Hereditas(Beijing) ›› 2023, Vol. 45 ›› Issue (11): 1018-1027.doi: 10.16288/j.yczz.23-236
• Review • Previous Articles Next Articles
Progress on the non-canonical mismatch repair in Mycobacterium and its role in antibiotic resistance
Shasha Xiang( ), Jianping Xie(
), Jianping Xie( )
)
- Institute of Modern Biopharmaceuticals, School of Life Sciences, Southwest University, Chongqing 400715, China
-
Received:2023-09-11Revised:2023-10-29Online:2023-11-20Published:2023-11-03 -
Contact:Jianping Xie E-mail:1977015429@qq.com;georgex@swu.edu.cn -
Supported by:National Natural Science Foundation of China(82072246)
Cite this article
Shasha Xiang, Jianping Xie. Progress on the non-canonical mismatch repair in Mycobacterium and its role in antibiotic resistance[J]. Hereditas(Beijing), 2023, 45(11): 1018-1027.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
Table 1
Summary of key components and functions of canonical mismatch repair and non-canonical mismatch repair"
| 修复类型 | 生物种类 | 关键成分 | 功能 |
|---|---|---|---|
| 细菌典型错配修复 | 大肠杆菌E. coli | MutS | 识别错配DNA |
| MutL | 下游介质 | ||
| MutH | 内切酶 | ||
| UvrD | 解旋酶 | ||
| Exo1/ExoX | 3′→5′核酸外切酶 | ||
| ExoVII/RecJ | 5′→3′核酸外切酶 | ||
| DNA聚合酶III | 聚合酶 | ||
| 甲基化酶Dam酶 | DNA甲基化酶 | ||
| SSB结合蛋白 | 稳定DNA的单链区域,增强DNA聚合酶的活性 | ||
| DNA连接酶 | 连接酶 | ||
| 真核生物典型错配修复 | 真核生物 | MutSα/MutSβ | 识别错配DNA |
| MutLα | 下游介质;核酸内切酶 | ||
| Exo1 | 外切酶 | ||
| DNA聚合酶δ | 聚合酶 | ||
| PCNA/RFC | 激活MutLα活性 | ||
| DNA连接酶 | 连接酶 | ||
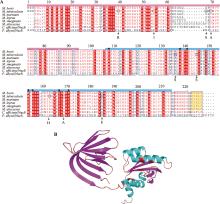
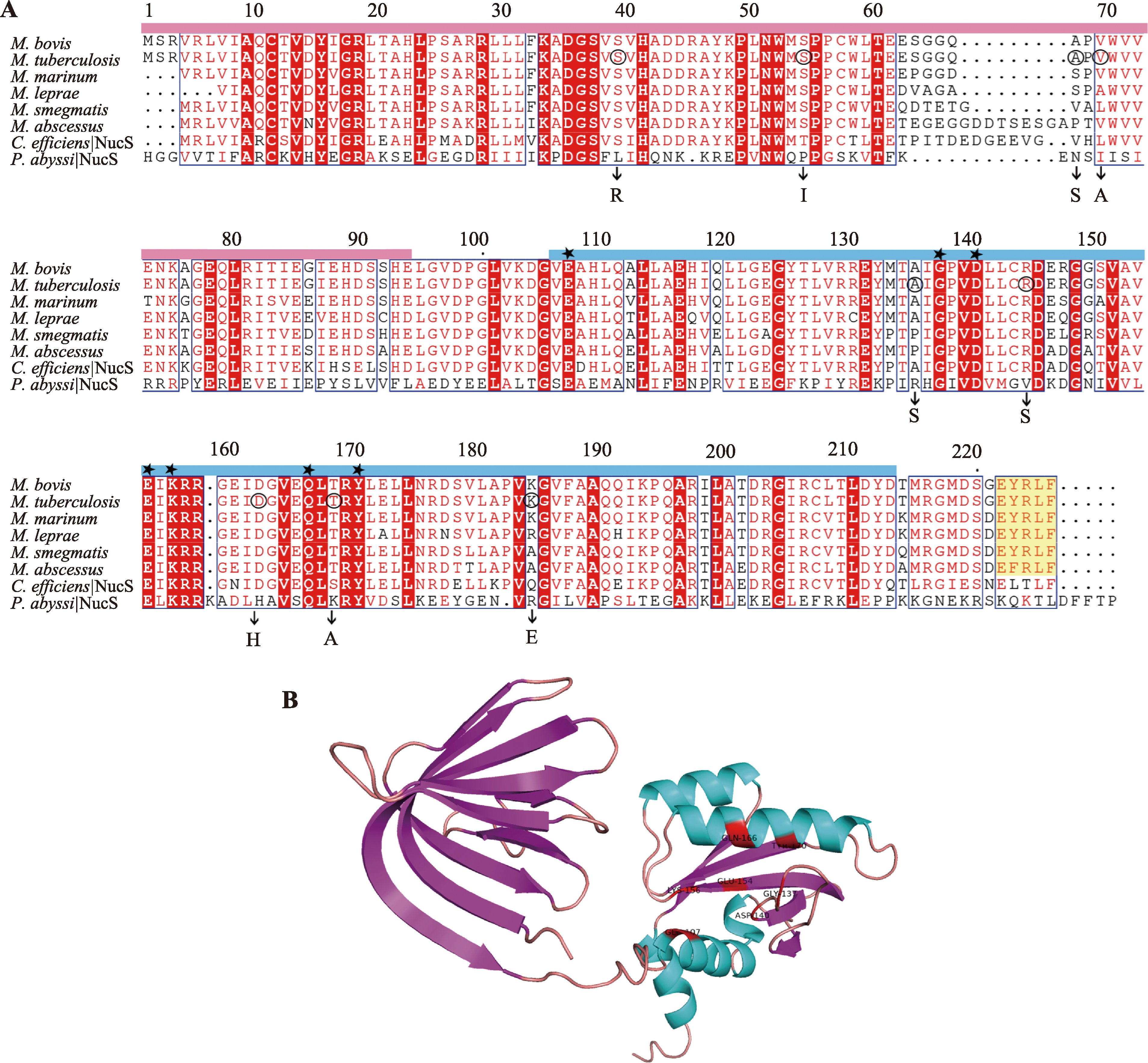
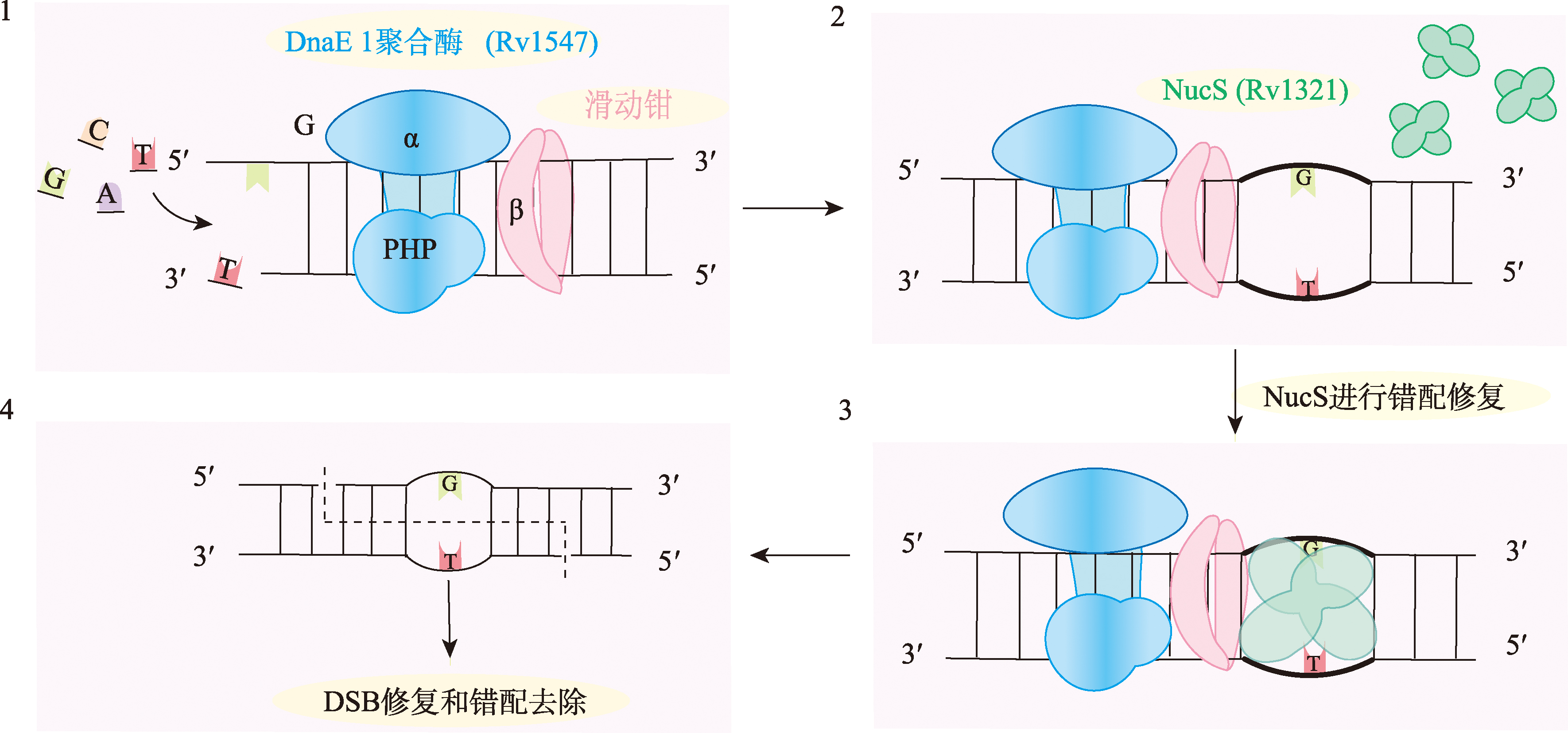
| 非典型错配修复 | 分枝杆菌 | NucS | 识别错配碱基,核酸内切酶 |
| DnaE1聚合酶 | DNA聚合酶 |
| [1] |
Chatterjee N, Walker GC. Mechanisms of DNA damage, repair, and mutagenesis. Environ Mol Mutagen, 2017, 58(5): 235-263.
doi: 10.1002/em.22087 pmid: 28485537 |
| [2] |
Olave MC, Graham RP. Mismatch repair deficiency: the what, how and why it is important. Genes Chromosomes Cancer, 2022, 61(6): 314-321.
doi: 10.1002/gcc.v61.6 |
| [3] |
Lenhart JS, Pillon MC, Guarné A, Biteen JS, Simmons LA. Mismatch repair in Gram-positive bacteria. Res Microbiol, 2016, 167(1): 4-12.
doi: 10.1016/j.resmic.2015.08.006 pmid: 26343983 |
| [4] |
Dos Vultos T, Mestre O, Tonjum T, Gicquel B. DNA repair in Mycobacterium tuberculosis revisited. FEMS Microbiol Rev, 2009, 33(3): 471-487.
doi: 10.1111/j.1574-6976.2009.00170.x pmid: 19385996 |
| [5] |
Fishel R. Mismatch repair. J Biol Chem, 2015, 290(44): 26395-26403.
doi: 10.1074/jbc.R115.660142 pmid: 26354434 |
| [6] |
Hsieh P, Yamane K. DNA mismatch repair: molecular mechanism, cancer, and ageing. Mech Ageing Dev, 2008, 129(7-8): 391-407.
doi: 10.1016/j.mad.2008.02.012 pmid: 18406444 |
| [7] |
Putnam CD. Evolution of the methyl directed mismatch repair system in Escherichia coli. DNA Repair (Amst), 2016, 38: 32-41.
doi: 10.1016/j.dnarep.2015.11.016 |
| [8] |
Barras F, Marinus MG. The great GATC: DNA methylation in E. coli. Trends Genet, 1989, 5(5): 139-43.
pmid: 2667217 |
| [9] |
Hu CK, Zhao YQ, Sun HY, Yang YX.Synergism of Dam, MutH, and MutS in methylation-directed mismatch repair in Escherichia coli. Mutat Res, 2017, 795: 31-33.
doi: 10.1016/j.mrfmmm.2016.12.002 |
| [10] |
Groothuizen FS, Sixma TK. The conserved molecular machinery in DNA mismatch repair enzyme structures. DNA Repair (Amst), 2016, 38: 14-23.
doi: 10.1016/j.dnarep.2015.11.012 |
| [11] |
Junop MS, Yang W, Funchain P, Clendenin W, Miller JH. In vitro and in vivo studies of MutS, MutL and MutH mutants: correlation of mismatch repair and DNA recombination. DNA Repair (Amst), 2003, 2(4): 387-405.
doi: 10.1016/S1568-7864(02)00245-8 |
| [12] |
Jeong C, Cho WK, Song KM, Cook C, Yoon TY, Ban C, Fishel R, Lee JB. MutS switches between two fundamentally distinct clamps during mismatch repair. Nat Struct Mol Biol, 2011, 18(3): 379-385.
doi: 10.1038/nsmb.2009 pmid: 21278758 |
| [13] |
Au KG, Welsh K, Modrich P. Initiation of methyl-directed mismatch repair. J Biol Chem, 1992, 267(17): 12142-12148.
pmid: 1601880 |
| [14] |
Yamaguchi M, Dao V, Modrich P. MutS and MutL activate DNA helicase II in a mismatch-dependent manner. J Biol Chem, 1998, 273(15): 9197-9201.
doi: 10.1074/jbc.273.15.9197 pmid: 9535910 |
| [15] |
Grilley M, Griffith J, Modrich P. Bidirectional excision in methyl-directed mismatch repair. J Biol Chem, 1993, 268(16): 11830-11837.
pmid: 8505311 |
| [16] |
Modrich P, Lahue R. Mismatch repair in replication fidelity, genetic recombination, and cancer biology. Annu Rev Biochem, 1996, 65: 101-133.
pmid: 8811176 |
| [17] |
Hsieh P. Molecular mechanisms of DNA mismatch repair. Mutat Res, 2001, 486(2): 71-87.
doi: 10.1016/S0921-8777(01)00088-X |
| [18] |
Liu DK, Keijzers G, Rasmussen LJ. DNA mismatch repair and its many roles in eukaryotic cells. Mutat Res Rev Mutat Res, 2017, 773: 174-187.
doi: 10.1016/j.mrrev.2017.07.001 pmid: 28927527 |
| [19] |
Schroering AG, Edelbrock MA, Richards TJ, Williams KJ. The cell cycle and DNA mismatch repair. Exp Cell Res, 2007, 313(2): 292-304.
pmid: 17157834 |
| [20] |
Iyer RR, Pluciennik A, Burdett V, Modrich PL. DNA mismatch repair: functions and mechanisms. Chem Rev, 2006, 106(2): 302-323.
pmid: 16464007 |
| [21] |
McCulloch SD, Gu LY, Li GM. Bi-directional processing of DNA loops by mismatch repair-dependent and -independent pathways in human cells. J Biol Chem, 2003, 278(6): 3891-3896.
doi: 10.1074/jbc.M210687200 pmid: 12458199 |
| [22] |
Manhart CM, Alani E. Roles for mismatch repair family proteins in promoting meiotic crossing over. DNA Repair (Amst), 2016, 38: 84-93.
doi: 10.1016/j.dnarep.2015.11.024 |
| [23] |
Plotz G, Raedle J, Brieger A, Trojan J, Zeuzem S. hMutSalpha forms an ATP-dependent complex with hMutLalpha and hMutLbeta on DNA. Nucleic Acids Res, 2002, 30(3): 711-718.
pmid: 11809883 |
| [24] |
Kadyrov FA, Dzantiev L, Constantin N, Modrich P. Endonucleolytic function of MutLalpha in human mismatch repair. Cell, 2006, 126(2): 297-308.
pmid: 16873062 |
| [25] |
Tran PT, Erdeniz N, Symington LS, Liskay RM. EXO1-A multi-tasking eukaryotic nuclease. DNA Repair (Amst), 2004, 3(12): 1549-1559.
doi: 10.1016/j.dnarep.2004.05.015 |
| [26] |
Nielsen FC, Jäger AC, Lützen A, Bundgaard JR, Rasmussen LJ. Characterization of human exonuclease 1 in complex with mismatch repair proteins, subcellular localization and association with PCNA. Oncogene, 2004, 23(7): 1457-1468.
pmid: 14676842 |
| [27] |
Longley MJ, Pierce AJ, Modrich P. DNA polymerase delta is required for human mismatch repair in vitro. J Biol Chem, 1997, 272(16): 10917-10921.
doi: 10.1074/jbc.272.16.10917 pmid: 9099749 |
| [28] |
Ban C, Junop M, Yang W. Transformation of MutL by ATP binding and hydrolysis: a switch in DNA mismatch repair. Cell, 1999, 97(1): 85-97.
pmid: 10199405 |
| [29] |
Mizrahi V, Andersen SJ. DNA repair in Mycobacterium tuberculosis. What have we learnt from the genome sequence?. Mol Microbiol, 1998, 29(6): 1331-1339.
pmid: 9781872 |
| [30] |
Ren B, Kühn J, Meslet-Cladiere L, Briffotaux J, Norais C, Lavigne R, Flament D, Ladenstein R, Myllykallio H. Structure and function of a novel endonuclease acting on branched DNA substrates. EMBO J, 2009, 28(16): 2479-2489.
doi: 10.1038/emboj.2009.192 pmid: 19609302 |
| [31] |
Castañeda-García A, Prieto AI, Rodríguez-Beltrán J, Alonso N, Cantillon D, Costas C, Pérez-Lago L, Zegeye ED, Herranz M, Plociński P, Tonjum T, García de Viedma D, Paget M, Waddell SJ, Rojas AM, Doherty AJ, Blázquez J. A non-canonical mismatch repair pathway in prokaryotes. Nat Commun, 2017, 8: 14246.
doi: 10.1038/ncomms14246 pmid: 28128207 |
| [32] |
Meslet-Cladiére L, Norais C, Kuhn J, Briffotaux J, Sloostra JW, Ferrari E, Hübscher U, Flament D, Myllykallio H. A novel proteomic approach identifies new interaction partners for proliferating cell nuclear antigen. J Mol Biol, 2007, 372(5): 1137-1148.
pmid: 17720188 |
| [33] |
Ishino S, Nishi Y, Oda S, Uemori T, Sagara T, Takatsu N, Yamagami T, Shirai T, Ishino Y. Identification of a mismatch-specific endonuclease in hyperthermophilic Archaea. Nucleic Acids Res, 2016, 44(7): 2977-2986.
doi: 10.1093/nar/gkw153 pmid: 27001046 |
| [34] |
Cebrián-Sastre E, Martín-Blecua I, Gullón S, Blázquez J, Castañeda-García A. Control of genome stability by EndoMS/NucS-mediated non-canonical mismatch repair. Cells, 2021, 10(6): 1314.
doi: 10.3390/cells10061314 |
| [35] |
Nakae S, Hijikata A, Tsuji T, Yonezawa K, Kouyama KI, Mayanagi K, Ishino S, Ishino Y, Shirai T. Structure of the EndoMS-DNA complex as mismatch restriction endonuclease. Structure, 2016, 24(11): 1960-1971.
doi: S0969-2126(16)30294-5 pmid: 27773688 |
| [36] |
Pingoud A, Fuxreiter M, Pingoud V, Wende W. Type II restriction endonucleases: structure and mechanism. Cell Mol Life Sci, 2005, 62(6): 685-707.
pmid: 15770420 |
| [37] |
Takemoto N, Numata I, Su'etsugu M, Miyoshi-Akiyama T. Bacterial EndoMS/NucS acts as a clamp-mediated mismatch endonuclease to prevent asymmetric accumulation of replication errors. Nucleic Acids Res, 2018, 46(12): 6152-6165.
doi: 10.1093/nar/gky481 pmid: 29878158 |
| [38] |
Kunkel TA, Bebenek K. DNA replication fidelity. Annu Rev Biochem, 2000, 69: 497-529.
pmid: 10966467 |
| [39] | Xie ZH. The fidelity mechanism of DNA synthesis. Hereditas(Beijing), 2012, 34(6): 679-686. |
| 谢兆辉. DNA合成的忠实性机制. 遗传, 2012, 34(6): 679-686. | |
| [40] |
Rock JM, Lang UF, Chase MR, Ford CB, Gerrick ER, Gawande R, Coscolla M, Gagneux S, Fortune SM, Lamers MH. DNA replication fidelity in Mycobacterium tuberculosis is mediated by an ancestral prokaryotic proofreader. Nat Genet, 2015, 47(6): 677-681.
doi: 10.1038/ng.3269 pmid: 25894501 |
| [41] |
Grabowski B, Kelman Z. Archeal DNA replication: eukaryal proteins in a bacterial context. Annu Rev Microbiol, 2003, 57: 487-516.
pmid: 14527289 |
| [42] |
Creze C, Ligabue A, Laurent S, Lestini R, Laptenok SP, Khun J, Vos MH, Czjzek M, Myllykallio H, Flament D. Modulation of the Pyrococcus abyssi NucS endonuclease activity by replication clamp at functional and structural levels. J Biol Chem, 2012, 287(19): 15648-15660.
doi: 10.1074/jbc.M112.346361 pmid: 22431731 |
| [43] |
Ishino S, Skouloubris S, Kudo H l'Hermitte-Stead C, Es-Sadik A, Lambry JC, Ishino Y, Myllykallio H. Activation of the mismatch-specific endonuclease EndoMS/NucS by the replication clamp is required for high fidelity DNA replication. Nucleic Acids Res, 2018, 46(12): 6206-6217.
doi: 10.1093/nar/gky460 pmid: 29846672 |
| [44] |
Castañeda-García A, Martín-Blecua I, Cebrián-Sastre E, Chiner-Oms A, Torres-Puente M, Comas I, Blázquez J. Specificity and mutagenesis bias of the mycobacterial alternative mismatch repair analyzed by mutation accumulation studies. Sci Adv, 2020, 6(7): eaay4453.
doi: 10.1126/sciadv.aay4453 |
| [45] |
Garibyan L, Huang T, Kim M, Wolff E, Nguyen A, Nguyen T, Diep A, Hu K, Iverson A, Yang H, Miller JH. Use of the rpoB gene to determine the specificity of base substitution mutations on the Escherichia coli chromosome. DNA Repair (Amst), 2003, 2(5): 593-608.
doi: 10.1016/S1568-7864(03)00024-7 |
| [46] |
Schofield MJ, Hsieh P. DNA mismatch repair: molecular mechanisms and biological function. Annu Rev Microbiol, 2003, 57: 579-608.
pmid: 14527292 |
| [47] |
Tham KC, Kanaar R, Lebbink JHG. Mismatch repair and homeologous recombination. DNA Repair (Amst), 2016, 38: 75-83.
doi: 10.1016/j.dnarep.2015.11.010 |
| [48] |
Tham KC, Hermans N, Winterwerp HHK, Cox MM, Wyman C, Kanaar R, Lebbink JHG. Mismatch repair inhibits homeologous recombination via coordinated directional unwinding of trapped DNA structures. Mol Cell, 2013, 51(3): 326-337.
doi: 10.1016/j.molcel.2013.07.008 |
| [49] |
Worth L Jr, Clark S, Radman M, Modrich P. Mismatch repair proteins MutS and MutL inhibit RecA-catalyzed strand transfer between diverged DNAs. Proc Natl Acad Sci USA, 1994, 91(8): 3238-3241.
pmid: 8159731 |
| [50] |
Spies M, Fishel R. Mismatch repair during homologous and homeologous recombination. Cold Spring Harb Perspect Biol, 2015, 7(3): a022657.
doi: 10.1101/cshperspect.a022657 |
| [51] |
Matic I, Radman M, Taddei F, Picard B, Doit C, Bingen E, Denamur E, Elion J. Highly variable mutation rates in commensal and pathogenic Escherichia coli. Science, 1997, 277(5333): 1833-1834.
doi: 10.1126/science.277.5333.1833 pmid: 9324769 |
| [52] |
Taddei F, Radman M, Maynard-Smith J, Toupance B, Gouyon PH, Godelle B. Role of mutator alleles in adaptive evolution. Nature, 1997, 387(6634): 700-702.
doi: 10.1038/42696 |
| [53] |
Gutierrez A, Laureti L, Crussard S, Abida H, Rodríguez- Rojas A, Blázquez J, Baharoglu Z, Mazel D, Darfeuille F, Vogel J, Matic I. β-Lactam antibiotics promote bacterial mutagenesis via an RpoS-mediated reduction in replication fidelity. Nat Commun, 2013, 4: 1610.
doi: 10.1038/ncomms2607 pmid: 23511474 |
| [54] |
Oliver A, Mena A. Bacterial hypermutation in cystic fibrosis, not only for antibiotic resistance. Clin Microbiol Infect, 2010, 16(7): 798-808.
doi: 10.1111/j.1469-0691.2010.03250.x |
| [55] |
Gagneux S. Ecology and evolution of Mycobacterium tuberculosis. Nat Rev Microbiol, 2018, 16(4): 202-213.
doi: 10.1038/nrmicro.2018.8 pmid: 29456241 |
| [56] |
Ford CB, Shah RR, Maeda MK, Gagneux S, Murray MB, Cohen T, Johnston JC, Gardy J, Lipsitch M, Fortune SM. Mycobacterium tuberculosis mutation rate estimates from different lineages predict substantial differences in the emergence of drug-resistant tuberculosis. Nat Genet, 2013, 45(7): 784-790.
doi: 10.1038/ng.2656 pmid: 23749189 |
| [57] |
Fressatti Cardoso R, Martín-Blecua I, Pietrowski Baldin V, Meneguello JE, Valverde JR, Blázquez J, Castañeda-García A. Noncanonical mismatch repair protein NucS modulates the emergence of antibiotic resistance in Mycobacterium abscessus. Microbiol Spectr, 2022, 10(6): e0222822.
doi: 10.1128/spectrum.02228-22 |
| [58] |
Daley CL, Iaccarino JM, Lange C, Cambau E, Wallace RJ Jr, Andrejak C, Böttger EC, Brozek J, Griffith DE, Guglielmetti L, Huitt GA, Knight SL, Leitman P, Marras TK, Olivier KN, Santin M, Stout JE, Tortoli E, van Ingen J, Wagner D, Winthrop KL. Treatment of nontuberculous mycobacterial pulmonary disease: an official ATS/ERS/ESCMID/IDSA clinical practice guideline. Eur Respir J, 2020, 56(1): 2000535.
doi: 10.1183/13993003.00535-2020 |
| [59] |
Boshoff HI, Myers TG, Copp BR, McNeil MR, Wilson MA, Barry CE 3rd. The transcriptional responses of Mycobacterium tuberculosis to inhibitors of metabolism: novel insights into drug mechanisms of action. J Biol Chem, 2004, 279(38): 40174-40184.
doi: 10.1074/jbc.M406796200 pmid: 15247240 |
| No related articles found! |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||