Hereditas(Beijing) ›› 2023, Vol. 45 ›› Issue (6): 464-471.doi: 10.16288/j.yczz.23-051
• Frontier Focus • Previous Articles Next Articles
Exon junction complex modulates the formation of the m6A epitranscriptome
Penghui Song1( ), Lijuan Ma2(
), Lijuan Ma2( ), Dong Yan1(
), Dong Yan1( )
)
- 1. State Key Laboratory of Genetic Engineering, School of Life Sciences, Fudan University, Shanghai 200438, China
2. CAS Center for Excellence in Molecular Plant Sciences, Chinese Academy of Sciences, University of Chinese Academy of Sciences, Shanghai 200032, China
-
Received:2023-03-09Revised:2023-05-17Online:2023-06-20Published:2023-05-22 -
Contact:Yan Dong E-mail:21110700039@m.fudan.edu.cn;malijuan@cemps.ac.cn;yandong@fudan.edu.cn -
Supported by:National Natural Science Foundation of China(31970786);National Natural Science Foundation of China(32270868)
Cite this article
Penghui Song, Lijuan Ma, Dong Yan. Exon junction complex modulates the formation of the m6A epitranscriptome[J]. Hereditas(Beijing), 2023, 45(6): 464-471.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
| [1] |
Hsu PJ, Shi HL, He C. Epitranscriptomic influences on development and disease. Genome Biol, 2017, 18(1): 197.
doi: 10.1186/s13059-017-1336-6 pmid: 29061143 |
| [2] |
Zhao LY, Song JH, Liu YB, Song CX, Yi CQ. Mapping the epigenetic modifications of DNA and RNA. Protein Cell, 2020, 11(11): 792-808.
doi: 10.1007/s13238-020-00733-7 |
| [3] | Yang Y, Chen YS, Sun BF, Yang YG. RNA methylation: regulations and mechanisms. Hereditas(Beijing), 2018, 40(11): 964-976. |
| 杨莹, 陈宇晟, 孙宝发, 杨运桂. RNA甲基化修饰调控和规律. 遗传, 2018, 40(11): 964-976. | |
| [4] |
Jonkhout N, Tran J, Smith MA, Schonrock N, Mattick JS, Novoa EM. The RNA modification landscape in human disease. RNA, 2017, 23(12): 1754-1769.
doi: 10.1261/rna.063503.117 pmid: 28855326 |
| [5] | Boccaletto P, Stefaniak F, Ray A, Cappannini A, Mukherjee S, Purta E, Kurkowska M, Shirvanizadeh N, Destefanis E, Groza P, Avşar G, Romitelli A, Pir P, Dassi E, Conticello SG, Aguilo F, Bujnicki JM.MODOMICS: a database of RNA modification pathways. 2021 update. Nucleic Acids Res, 2022, 50(D1): D231-D235. |
| [6] |
Davis FF, Allen FW. Ribonucleic acids from yeast which contain a fifth nucleotide. J Biol Chem, 1957, 227(2): 907-915.
pmid: 13463012 |
| [7] |
Suzuki T. The expanding world of tRNA modifications and their disease relevance. Nat Rev Mol Cell Biol, 2021, 22(6): 375-392.
doi: 10.1038/s41580-021-00342-0 |
| [8] |
Sendinc E, Shi Y. RNA m6A methylation across the transcriptome. Mol Cell, 2023, 83(3): 428-441.
doi: 10.1016/j.molcel.2023.01.006 pmid: 36736310 |
| [9] |
Zhang X, Jia GF. RNA epigenetic modification: N6-methyladenosine. Hereditas(Beijing), 2016, 38(4): 275-288.
doi: 10.16288/j.yczz.16-049 pmid: 27103452 |
|
张笑, 贾桂芳. RNA表观遗传修饰:N6-甲基腺嘌呤. 遗传, 2016, 38(4): 275-288.
doi: 10.16288/j.yczz.16-049 pmid: 27103452 |
|
| [10] |
Bokar JA, Shambaugh ME, Polayes D, Matera AG, Rottman FM. Purification and cDNA cloning of the AdoMet- binding subunit of the human mRNA (N6-adenosine)- methyltransferase. RNA, 1997, 3(11): 1233-1247.
pmid: 9409616 |
| [11] |
Jia GF, Fu Y, Zhao X, Dai Q, Zheng GQ, Yang Y, Yi CQ, Lindahl T, Pan T, Yang YG, He C. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol, 2011, 7(12): 885-887.
doi: 10.1038/nchembio.687 pmid: 22002720 |
| [12] |
Zheng GQ, Dahl JA, Niu YM, Fedorcsak P, Huang CM, Li CJ, Vågbø CB, Shi Y, Wang WL, Song SH, Lu ZK, Bosmans RPG, Dai Q, Hao YJ, Yang X, Zhao WM, Tong WM, Wang XJ, Bogdan F, Furu K, Fu Y, Jia GF, Zhao X, Liu J, Krokan HE, Klungland A, Yang YG, He C. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell, 2013, 49(1): 18-29.
doi: 10.1016/j.molcel.2012.10.015 pmid: 23177736 |
| [13] |
He PC, He C. m6A RNA methylation: from mechanisms to therapeutic potential. EMBO J, 2021, 40(3): e105977.
doi: 10.15252/embj.2020105977 |
| [14] |
Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR. Comprehensive analysis of mRNA methylation reveals enrichment in 3' UTRs and near stop codons. Cell, 2012, 149(7): 1635-1646.
doi: 10.1016/j.cell.2012.05.003 pmid: 22608085 |
| [15] |
Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, Cesarkas K, Jacob-Hirsch J, Amariglio N, Kupiec M, Sorek R, Rechavi G. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature, 2012, 485(7397): 201-206.
doi: 10.1038/nature11112 |
| [16] |
Schwartz S, Agarwala SD, Mumbach MR, Jovanovic M, Mertins P, Shishkin A, Tabach Y, Mikkelsen TS, Satija R, Ruvkun G, Carr SA, Lander ES, FinkGR, Regev A. High-resolution mapping reveals a conserved, widespread, dynamic mRNA methylation program in yeast meiosis. Cell, 2013, 155(6): 1409-1421.
doi: 10.1016/j.cell.2013.10.047 pmid: 24269006 |
| [17] |
Yang Y, Hsu PJ, Chen YS, Yang YG. Dynamic transcriptomic m6A decoration: writers, erasers, readers and functions in RNA metabolism. Cell Res, 2018, 28(6): 616-624.
doi: 10.1038/s41422-018-0040-8 |
| [18] |
Meyer KD, Jaffrey SR. Rethinking m6A readers, writers, and erasers. Annu Rev Cell Dev Biol, 2017, 33: 319-342.
doi: 10.1146/annurev-cellbio-100616-060758 pmid: 28759256 |
| [19] |
van Tran N, Ernst FGM, Hawley BR, Zorbas C, Ulryck N, Hackert P, Bohnsack KE, Bohnsack MT, Jaffrey SR, Graille M, Lafontaine DLJ.The human 18S rRNA m6A methyltransferase METTL 5 is stabilized by TRMT112. Nucleic Acids Res, 2019, 47(15): 7719-7733.
doi: 10.1093/nar/gkz619 |
| [20] |
Ma HH, Wang XY, Cai JB, Dai Q, Natchiar SK, Lv R, Chen K, Lu ZK, Chen H, Shi YG, Lan F, Fan J, Klaholz BP, Pan T, Shi Y, He C. N6-Methyladenosine methyltransferase ZCCHC4 mediates ribosomal RNA methylation. Nat Chem Biol, 2019, 15(1): 88-94.
doi: 10.1038/s41589-018-0184-3 |
| [21] |
Chen H, Gu L, Orellana EA, Wang YY, Guo JJ, Liu Q, Wang LF, Shen ZF, Wu H, Gregory RI, Xing Y, Shi Y. METTL4 is an snRNA m6Am methyltransferase that regulates RNA splicing. Cell Res, 2020, 30(6): 544-547.
doi: 10.1038/s41422-019-0270-4 pmid: 31913360 |
| [22] |
Goh YT, Koh CWQ, Sim DY, Roca X, Goh WSS. METTL4 catalyzes m6Am methylation in U2 snRNA to regulate pre-mRNA splicing. Nucleic Acids Res, 2020, 48(16): 9250-9261.
doi: 10.1093/nar/gkaa684 pmid: 32813009 |
| [23] |
Warda AS, Kretschmer J, Hackert P, Lenz C, Urlaub H, Höbartner C, Sloan KE, Bohnsack MT. Human METTL16 is a N6-methyladenosine (m6A) methyltransferase that targets pre-mRNAs and various non-coding RNAs. EMBO Rep, 2017, 18(11): 2004-2014.
doi: 10.15252/embr.201744940 |
| [24] |
Pendleton KE, Chen BB, Liu KQ, Hunter OV, Xie Y, Tu BP, Conrad NK. The U6 snRNA m6A methyltransferase METTL16 regulates SAM synthetase intron retention. Cell, 2017, 169(5): 824-835.e814.
doi: S0092-8674(17)30530-5 pmid: 28525753 |
| [25] |
Mendel M, Delaney K, Pandey RR, Chen KM, Wenda JM, Vågbø CB, Steiner FA, Homolka D, Pillai RS. Splice site m6A methylation prevents binding of U2AF35 to inhibit RNA splicing. Cell, 2021, 184(12): 3125-3142.e3125.
doi: 10.1016/j.cell.2021.03.062 |
| [26] |
Liu JZ, Yue YN, Han DL, Wang X, Fu Y, Zhang L, Jia GF, Yu M, Lu ZK, Deng X, Chen WZ, He C. A METTL3- METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat Chem Biol, 2014, 10(2): 93-95.
doi: 10.1038/nchembio.1432 |
| [27] |
Bokar JA, Rath-Shambaugh ME, Ludwiczak R, Narayan P, Rottman F. Characterization and partial purification of mRNA N6-adenosine methyltransferase from HeLa cell nuclei. Internal mRNA methylation requires a multisubunit complex. J Biol Chem, 1994, 269(26): 17697-17704.
pmid: 8021282 |
| [28] |
Wang P, Doxtader KA, Nam YS. Structural basis for cooperative function of Mettl3 and Mettl14 methyltransferases. Mol Cell, 2016, 63(2): 306-317.
doi: S1097-2765(16)30227-1 pmid: 27373337 |
| [29] |
Wang X, Feng J, Xue Y, Guan ZY, Zhang DL, Liu Z, Gong Z, Wang Q, Huang JB, Tang C, Zou TT, Yin P. Structural basis of N6-adenosine methylation by the METTL3- METTL14 complex. Nature, 2016, 534(7608): 575-578.
doi: 10.1038/nature18298 |
| [30] |
Śledź P, Jinek M. Structural insights into the molecular mechanism of the m6A writer complex. Elife, 2016, 5: e18434.
doi: 10.7554/eLife.18434 |
| [31] |
Yankova E, Blackaby W, Albertella M, Rak J, De Braekeleer E, Tsagkogeorga G, Pilka ES, Aspris D, Leggate D, Hendrick AG, Webster NA, Andrews B, Fosbeary R, Guest P, Irigoyen N, Eleftheriou M, Gozdecka M, Dias JML, Bannister AJ, Vick B, Jeremias I, Vassiliou GS, Rausch O, Tzelepis K, Kouzarides T. Small-molecule inhibition of METTL3 as a strategy against myeloid leukaemia. Nature, 2021, 593(7860): 597-601.
doi: 10.1038/s41586-021-03536-w |
| [32] |
Zhong SL, Li HY, Bodi Z, Button J, Vespa L, Herzog M, Fray RG. MTA is an Arabidopsis messenger RNA adenosine methylase and interacts with a homolog of a sex-specific splicing factor. Plant Cell, 2008, 20(5): 1278-1288.
doi: 10.1105/tpc.108.058883 pmid: 18505803 |
| [33] |
Ping XL, Sun BF, Wang L, Xiao W, Yang X, Wang WJ, Adhikari S, Shi Y, Lv Y, Chen YS, Zhao X, Li A, Yang Y, Dahal U, Lou XM, Liu X, Huang J, Yuan WP, Zhu XF, Cheng T, Zhao YL, Wang XQ, Rendtlew Danielsen JM, Liu F, Yang YG. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res, 2014, 24(2): 177-189.
doi: 10.1038/cr.2014.3 pmid: 24407421 |
| [34] |
Horiuchi K, Kawamura T, Iwanari H, Ohashi R, Naito M, Kodama T, Hamakubo T: Identification of Wilms' tumor 1-associating protein complex and its role in alternative splicing and the cell cycle. J Biol Chem, 2013, 288(46): 33292-33302.
doi: 10.1074/jbc.M113.500397 pmid: 24100041 |
| [35] |
Yue YN, Liu J, Cui XL, Cao J, Luo GZ, Zhang ZZ, Cheng T, Gao MS, Shu X, Ma HH, Wang FQ, Wang XX, Shen S, Wang YZ, Feng XH, He C, Liu JZ. VIRMA mediates preferential m6A mRNA methylation in 3'UTR and near stop codon and associates with alternative polyadenylation. Cell Discov, 2018, 4: 10.
doi: 10.1038/s41421-018-0019-0 |
| [36] |
Růžička K, Zhang M, Campilho A, Bodi Z, Kashif M, Saleh M, Eeckhout D, El-Showk S, Li HY, Zhong SL, De Jaeger G, Mongan NP, Hejátko J, Helariutta Y, Fray RG. Identification of factors required for m6A mRNA methylation in Arabidopsis reveals a role for the conserved E3 ubiquitin ligase HAKAI. New Phytol, 2017, 215(1): 157-172.
doi: 10.1111/nph.14586 pmid: 28503769 |
| [37] |
Wen J, Lv RT, Ma HH, Shen HJ, He CX, Wang JH, Jiao FF, Liu H, Yang PY, Tan L, Lan F, Shi YG, He C, Shi Y, Diao JB. Zc3h13 regulates nuclear RNA m6A methylation and mouse embryonic stem cell self-renewal. Mol Cell, 2018, 69(6): 1028-1038.e1026.
doi: 10.1016/j.molcel.2018.02.015 |
| [38] |
Guo J, Tang HW, Li J, Perrimon N, Yan D. Xio is a component of the Drosophila sex determination pathway and RNA N6-methyladenosine methyltransferase complex. Proc Natl Acad Sci USA, 2018, 115(14): 3674-3679.
doi: 10.1073/pnas.1720945115 |
| [39] |
Knuckles P, Lence T, Haussmann IU, Jacob D, Kreim N, Carl SH, Masiello I, Hares T, Villasenor R, Hess D, Andrade-Navarro MA, Biggiogera M, Helm M, Soller M, Bühler M, Roignant JY. Zc3h13/Flacc is required for adenosine methylation by bridging the mRNA-binding factor Rbm15/Spenito to the m6A machinery component Wtap/Fl(2)d. Genes Dev, 2018, 32(5-6): 415-429.
doi: 10.1101/gad.309146.117 |
| [40] |
Lence T, Akhtar J, Bayer M, Schmid K, Spindler L, Ho CH, Kreim N, Andrade-Navarro MA, Poeck B, Helm M, Roignant JY. m6A modulates neuronal functions and sex determination in Drosophila. Nature, 2016, 540(7632): 242-247.
doi: 10.1038/nature20568 |
| [41] |
Wang YH, Zhang LF, Ren H, Ma LJ, Guo J, Mao DC, Lu ZW, Lu LJ, Yan D. Role of Hakai in m6A modification pathway in Drosophila. Nat Commun, 2021, 12(1): 2159.
doi: 10.1038/s41467-021-22424-5 |
| [42] |
Bawankar P, Lence T, Paolantoni C, Haussmann IU, Kazlauskiene M, Jacob D, Heidelberger JB, Richter FM, Nallasivan MP, Morin V, Kreim N, Beli P, Helm M, Jinek M, Soller M, Roignant JY. Hakai is required for stabilization of core components of the m6A mRNA methylation machinery. Nat Commun, 2021, 12(1): 3778.
doi: 10.1038/s41467-021-23892-5 pmid: 34145251 |
| [43] |
Su SC, Li SS, Deng T, Gao MS, Yin Y, Wu BX, Peng C, Liu JZ, Ma JB, Zhang KM. Cryo-EM structures of human m6A writer complexes. Cell Res, 2022, 32(11): 982-994.
doi: 10.1038/s41422-022-00725-8 |
| [44] |
Patil DP, Chen CK, Pickering BF, Chow A, Jackson C, Guttman M, Jaffrey SR. m6A RNA methylation promotes XIST-mediated transcriptional repression. Nature, 2016, 537(7620): 369-373.
doi: 10.1038/nature19342 |
| [45] |
Huang HL, Weng HY, Zhou KR, Wu T, Zhao BS, Sun ML, Chen ZH, Deng XL, Xiao G, Auer F, Klemm L, Wu HZ, Zuo ZX, Qin X, Dong YZ, Zhou YL, Qin HJ, Tao S, Du J, Liu J, Lu ZK, Yin H, Mesquita A, Yuan CL, Hu YC, Sun WJ, Su R, Dong L, Shen C, Li CY, Qing Y, Jiang X, Wu XW, Sun M, Guan JL, Qu LH, Wei MJ, Müschen M, Huang G, He C, Yang JH, Chen JJ. Histone H3 trimethylation at lysine 36 guides m6A RNA modification co-transcriptionally. Nature 2019, 567(7748): 414-419.
doi: 10.1038/s41586-019-1016-7 |
| [46] |
Slobodin B, Han RQ, Calderone V, Vrielink JAF, Loayza-Puch F, Elkon R, Agami R. Transcription impacts the efficiency of mRNA translation via co-transcriptional N6-adenosine methylation. Cell, 2017, 169(2): 326-337.e312.
doi: S0092-8674(17)30358-6 pmid: 28388414 |
| [47] |
Barbieri I, Tzelepis K, Pandolfini L, Shi JW, Millán- Zambrano G, Robson SC, Aspris D, Migliori V, Bannister AJ, Han N, De Braekeleer E, Ponstingl H, Hendrick A, Vakoc CR, Vassiliou GS, Kouzarides T. Promoter-bound METTL3 maintains myeloid leukaemia by m6A-dependent translation control. Nature, 2017, 552(7683): 126-131.
doi: 10.1038/nature24678 |
| [48] |
Bertero A, Brown S, Madrigal P, Osnato A, Ortmann D, Yiangou L, Kadiwala J, Hubner NC, de Los Mozos IR, Sadée C, Lenaerts AS, Nakanoh S, Grandy R, Farnell E, Ule J, Stunnenberg HG, Mendjan S, Vallier L. The SMAD2/3 interactome reveals that TGFbeta controls m6A mRNA methylation in pluripotency. Nature, 2018, 555(7695): 256-259.
doi: 10.1038/nature25784 |
| [49] |
He PC, Wei JB, Dou XY, Harada BT, Zhang ZJ, Ge RQ, Liu C, Zhang LS, Yu XB, Wang S, Lyu R, Zou ZY, Chen MJ, He C. Exon architecture controls mRNA m6A suppression and gene expression. Science, 2023, 379(6633): 677-682.
doi: 10.1126/science.abj9090 |
| [50] |
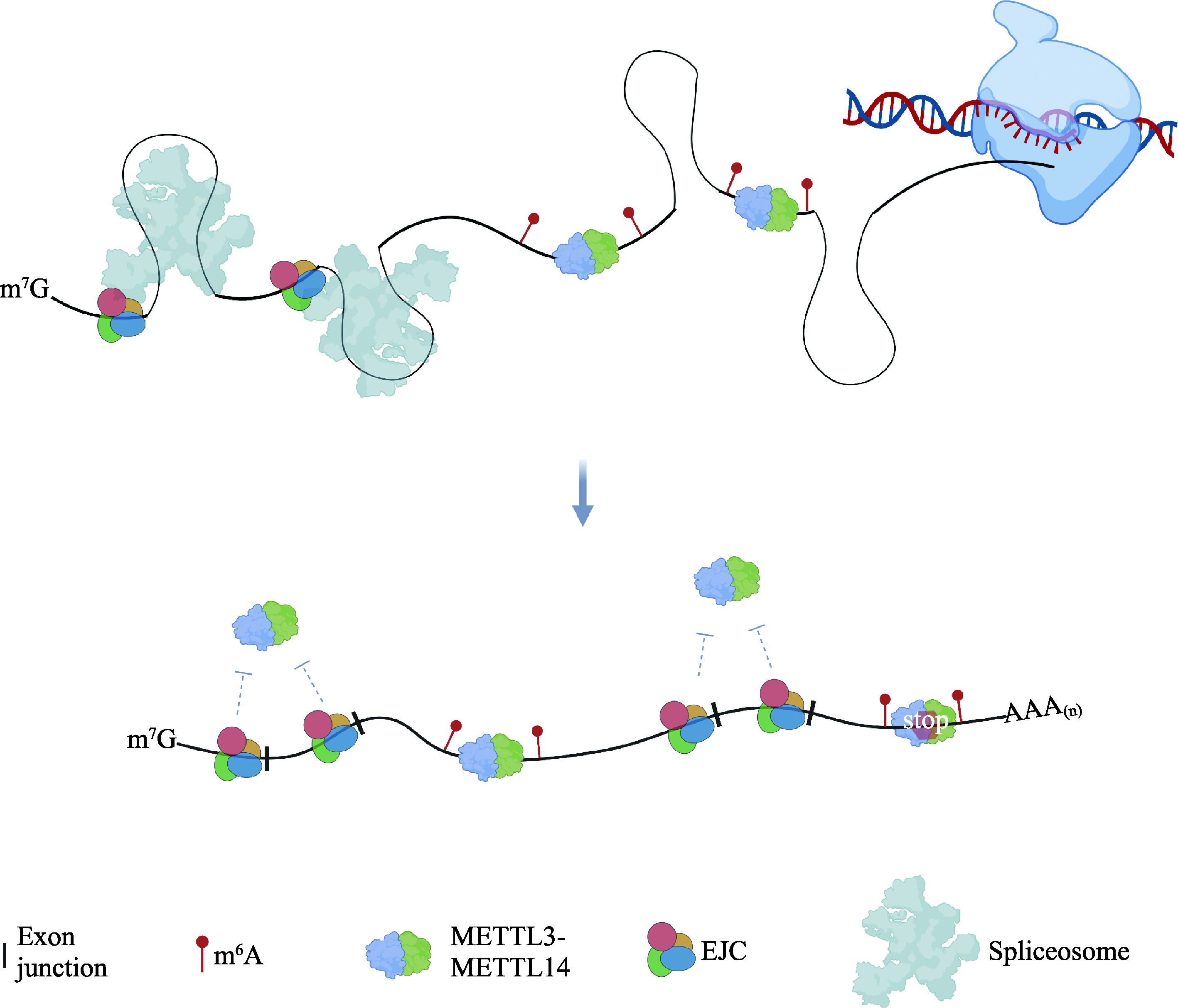
Le Hir H, Saulière J, Wang Z. The exon junction complex as a node of post-transcriptional networks. Nat Rev Mol Cell Biol, 2016, 17(1): 41-54.
doi: 10.1038/nrm.2015.7 |
| [51] |
Boehm V, Gehring NH. Exon junction complexes: supervising the gene expression assembly line. Trends Genet, 2016, 32(11): 724-735.
doi: S0168-9525(16)30104-4 pmid: 27667727 |
| [52] | Zaccara S, Ries RJ, Jaffrey SR. Reading, writing and erasing mRNA methylation. Nat Rev Mol Cell Biol, 2019, 20(10): 608-624. |
| [53] |
Singh G, Kucukural A, Cenik C, Leszyk JD, Shaffer SA, Weng ZP, Moore MJ. The cellular EJC interactome reveals higher-order mRNP structure and an EJC-SR protein nexus. Cell, 2012, 151(4): 750-764.
doi: S0092-8674(12)01220-2 pmid: 23084401 |
| [54] |
Mabin JW, Woodward LA, Patton RD, Yi ZX, Jia MX, Wysocki VH, Bundschuh R, Singh G. The exon junction complex undergoes a compositional switch that alters mRNP structure and nonsense-mediated mRNA decay Activity. Cell Rep, 2018, 25(9): 2431-2446.e2437.
doi: S2211-1247(18)31809-6 pmid: 30466796 |
| [55] |
Yang X, Triboulet R, Liu Q, Sendinc E, Gregory RI. Exon junction complex shapes the m6A epitranscriptome. Nat Commun, 2022, 13(1): 7904.
doi: 10.1038/s41467-022-35643-1 pmid: 36550132 |
| [56] |
Uzonyi A, Dierks D, Nir R, Kwon OS, Toth U, Barbosa I, Burel C, Brandis A, Rossmanith W, Le Hir H, Slobodin B, Schwartz S. Exclusion of m6A from splice-site proximal regions by the exon junction complex dictates m6A topologies and mRNA stability. Mol Cell, 2023, 83(2): 237-251.e237.
doi: 10.1016/j.molcel.2022.12.026 pmid: 36599352 |
| [57] |
Haussmann IU, Bodi Z, Sanchez-Moran E, Mongan NP, Archer N, Fray RG, Soller M. m6A potentiates Sxl alternative pre-mRNA splicing for robust Drosophila sex determination. Nature, 2016, 540(7632): 301-304.
doi: 10.1038/nature20577 |
| [1] | Shan He, Jian Zhao, Xiaofeng Song. Effects of N6-methyladenosine modification on the function of the female reproductive system [J]. Hereditas(Beijing), 2023, 45(6): 472-487. |
| [2] | Juan Wang, Yuening Yang, Weilan Piao, Hua Jin. Uridylation: a vital way for cellular RNA surveillance [J]. Hereditas(Beijing), 2022, 44(6): 449-465. |
| [3] | Xuqing Liu,Yubang Gao,Liangzhen Zhao,Yuchen Cai,Huiyuan Wang,Miao Miao,Lianfeng Gu,Hangxiao Zhang. Biogenesis, research methods, and functions of circular RNAs [J]. Hereditas(Beijing), 2019, 41(6): 469-485. |
| [4] | Peng Xue, Tao Jiang, Xingjia Shen. Advances in m 6A modification and its regulation of viral replication [J]. Hereditas(Beijing), 2019, 41(5): 404-412. |
| [5] | Jinchuan Wei,Tianyi Xu,Jing Wu,Xiaofeng Song. Molecular mechanisms of recursive splicing events in long introns of eukaryotes [J]. Hereditas(Beijing), 2019, 41(2): 89-97. |
| [6] | Qiying Leng, Jiahui Zheng, Haidong Xu, Patricia Adu-Asiamah, Ying Zhang, Bingwang Du, Li Zhang. Cloning and expression analysis of chicken circular transcript of insulin degrading enzyme gene [J]. Hereditas(Beijing), 2019, 41(12): 1129-1137. |
| [7] | Jianshu Zhuo, Xiaoyan Jing, Xin Du, Xiuqin Yang. Generation of Chimeric RNAs by cis-splicing of adjacent genes (cis-SAGe) in mammals [J]. Hereditas(Beijing), 2018, 40(2): 145-154. |
| [8] | Yongxin Zou,Yaoqin Gong. Aberrant RNA splicing as the molecular basis of some pathogenic variants [J]. Hereditas(Beijing), 2017, 39(3): 200-207. |
| [9] | Jiao Li, Yuqi Guo, Weiling Cui, Aihua Xu, Zengyuan Tian. Response of maize serine/arginine-rich protein gene family in seedlings to drought stress [J]. HEREDITAS(Beijing), 2014, 36(7): 697-706. |
| [10] | Jinxuan Zhao, Fang Wang, Zhengrong Xu, Yimei Fan. The epigenetic effect on pre-mRNA alternative splicing [J]. HEREDITAS, 2014, 36(3): 248-255. |
| [11] | LI Yu-Li, YU Jun, SONG Shu-Hui. Recent progresses in RNA N6-methyladenosine research [J]. HEREDITAS, 2013, 35(12): 1340-1351. |
| [12] | LUO Yan, WANG Ying. Alternative splicing of SmU2AFin Solanum melongena L. [J]. HEREDITAS, 2008, 30(11): 1499-1505. |
| [13] | WANG Wei, LAI Mao-De. Alternative Splicing of Insulin Receptor mRNA in Cancer and Type 2 Diabetes Mellitus: A Review [J]. HEREDITAS, 2006, 28(2): 226-230. |
| [14] | ZHOU Li-Wei, MA Fei, LI Qing-Wei. Progress in the Study on Alternative Splicing and Functions of Kininogen Genes [J]. HEREDITAS, 2006, 28(12): 1649-1649~1655. |
| [15] | XU Xian-Guo, WU Jun-Jie, HONG Xiao-Zhen, ZHU Fa-Ming, YAN Li-Xing. Identification of Nine Novel Alternative Splicing Isoforms of RHD mRNA [J]. HEREDITAS, 2006, 28(10): 1213-1218. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||