorana HG . Enzymatic synthesis of deoxyribonucleic acid, XVI. oligonucleotides as templates and the mechanism of their replication. Proc Natl Acad Sci USA, 1964,51(2):315-323. |
| [5] | Shao CC, Lin MX, Zhou ZH, Zhou YQ, Shen YW, Xue AM, Zhou HG, Tang QQ, Xie JH . Mutation analysis of 19 autosomal short tandem repeats in Chinese Han population from Shanghai. Int J Legal Med, 2016,130(6):1439-1444. |
| [6] | Li HX, Peng D, Wang Y, Wu RG, Zhang YM, Li R, Sun HY . Evaluation of genetic parameters of 23 autosomal STR loci in a southern Chinese Han population. Ann Hum Biol, 2018,45(4):359-364. |
| [7] | Wang HD, Kand B, Su N, He M, Zhang B, Guo YX, Zhu BF, Liao SX, Zeng ZS . Evaluation of the genetic parameters and mutation analysis of 22 STR loci in the central Chinese Han population, Int J Legal Med, 2017,131(1):103-105. |
| [8] | Qu N, Zhang XC, Liang H, Ou XL . Analysis of genetic polymorphisms and mutations at 23 autosomal STR loci in Guangdong Han population. Forensic Sci Int Genet, 2019,38:e16-e17. |
| [9] | Slooten K, Ricciardi F . Estimation of mutation probabilities for autosomal STR markers. Forensic Sci Int Genet, 2013,7(3):337-344. |
| [10] | Brinkmann B, Klintschar M, Neuhuber F, Hühne J, Rolf B . Mutation rate in human microsatellites: influence of the structure and length of the tandem repeat. Am J Hum Genet, 1998,62(6):1408-1415. |
| [11] | Weber JL, Wong C . Mutation of human short tandem repeats. Hum Mol Genet, 1993,2(8):1123-1128. |
| [12] | Jin B, Su Q, Luo HB, Li YB, Wu J, Yan J, Hou YP, Liang WB, Zhang L . Mutational analysis of 33 autosomal short tandem repeat (STR) loci in southwest Chinese Han population based on trio parentage testing. Forensic Sci Int Genet, 2016,23:86-90. |
| [13] | Taylor MJ, Fisk NM . Prenatal diagnosis in multiple pregnancy. Baillieres Best Pract Res Clin Obstet Gynaecol, 2000,14(4):663-675. |
| [14] | Wurmb-Schwark NV, Schwark T, Christiansen L, Lorenz D, Oehmichen M . The use of different multiplex PCRs for twin zygosity determination and its application in forensic trace analysis. Leg Med(Tokyo), 2004,6(2):125-130. |
| [15] | Xu YJ, Li TT, Pu T, Cao RX, Long F, Chen S, Sun K, Xu R . Copy number variants and exome sequencing analysis in six pairs of Chinese monozygotic twins discordant for congenital heart disease. Twin Res Hum Genet, 2017,20(6):521-532. |
| [16] | Weber Lehmann J, Schilling E, Gradl G, Richter DC, Wiehler J, Rolf B . Finding the needle in the haystack: differentiating “identical” twins in paternity testing and forensics by ultra-deep next generation sequencing. Forensic Sci Int Genet, 2014,9:42-46. |
| [17] | Wang LF, Yang Y, Zhang XN, Quan XL, Wu YM . Tri-allelic pattern of short tandem repeats identifies the murderer among identical twins and suggests an embryonic mutational origin. Forensic Sci Int Genet, 2015,16:239-245. |
| [18] | Jónsson H, Sulem P, Kehr B, Kristmundsdottir S, Zink F, Hjartarson E, Hardarson MT, Hjorleifsson KE, Eggertsson HP, Gudjonsson SA, Ward LD, Arnadottir GA, Helgason EA, Helgason H, Gylfason A, Jonasdottir A, Jonasdottir A, Rafnar T, Frigge M, Stacey SN, Magnusson OT, Thorsteinsdottir U, Masson G, Kong A, Halldorsson BV, Helgason A, Gudbjartsson DF, Stefansson K . Parental influence on human germline de novo mutations in 1,548 trios from Iceland. Nature, 2017,549(7673):519-522. |
| [19] | Valdes AM, Slatkin M, Freimer NB . Allele frequencies at microsatellite loci: the stepwise mutation model revisited. Genetics, 1993,133(3):737-749. |
| [20] | Klintschar M, Dauber EM, Ricci U, Cerri N, Immel UD, Kleiber M, Mayr WR . Haplotype studies support slippage as the mechanism of germline mutations in short tandem repeats. Electrophoresis, 2010,25(20):3344-3348. |
| [21] | Lan Q, Wang HD, Shen CM, Guo YX, Yin CY, Xie T, Fang YT, Zhou YS, Zhu BF . Mutability analysis towards 21 STR loci included in the AGCU 21+1 kit in Chinese Han population. Int J Legal Med, 2018,132(5):1287-1291. |
| [22] | Ge JY, Budowle B, Aranda XG, Planz JV, Eisenberg AJ, Chakraborty R . Mutation rates at Y chromosome short tandem repeats in Texas populations, Forensic Sci Int Genet, 2009,3(3):179-184. |
| [23] | Xu W, Wang YQ, Zhang DD, Wang DX, Zhou L, Ye XL, Zhu CG, Shi YZ . Mutation analysis of 21 autosomal short tandem repeats in Han population from Hunan, China. Ann Hum Biol. 2019,46(3):254-260. |
| [24] | Dupuy BM, Stenersen M, Egeland T, Olaisen B . Y-chromosomal microsatellite mutation rates: differences in mutation rate between and within loci, Hum Mutat, 2004,23(2):117-124. |
| [25] | Xie BB, Chen L, Yang YR, Lv YX, Chen J, Shi Y, Chen C, Zhao HY, Yu ZL, Liu YC, Fang XD, Yan JW . Genetic distribution of 39 STR loci in 1027 unrelated Han individuals from northern China. Forensic Sci Int Genet, 2015,19:205-206. |
| [26] | Chakraborty R, Stivers DN, Zhong YX . Estimation of mutation rates from parentage exclusion data: applications to STR and VNTR loci. Mutat Res, 1996,354(1):41-48. |
| [27] | Vicard P, Dawid AP . A statistical treatment of biases affecting the estimation of mutation rates. Mutat Res, 2004,547(1-2):19-33. |
| [28] | Yan JW, Liu YC, Tang H, Zhang QX, Huo ZY, Hu SN, Yu J . Mutations at 17 STR loci in Chinese population. Forensic Sci Int, 2006,162(1-3):53-54. |
| [29] | Lu DJ, Liu QL, Wu WW, Zhao H . Mutation analysis of 24 short tandem repeats in Chinese Han population. Int J Legal Med, 2012,126(2):331-335. |
| [30] | Qian XQ, Yin CY, Ji Q, Li K, Fan HT, Yu YF, Bu FL, Hu LL, Wang JW, Mu HF, Haigh S, Chen F . Mutation rate analysis at 19 autosomal microsatellites. Electrophoresis, 2015,36(14):1633-1639. |
| [附录] | 附录涉及的参考文献(References): |
| [1] | Yan JW, Liu YC, Tang H, Zhang QX, Huo ZY, Hu SN, Yu J . Mutations at 17 STR loci in Chinese population. Forensic Sci Int, 2006,162(1-3):53-54. |
| [2] | Qian XQ, Yin CY, Ji Q, Li K, Fan HT, Yu YF, Bu FL, Hu LL, Wang JW, Mu HF . Mutation rate analysis at 19 autosomal microsatellites. Electrophoresis, 2015,36(14):1633-1639. |
| [3] | Li HX, Peng D, Wang Y, Wu RG, Zhang YM, Li R, Sun HY . Evaluation of genetic parameters of 23 autosomal STR loci in a Southern Chinese Han population, Ann Hum Biol, 2018,45(4):359-364. |
| [4] | Wang. HD, Kang B, Su N, He M, Zhang B, Guo YX, Zhu BF, Liao. SX, Zeng ZS. Evaluation of the genetic parameters and mutation analysis of 22 STR loci in the central Chinese Han population. Int J Legal Med, 2016,131(1):103-105. |
| [5] | Shao CC, Lin MX, Zhou ZH, Zhou YQ, Shen YW, Xue AM, Zhou HG, Tang QQ, Xie JH . Mutation analysis of 19 autosomal short tandem repeats in Chinese Han population from Shanghai. Int J Legal Med, 2016,130(6):1439-1444. |
| [6] | Yang MQ, Ren Z, Ji JY, Zhou H, Zhang HL, Dai JL, Wang J, Huang J . Population genetic data and mutations of 22 autosomal STR loci in Guizhou Han population. Forensic Sci Int Genet, 2017,29:e29-e30. |
| [7] | Sun LJ, Shi K, Tan L, Zhang Q, Fu LH, Zhang XJ, Fu GP, Li SJ, Cong B . Analysis of genetic polymorphisms and mutations at 19 STR loci in Hebei Han population. Forensic Sci Int Genet, 2017,31:e50-e51. |
| [8] | Zhang XF, Liu LL, Xie RF, Wang GY, Shi Y, Gu T, Hu LP, Nie SJ . Population data and mutation rates of 20 autosomal STR loci in a Chinese Han population from Yunnan Province, Southwest China. Int J Legal Med, 2018,132(4):1083-1085. |
| [9] | Qu N, Zhang XC, Liang H, XL Ou . Analysis of genetic polymorphisms and mutations at 23 autosomal STR loci in Guangdong Han population. Forensic Sci Int Genet, 2019,38:e16-e17. |
| [1] | Xueqian Wang, Qingzhen Zhang, Peng Cheng, Tingting Dong, Weiguo Li, Zhe Zhou, Shengqi Wang. Genetic polymorphisms of 66 InDel loci in the Chinese Han population [J]. Hereditas(Beijing), 2022, 44(4): 335-345. |
| [2] | Ming Liu, Yi Li, Yafang Yang, Yuwen Yan, Fan Liu, Caixia Li, Faming Zeng, Wenting Zhao. Human facial shape related SNP analysis in Han Chinese populations [J]. Hereditas(Beijing), 2020, 42(7): 680-690. |
| [3] | JIN Yun-Kai, LIU Chun-Lin, RUAN Ying. 6b genes: the important effective factors relative to tumor formation in plants [J]. HEREDITAS, 2011, 33(11): 1212-1218. |
| [4] | HAN Chun-Mei, ZHANG Jia-Bao, GAO Qing-Hua, CHEN Qing-Bo. Study on Parentage Testing in JIRONG Rabbit by Microsatellite Markers [J]. HEREDITAS, 2005, 27(6): 903-907. |
| [5] | JIA Zhen-Jun, WU Jin, LI Hui, HOU Yi-Ping, ZHANG Wei-Juan, ZHOU Xue-Ping, DENG Jian-Qiang, SHI Mei-Sen, ZHANG Ji, LI Ying-Bi. Study of Five STR Loci in A Chinese Han Population [J]. HEREDITAS, 2005, 27(3): 343-348. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||