Hereditas(Beijing) ›› 2023, Vol. 45 ›› Issue (11): 1028-1038.doi: 10.16288/j.yczz.23-193
• Review • Previous Articles Next Articles
Progress on mechanisms of antibiotic resistance in Clostridioides difficile
- Key Laboratory of Clinical Pharmacology of Antibiotics,Institute of Antibiotics, Huashan Hospital, Fudan University, Shanghai 200040, China
-
Received:2023-08-18Revised:2023-10-27Online:2023-11-20Published:2023-11-03 -
Contact:Haihui Huang E-mail:txu20@fudan.edu.cn;huanghaihui@fudan -
Supported by:Natural Science Foundation of Shanghai(21ZR1410800)
Cite this article
Teng Xu, Haihui Huang. Progress on mechanisms of antibiotic resistance in Clostridioides difficile[J]. Hereditas(Beijing), 2023, 45(11): 1028-1038.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
| [1] |
Antimicrobial Resistance Collaborators. Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet, 2022, 399(10325): 629-655.
doi: 10.1016/S0140-6736(21)02724-0 pmid: 35065702 |
| [2] |
Peng Z, Jin DZ, Kim HB, Stratton CW, Wu B, Tang YW, Sun XM. Update on antimicrobial resistance in clostridium difficile: resistance mechanisms and antimicrobial susceptibility testing. J Clin Microbiol, 2017, 55(7): 1998-2008.
doi: 10.1128/JCM.02250-16 pmid: 28404671 |
| [3] | Owens RC, Donskey CJ, Gaynes RP, Loo VG, Muto CA. Antimicrobial-associated risk factors for clostridium difficile infection. Clin Infect Dis, 2008, 46 Suppl 1: S19-S31. |
| [4] | Khoruts A, Staley C, Sadowsky MJ. Faecal microbiota transplantation for clostridioides difficile: mechanisms and pharmacology. Nat Rev Gastroenterol Hepatol, 2021, 18(1): 67-80. |
| [5] |
Guh AY, Mu Y, Winston LG, Johnston H, Olson D, Farley MM, Wilson LE, Holzbauer SM, Phipps EC, Dumyati GK, Beldavs ZG, Kainer MA, Karlsson M, Gerding DN, McDonald LC, Emerging Infections Program Clostridioides difficile Infection Working Group. Trends in U.S. burden of clostridioides difficile infection and outcomes. N Engl J Med, 2020, 382(14): 1320-1330.
doi: 10.1056/NEJMoa1910215 |
| [6] |
Johnson S, Lavergne V, Skinner AM, Gonzales-Luna AJ, Garey KW, Kelly CP, Wilcox MH. Clinical practice guideline by the infectious diseases society of america (IDSA) and society for healthcare epidemiology of america (SHEA): 2021 focused update guidelines on management of clostridioides difficile infection in adults. Clin Infect Dis, 2021, 73(5): e1029-e1044.
doi: 10.1093/cid/ciab549 pmid: 34164674 |
| [7] | van Prehn J, Reigadas E, Vogelzang EH, Bouza E, Hristea A, Guery B, Krutova M, Norén T, Allerberger F, Coia JE, Goorhuis A, van Rossen TM, Ooijevaar RE, Burns K, Scharvik Olesen BR, Tschudin-Sutter S, Wilcox MH, Vehreschild MJGT, Fitzpatrick F, Kuijper EJ, Guideline Committee of the European Study Group on Clostridioides difficile. European society of clinical microbiology and infectious diseases: 2021 update on the treatment guidance document for clostridioides difficile infection in adults. Clin Microbiol Infect, 2021, 27 Suppl 2: S1-S21. |
| [8] | Petrosillo N, Granata G, Cataldo MA. Novel antimicrobials for the treatment of clostridium difficile infection. Front Med (Lausanne), 2018, 5: 96. |
| [9] |
Kechagias KS, Chorepsima S, Triarides NA, Falagas ME. Tigecycline for the treatment of patients with clostridium difficile infection: an update of the clinical evidence. Eur J Clin Microbiol Infect Dis, 2020, 39(6): 1053-1058.
doi: 10.1007/s10096-019-03756-z |
| [10] |
Major G, Bradshaw L, Boota N, Sprange K, Diggle M, Montgomery A, Jawhari A, Spiller RC, RAPID Collaboration Group. Follow-on rifaximin for the prevention of recurrence following standard treatment of infection with clostridium difficile (RAPID): a randomised placebo controlled trial. Gut, 2019, 68(7): 1224-1231.
doi: 10.1136/gutjnl-2018-316794 |
| [11] |
Ondo WG, Vuong KD, Wang Q. Restless legs syndrome in monozygotic twins: clinical correlates. Neurology, 2000, 55(9): 1404-1406.
pmid: 11087794 |
| [12] |
Tang CJ, Cui LB, Xu YQ, Xie L, Sun PF, Liu CC, Xia WY, Liu GY. The incidence and drug resistance of Clostridium difficile infection in Mainland China: a systematic review and meta-analysis. Sci Rep, 2016, 6: 37865.
doi: 10.1038/srep37865 pmid: 27897206 |
| [13] |
Jin DZ, Luo Y, Huang C, Cai J, Ye JL, Zheng Y, Wang LQ, Zhao P, Liu AB, Fang WJ, Wang XJ, Xia SC, Jiang JM, Tang YW. Molecular epidemiology of clostridium difficile infection in hospitalized patients in eastern china. J Clin Microbiol, 2017, 55(3): 801-810.
doi: 10.1128/JCM.01898-16 pmid: 27974547 |
| [14] |
Gao Q, Huang HH. Update on antimicrobial resistance in clostridium difficile. Hereditas(Beijing), 2015, 37(5): 458-464.
doi: 10.16288/j.yczz.15-131 pmid: 25998434 |
|
高琼, 黄海辉. 艰难梭菌耐药性及耐药机制研究进展. 遗传, 2015, 37(5): 458-464.
doi: 10.16288/j.yczz.15-131 pmid: 25998434 |
|
| [15] |
Pépin J, Valiquette L, Gagnon S, Routhier S, Brazeau I.Outcomes of clostridium difficile-associated disease treated with metronidazole or vancomycin before and after the emergence of NAP1/027. Am J Gastroenterol, 2007, 102(12): 2781-2788.
doi: 10.1111/j.1572-0241.2007.01539.x pmid: 17900327 |
| [16] |
Dingsdag SA, Hunter N. Metronidazole: an update on metabolism, structure-cytotoxicity and resistance mechanisms. J Antimicrob Chemother, 2018, 73(2): 265-279.
doi: 10.1093/jac/dkx351 pmid: 29077920 |
| [17] |
Lynch T, Chong P, Zhang J, Hizon R, Du T, Graham MR, Beniac DR, Booth TF, Kibsey P, Miller M, Gravel D, Mulvey MR, Canadian Nosocomial Infection Surveillance Program (CNISP). Characterization of a stable, metronidazole-resistant clostridium difficile clinical isolate. PLoS One, 2013, 8(1): e53757.
doi: 10.1371/journal.pone.0053757 |
| [18] |
Wellinghausen N, Chatterjee I, Berger A, Niederfuehr A, Proctor RA, Kahl BC. Characterization of clinical enterococcus faecalis small-colony variants. J Clin Microbiol, 2009, 47(9): 2802-2811.
doi: 10.1128/JCM.00485-09 pmid: 19605585 |
| [19] |
Kahl BC, Becker K, Löffler B. Clinical significance and pathogenesis of staphylococcal small colony variants in persistent infections. Clin Microbiol Rev, 2016, 29(2): 401-427.
doi: 10.1128/CMR.00069-15 pmid: 26960941 |
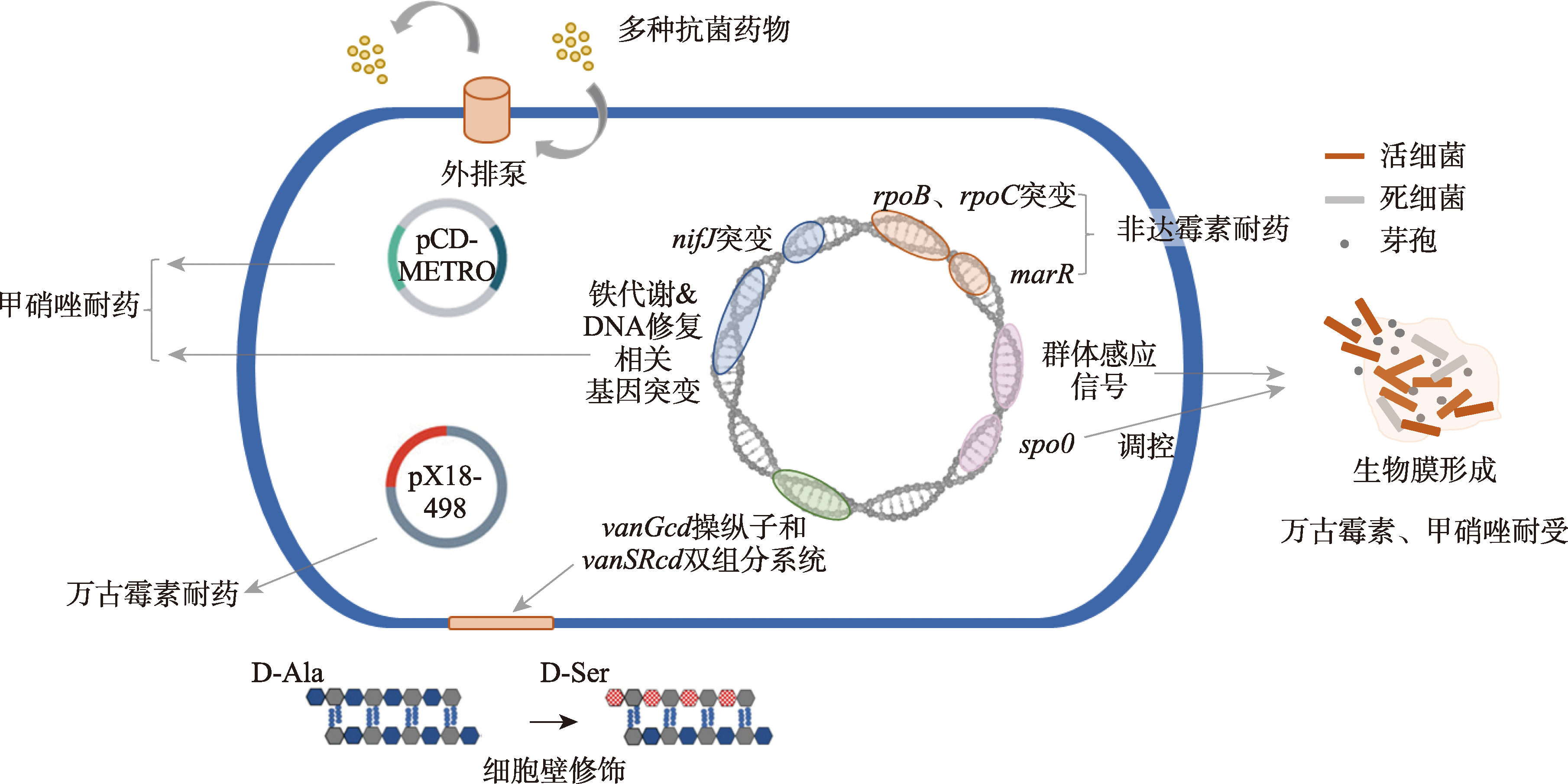
| [20] | Deshpande A, Wu XQ, Huo WW, Palmer KL, Hurdle JG. Chromosomal resistance to metronidazole in clostridioides difficile can be mediated by epistasis between iron homeostasis and oxidoreductases. Antimicrob Agents Chemother, 2020, 64(8): e00415-e00420. |
| [21] |
Moura I, Monot M, Tani C, Spigaglia P, Barbanti F, Norais N, Dupuy B, Bouza E, Mastrantonio P. Multidisciplinary analysis of a nontoxigenic clostridium difficile strain with stable resistance to metronidazole. Antimicrob Agents Chemother, 2014, 58(8): 4957-4960.
doi: 10.1128/AAC.02350-14 pmid: 24913157 |
| [22] |
Arcay RM, Suárez-Bode L, López-Causapé C, Oliver A, Mena A. Emergence of high-level and stable metronidazole resistance in Clostridioides difficile. Int J Antimicrob Agents, 2020, 55(1): 105830.
doi: 10.1016/j.ijantimicag.2019.10.011 |
| [23] |
Olaitan AO, Dureja C, Youngblom MA, Topf MA, Shen WJ, Gonzales-Luna AJ, Deshpande A, Hevener KE, Freeman J, Wilcox MH, Palmer KL, Garey KW, Pepperell CS, Hurdle JG. Decoding a cryptic mechanism of metronidazole resistance among globally disseminated fluoroquinolone-resistant clostridioides difficile. Nat Commun, 2023, 14(1): 4130.
doi: 10.1038/s41467-023-39429-x pmid: 37438331 |
| [24] |
Boekhoud IM, Hornung BVH, Sevilla E, Harmanus C, Bos-Sanders IMJG, Terveer EM, Bolea R, Corver J, Kuijper EJ, Smits WK. Plasmid-mediated metronidazole resistance in clostridioides difficile. Nat Commun, 2020, 11(1): 598.
doi: 10.1038/s41467-020-14382-1 pmid: 32001686 |
| [25] |
Chong PM, Lynch T, Mccorrister S, Kibsey P, Miller M, Gravel D, Westmacott GR, Mulvey MR, Canadian Nosocomial Infection Surveillance Program (CNISP). Proteomic analysis of a NAP1 clostridium difficile clinical isolate resistant to metronidazole. PLoS One, 2014, 9(1): e82622.
doi: 10.1371/journal.pone.0082622 |
| [26] |
Boekhoud IM, Sidorov I, Nooij S, Harmanus C, Bos-Sanders IMJG, Viprey V, Spittal W, Clark E, Davies K, Freeman J, Kuijper EJ, Smits WK, COMBACTE-CDI Consortium. Haem is crucial for medium-dependent metronidazole resistance in clinical isolates of Clostridioides difficile. J Antimicrob Chemother, 2021, 76(7): 1731-1740.
doi: 10.1093/jac/dkab097 pmid: 33876817 |
| [27] |
Gonzales-Luna AJ, Olaitan AO, Shen WJ, Deshpande A, Carlson TJ, Dotson KM, Lancaster C, Begum K, Alam MJ, Hurdle JG, Garey KW. Reduced susceptibility to metronidazole is associated with initial clinical failure in clostridioides difficile infection. Open Forum Infect Dis, 2021, 8(8): ofab365.
doi: 10.1093/ofid/ofab365 |
| [28] |
Zhao HL, Nickle DC, Zeng Z, Law PYT, Wilcox MH, Chen L, Peng Y, Meng J, Deng ZQ, Albright A, Zhong HZ, Xu X, Zhu SD, Shen JD, Blanchard RL, Dorr MB, Shaw PM, Li JH. Global landscape of clostridioides difficile phylogeography, antibiotic susceptibility, and toxin polymorphisms by post-hoc whole-genome sequencing from the MODIFY I/II studies. Infect Dis Ther, 2021, 10(2): 853-870.
doi: 10.1007/s40121-021-00426-6 |
| [29] |
Ahmed MO, Baptiste KE. Vancomycin-Resistant Enterococci: A review of antimicrobial resistance mechanisms and perspectives of human and animal health. Microb Drug Resist, 2018, 24(5): 590-606.
doi: 10.1089/mdr.2017.0147 pmid: 29058560 |
| [30] | Ramírez-Vargas G, Quesada-Gómez C, Acuña-Amador L, López-Ureña D, Murillo T, Del Mar Gamboa-Coronado M, Chaves-Olarte E, Thomson N, Rodríguez-Cavallini E, Rodríguez C. A clostridium difficile lineage endemic to costa rican hospitals is multidrug resistant by acquisition of chromosomal mutations and novel mobile genetic elements. Antimicrob Agents Chemother, 2017, 61(4): e02054-16. |
| [31] |
Shen WJ, Deshpande A, Hevener KE, Endres BT, Garey KW, Palmer KL, Hurdle JG. Constitutive expression of the cryptic vanGCd operon promotes vancomycin resistance in clostridioides difficile clinical isolates. J Antimicrob Chemother, 2020, 75(4): 859-867.
doi: 10.1093/jac/dkz513 |
| [32] |
Saldanha GZ, Pires RN, Rauber AP, De Lima-Morales D, Falci DR, Caierão J, Pasqualotto AC, Martins AF. Genetic relatedness, virulence factors and antimicrobial resistance of C. difficile strains from hospitalized patients in a multicentric study in Brazil. J Glob Antimicrob Resist, 2020, 22: 117-121.
doi: S2213-7165(20)30008-4 pmid: 32006751 |
| [33] |
Depardieu F, Mejean V, Courvalin P. Competition between VanU(G) repressor and VanR(G) activator leads to rheostatic control of vanG vancomycin resistance operon expression. PLoS Genet, 2015, 11(4): e1005170.
doi: 10.1371/journal.pgen.1005170 |
| [34] |
Sebaihia M, Wren BW, Mullany P, Fairweather NF, Minton N, Stabler R, Thomson NR, Roberts AP, Cerdeño-Tárraga AM, Wang HM, Holden MTG, Wright A, Churcher C, Quail MA, Baker S, Bason N, Brooks K, Chillingworth T, Cronin A, Davis P, Dowd L, Fraser A, Feltwell T, Hance Z, Holroyd S, Jagels K, Moule S, Mungall K, Price C, Rabbinowitsch E, Sharp S, Simmonds M, Stevens K, Unwin L, Whithead S, Dupuy B, Dougan G, Barrell B, Parkhill J. The multidrug-resistant human pathogen clostridium difficile has a highly mobile, mosaic genome. Nat Genet, 2006, 38(7): 779-786.
doi: 10.1038/ng1830 pmid: 16804543 |
| [35] |
Wickramage I, Peng Z, Chakraborty S, Harmanus C, Kuijper EJ, Alrabaa S, Smits WK, Sun XM. The vanRCd mutation 343A>G, resulting in a Thr115Ala substitution, is associated with an elevated minimum inhibitory concentration (MIC) of vancomycin in clostridioides difficile clinical isolates from florida. Microbiol Spectr, 2023, 11(3): e0377722.
doi: 10.1128/spectrum.03777-22 |
| [36] |
Liu XD. ABC family transporters. Adv Exp Med Biol, 2019, 1141: 13-100.
doi: 10.1007/978-981-13-7647-4_2 pmid: 31571164 |
| [37] |
Rafii F, Park M. Detection and characterization of an ABC transporter in clostridium hathewayi. Arch Microbiol, 2008, 190(4): 417-426.
doi: 10.1007/s00203-008-0385-3 pmid: 18504552 |
| [38] |
Rafii F, Park M, Carman RJ. Characterization of an ATP-binding cassette from clostridium perfringens with homology to an ABC transporter from clostridium hathewayi. Anaerobe, 2009, 15(4): 116-121.
pmid: 19655423 |
| [39] |
Mcbride SM, Sonenshein AL. The dlt operon confers resistance to cationic antimicrobial peptides in clostridium difficile. Microbiology (Reading), 2011, 157(Pt 5): 1457-1465.
doi: 10.1099/mic.0.045997-0 pmid: 21330441 |
| [40] |
Ngernsombat C, Sreesai S, Harnvoravongchai P, Chankhamhaengdecha S, Janvilisri T. CD 2068 potentially mediates multidrug efflux in clostridium difficile. Sci Rep, 2017, 7(1): 9982.
doi: 10.1038/s41598-017-10155-x |
| [41] |
Dannheim H, Riedel T, Neumann-Schaal M, Bunk B, Schober I, Spröer C, Chibani CM, Gronow S, Liesegang H, Overmann J, Schomburg D.Manual curation and reannotation of the genomes of Clostridium difficile 630Δerm and C. difficile 630. J Med Microbiol, 2017, 66(3): 286-293.
doi: 10.1099/jmm.0.000427 pmid: 28357980 |
| [42] |
Zhou QS, Rao FQ, Chen ZH, Cheng YM, Zhang QF, Zhang J, Guan ZZ, He Y, Yu WF, Cui GZ, Qi XL, Hong W. The cwp 66 gene affects cell adhesion, stress tolerance, and antibiotic resistance in clostridioides difficile. Microbiol Spectr, 2022, 10(2): e0270421.
doi: 10.1128/spectrum.02704-21 |
| [43] |
Yan J, Bassler BL. Surviving as a community: antibiotic tolerance and persistence in bacterial biofilms. Cell Host Microbe, 2019, 26(1): 15-21.
doi: S1931-3128(19)30291-4 pmid: 31295420 |
| [44] |
Jarrad AM, Blaskovich MAT, Prasetyoputri A, Karoli T, Hansford KA, Cooper MA. Detection and investigation of eagle effect resistance to vancomycin in clostridium difficile with an ATP-Bioluminescence assay. Front Microbiol, 2018, 9: 1420.
doi: 10.3389/fmicb.2018.01420 pmid: 30013531 |
| [45] |
Wu XQ, Cherian PT, Lee RE, Hurdle JG. The membrane as a target for controlling hypervirulent clostridium difficile infections. J Antimicrob Chemother, 2013, 68(4): 806-815.
doi: 10.1093/jac/dks493 pmid: 23264511 |
| [46] |
Ðapa T, Leuzzi R, Ng YK, Baban ST, Adamo R, Kuehne SA, Scarselli M, Minton NP, Serruto D, Unnikrishnan M. Multiple factors modulate biofilm formation by the anaerobic pathogen clostridium difficile. J Bacteriol, 2013, 195(3): 545-555.
doi: 10.1128/JB.01980-12 pmid: 23175653 |
| [47] |
Tijerina-Rodríguez L, Villarreal-Treviño L, Baines SD, Morfín-Otero R, Camacho-Ortíz A, Flores-Treviño S, Maldonado-Garza H, Rodríguez-Noriega E, Garza- González E. High sporulation and overexpression of virulence factors in biofilms and reduced susceptibility to vancomycin and linezolid in recurrent clostridium [clostridioides] difficile infection isolates. PLoS One, 2019, 14(7): e0220671.
doi: 10.1371/journal.pone.0220671 |
| [48] |
Pu M, Cho JM, Cunningham SA, Behera GK, Becker S, Amjad T, Greenwood-Quaintance KE, Mendes-Soares H, Jones-Hall Y, Jeraldo PR, Chen J, Dunny G, Patel R, Kashyap PC. Plasmid acquisition alters vancomycin susceptibility in clostridioides difficile. Gastroenterology, 2021, 160(3): 941-945.e8.
doi: 10.1053/j.gastro.2020.10.046 pmid: 33197449 |
| [49] |
Lin W, Das K, Degen D, Mazumder A, Duchi D, Wang DY, Ebright YW, Ebright RY, Sineva E, Gigliotti M, Srivastava A, Mandal S, Jiang Y, Liu Y, Yin RH, Zhang ZN, Eng ET, Thomas D, Donadio S, Zhang HB, Zhang CS, Kapanidis AN, Ebright RH. Structural basis of transcription inhibition by fidaxomicin (lipiarmycin A3). Mol Cell, 2018, 70(1): 60-71.e15.
doi: S1097-2765(18)30171-0 pmid: 29606590 |
| [50] |
Goldstein EJC, Citron DM, Sears P, Babakhani F, Sambol SP, Gerding DN. Comparative susceptibilities to fidaxomicin (OPT-80) of isolates collected at baseline, recurrence, and failure from patients in two phase III trials of fidaxomicin against clostridium difficile infection. Antimicrob Agents Chemother, 2011, 55(11): 5194-5199.
doi: 10.1128/AAC.00625-11 pmid: 21844318 |
| [51] |
Babakhani F, Gomez A, Robert N, Sears P. Killing kinetics of fidaxomicin and its major metabolite, OP-1118, against clostridium difficile. J Med Microbiol, 2011, 60(Pt 8): 1213-1217.
doi: 10.1099/jmm.0.029470-0 pmid: 21349983 |
| [52] |
Schwanbeck J, Riedel T, Laukien F, Schober I, Oehmig I, Zimmermann O, Vermann J, Groß U, Zautner A E, Bohne W. Characterization of a clinical clostridioides difficile isolate with markedly reduced fidaxomicin susceptibility and a V1143D mutation in rpoB. J Antimicrob Chemother, 2019, 74(1): 6-10.
doi: 10.1093/jac/dky375 pmid: 30247587 |
| [53] |
Kuehne SA, Dempster AW, Collery MM, Joshi N, Jowett J, Kelly ML, Cave R, Longshaw CM, Minton NP. Characterization of the impact of rpoB mutations on the in vitro and in vivo competitive fitness of clostridium difficile and susceptibility to fidaxomicin. J Antimicrob Chemother, 2018, 73(4): 973-980.
doi: 10.1093/jac/dkx486 pmid: 29253242 |
| [54] |
Louie TJ, Miller MA, Mullane KM, Weiss K, Lentnek A, Golan Y, Gorbach S, Sears P, Shue YK, OPT-80-003 Clinical Study Group. Fidaxomicin versus vancomycin for clostridium difficile infection. N Engl J Med, 2011, 364(5): 422-431.
doi: 10.1056/NEJMoa0910812 |
| [55] |
Finegold SM, Molitoris D, Vaisanen ML, Song YL, Liu CX, Bolaños M. In vitro activities of OPT-80 and comparator drugs against intestinal bacteria. Antimicrob Agents Chemother, 2004, 48(12): 4898-4902.
pmid: 15561877 |
| [56] |
Leeds JA, Sachdeva M, Mullin S, Barnes SW, Ruzin A. In vitro selection, via serial passage, of clostridium difficile mutants with reduced susceptibility to fidaxomicin or vancomycin. J Antimicrob Chemother, 2014, 69(1): 41-44.
doi: 10.1093/jac/dkt302 pmid: 23887866 |
| [57] |
Carman RJ, Boone JH, Grover H, Wickham KN, Chen L. In vivo selection of rifamycin-resistant clostridium difficile during rifaximin therapy. Antimicrob Agents Chemother, 2012, 56(11): 6019-6020.
doi: 10.1128/AAC.00974-12 pmid: 22908175 |
| [58] |
Ng QX, Loke W, Foo NX, Mo Y, Yeo WS, Soh AYS. A systematic review of the use of rifaximin for clostridium difficile infections. Anaerobe, 2019, 55: 35-39.
doi: S1075-9964(18)30183-5 pmid: 30391527 |
| [59] | Beauduy C, Macdougall C. Update on management of clostridium difficile infection. Hosp Pharm, 2013, 48(s1): S7-S13. |
| [60] | 程敬伟, 刘文恩, 马小军, 肖盟, 张丽, 张莉萍, 赵建宏, 卓超. 中国成人艰难梭菌感染诊断和治疗专家共识. 协和医学杂志, 2017, 8(Z1): 131-138. |
| [61] | Baines SD, Wilcox MH.Antimicrobial resistance and reduced susceptibility in clostridium difficile: potential consequences for induction, treatment, and recurrence of c. difficile infection. Antibiotics (Basel), 2015, 4(3): 267-298. |
| [62] |
Sholeh M, Krutova M, Forouzesh M, Mironov S, Sadeghifard N, Molaeipour L, Maleki A, Kouhsari E. Antimicrobial resistance in clostridioides (clostridium) difficile derived from humans: a systematic review and meta-analysis. Antimicrob Resist Infect Control, 2020, 9(1): 158.
doi: 10.1186/s13756-020-00815-5 |
| [63] |
Gasparrini AJ, Markley JL, Kumar H, Wang B, Fang LT, Irum S, Symister CT, Wallace M, Burnham CAD, Andleeb S, Tolia NH, Wencewicz TA, Dantas G. Tetracycline- inactivating enzymes from environmental, human commensal, and pathogenic bacteria cause broad-spectrum tetracycline resistance. Commun Biol, 2020, 3(1): 241.
doi: 10.1038/s42003-020-0966-5 pmid: 32415166 |
| [64] | Dingle KE, Didelot X, Quan TP, Eyre DW, Stoesser N, Marwick CA, Coia J, Brown D, Buchanan S, Ijaz UZ, Goswami C, Douce G, Fawley WN, Wilcox MH, Peto TEA, Walker AS, Crook DW.A role for tetracycline selection in recent evolution of agriculture- associated clostridium difficile PCR ribotype 078. mBio, 2019, 10(2): e02790-18. |
| [65] |
Collins DA, Riley TV. Ridinilazole: a novel, narrow- spectrum antimicrobial agent targeting Clostridium (Clostridioides) difficile. Lett Appl Microbiol, 2022, 75(3): 526-536.
doi: 10.1111/lam.13664 |
| [66] |
Thorpe CM, Kane AV, Chang J, Tai A, Vickers RJ, Snydman DR. Enhanced preservation of the human intestinal microbiota by ridinilazole, a novel clostridium difficile-targeting antibacterial, compared to vancomycin. PLoS One, 2018, 13(8): e0199810.
doi: 10.1371/journal.pone.0199810 |
| [67] |
Hind C, Clifford M, Woolley C, Harmer J, McGee LMC, Tyson-Hirst I, Tait HJ, Brooke DP, Dancer SJ, Hunter IS, Suckling CJ, Beveridge R, Parkinson JA, Sutton JM, Scott FJ.Insights into the spectrum of activity and mechanism of action of MGB-BP-3. ACS Infect Dis, 2022, 8(12): 2552-2563.
doi: 10.1021/acsinfecdis.2c00445 pmid: 36444998 |
| [68] |
Murray B, Wolfe C, Marra A, Pillar C, Shinabarger D. In vitro activity of the novel antibacterial agent ibezapolstat (ACX-362E) against clostridioides difficile. J Antimicrob Chemother, 2020, 75(8): 2149-2155.
doi: 10.1093/jac/dkaa134 pmid: 32285102 |
| [69] |
Alshrari AS, Hudu SA, Elmigdadi F, Imran M. The urgent threat of clostridioides difficile infection: a glimpse of the drugs of the future, with related patents and prospects. Biomedicines, 2023, 11(2): 426.
doi: 10.3390/biomedicines11020426 |
| [70] |
Kullar R, Tran MCN, Goldstein EJC. Investigational treatment agents for recurrent clostridioides difficile infection (rCDI). J Exp Pharmacol, 2020, 12: 371-384.
doi: 10.2147/JEP.S242959 |
| [71] |
Butler MS, Gigante V, Sati H, Paulin S, Al-Sulaiman L, Rex JH, Fernandes P, Arias CA, Paul M, Thwaites GE, Czaplewski L, Alm RA, Lienhardt C, Spigelman M, Silver LL, Ohmagari N, Kozlov R, Harbarth S, Beyer P. Analysis of the clinical pipeline of treatments for drug-resistant bacterial infections: despite progress, more action is needed. Antimicrob Agents Chemother, 2022, 66(3): e0199121.
doi: 10.1128/aac.01991-21 |
| [1] | Hongju Zhu,Wenge Liu. Progress on salt resistance in autopolyploid plants [J]. Hereditas(Beijing), 2018, 40(4): 315-326. |
| [2] | Qiong Gao, Haihui Huang. Update on antimicrobial resistance in Clostridium difficile [J]. HEREDITAS(Beijing), 2015, 37(5): 458-464. |
| [3] | Chunhui Chen, Xiaogang Xu. Genetic characteristics of vancomycin resistance gene cluster in Enterococcus spp [J]. HEREDITAS(Beijing), 2015, 37(5): 452-457. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||