Hereditas(Beijing) ›› 2024, Vol. 46 ›› Issue (10): 779-794.doi: 10.16288/j.yczz.24-203
• Review • Previous Articles Next Articles
Advances in high throughput sequencing methods for DNA damage and repair
- Key Laboratory of Multi-Cell Systems, Shanghai Institute of Biochemistry and Cell Biology, Center for Excellence in Molecular Cell Science, Chinese Academy of Sciences, Shanghai 200031, China
-
Received:2024-07-04Revised:2024-09-21Online:2024-09-23Published:2024-09-23 -
Contact:Wei Wu E-mail:liangyu2022@sibcb.ac.cn;wuw@sibcb.ac.cn -
Supported by:National Natural Science Foundation of China(32370575)
Cite this article
Yu Liang, Wei Wu. Advances in high throughput sequencing methods for DNA damage and repair[J]. Hereditas(Beijing), 2024, 46(10): 779-794.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
| [1] | Pommier Y, Nussenzweig A, Takeda S, Austin C. Human topoisomerases and their roles in genome stability and organization. Nat Rev Mol Cell Biol, 2022, 23(6): 407-427. |
| [2] |
Chatterjee N, Walker GC. Mechanisms of DNA damage, repair, and mutagenesis. Environ Mol Mutagen, 2017, 58(5): 235-263.
doi: 10.1002/em.22087 pmid: 28485537 |
| [3] |
Tubbs A, Nussenzweig A. Endogenous DNA damage as a source of genomic instability in cancer. Cell, 2017, 168(4): 644-656.
doi: S0092-8674(17)30005-3 pmid: 28187286 |
| [4] | Iyama T, Wilson DM 3rd. DNA repair mechanisms in dividing and non-dividing cells. DNA Repair (Amst), 2013, 12(8): 620-636. |
| [5] |
Caldecott KW, Ward ME, Nussenzweig A. The threat of programmed DNA damage to neuronal genome integrity and plasticity. Nat Genet, 2022, 54(2): 115-120.
doi: 10.1038/s41588-021-01001-y pmid: 35145299 |
| [6] | Groelly FJ, Fawkes M, Dagg RA, Blackford AN, Tarsounas M. Targeting DNA damage response pathways in cancer. Nat Rev Cancer, 2023, 23(2): 78-94. |
| [7] | Liang Y, Yuan QQ, Zheng QJ, Mei ZL, Song YW, Yan H, Yang JJ, Wu SH, Yuan J, Wu W. DNA damage atlas: an atlas of DNA damage and repair. Nucleic Acids Res, 2024, 52(D1): D1218-D1226. |
| [8] | Vergara X, Schep R, Medema RH, van Steensel B. From fluorescent foci to sequence: Illuminating DNA double strand break repair by high-throughput sequencing technologies. DNA Repair (Amst), 2022, 118: 103388. |
| [9] | Pfeifer GP, Jin SG. Methods and applications of genome-wide profiling of DNA damage and rare mutations. Nat Rev Genet, 2024. |
| [10] | Prasad S, Gupta SC, Pandey MK, Tyagi AK, Deb L. Oxidative stress and cancer: advances and challenges. Oxid Med Cell Longev, 2016, 2016: 5010423. |
| [11] | Hussain T, Tan B, Yin YL, Blachier F, Tossou MCB, Rahu N. Oxidative stress and inflammation: what polyphenols can do for us? Oxid Med Cell Longev, 2016, 2016: 7432797. |
| [12] |
Kim GH, Kim JE, Rhie SJ, Yoon S. The role of oxidative stress in neurodegenerative diseases. Exp Neurobiol, 2015, 24(4): 325-340.
doi: 10.5607/en.2015.24.4.325 pmid: 26713080 |
| [13] | David SS, O'Shea VL, Kundu S. Base-excision repair of oxidative DNA damage. Nature, 2007, 447(7147): 941-950. |
| [14] |
Krokan HE, Nilsen H, Skorpen F, Otterlei M, Slupphaug G. Base excision repair of DNA in mammalian cells. FEBS Lett, 2000, 476(1-2): 73-77.
doi: 10.1016/s0014-5793(00)01674-4 pmid: 10878254 |
| [15] | Gorini F, Scala G, Ambrosio S, Majello B, Amente S. OxiDIP-Seq for genome-wide mapping of damaged DNA containing 8-oxo-2'-deoxyguanosine. Bio Protoc, 2022, 12(21): e4540. |
| [16] | Fang YX, Zou P. Genome-wide mapping of oxidative DNA damage via engineering of 8-oxoguanine DNA glycosylase. Biochemistry, 2020, 59(1): 85-89. |
| [17] |
Ding Y, Fleming AM, Burrows CJ. Sequencing the mouse genome for the oxidatively modified base 8-oxo-7,8-dihydroguanine by OG-Seq. J Am Chem Soc, 2017, 139(7): 2569-2572.
doi: 10.1021/jacs.6b12604 pmid: 28150947 |
| [18] |
An J, Yin MD, Yin JY, Wu SZ, Selby CP, Yang YY, Sancar A, Xu GL, Qian MX, Hu JC. Genome-wide analysis of 8-oxo-7,8-dihydro-2'-deoxyguanosine at single-nucleotide resolution unveils reduced occurrence of oxidative damage at G-quadruplex sites. Nucleic Acids Res, 2021, 49(21): 12252-12267.
doi: 10.1093/nar/gkab1022 pmid: 34788860 |
| [19] |
Wu JZ, McKeague M, Sturla SJ. Nucleotide-resolution genome-wide mapping of oxidative DNA damage by Click-Code-Seq. J Am Chem Soc, 2018, 140(31): 9783-9787.
doi: 10.1021/jacs.8b03715 pmid: 29944356 |
| [20] |
El-Sagheer AH, Sanzone AP, Gao R, Tavassoli A, Brown T. Biocompatible artificial DNA linker that is read through by DNA polymerases and is functional in Escherichia coli. Proc Natl Acad Sci USA, 2011, 108(28): 11338-11343.
doi: 10.1073/pnas.1101519108 pmid: 21709264 |
| [21] |
Xiao SJ, Fleming AM, Burrows CJ. Sequencing for oxidative DNA damage at single-nucleotide resolution with click-code-seq v2.0. Chem Commun (Camb), 2023, 59(58): 8997-9000.
doi: 10.1039/d3cc02699j pmid: 37401666 |
| [22] |
Poetsch AR, Boulton SJ, Luscombe NM. Genomic landscape of oxidative DNA damage and repair reveals regioselective protection from mutagenesis. Genome Biol, 2018, 19(1): 215.
doi: 10.1186/s13059-018-1582-2 pmid: 30526646 |
| [23] | Ray S, Abugable AA, Parker J, Liversidge K, Palminha NM, Liao CY, Acosta-Martin AE, Souza CDS, Jurga M, Sudbery I, El-Khamisy SF. A mechanism for oxidative damage repair at gene regulatory elements. Nature, 2022, 609(7929): 1038-1047. |
| [24] | Mao P, Brown AJ, Malc EP, Mieczkowski PA, Smerdon MJ, Roberts SA, Wyrick JJ. Genome-wide maps of alkylation damage, repair, and mutagenesis in yeast reveal mechanisms of mutational heterogeneity. Genome Res, 2017, 27(10): 1674-1684. |
| [25] |
Ding J, Taylor MS, Jackson AP, Reijns MAM. Genome-wide mapping of embedded ribonucleotides and other noncanonical nucleotides using emRiboSeq and EndoSeq. Nat Protoc, 2015, 10(9): 1433-1444.
doi: 10.1038/nprot.2015.099 pmid: 26313479 |
| [26] |
Krokan HE, Drabløs F, Slupphaug G. Uracil in DNA--occurrence, consequences and repair. Oncogene, 2002, 21(58): 8935-8948.
pmid: 12483510 |
| [27] | Galashevskaya A, Sarno A, Vågbø CB, Aas PA, Hagen L, Slupphaug G, Krokan HE. A robust, sensitive assay for genomic uracil determination by LC/MS/MS reveals lower levels than previously reported. DNA Repair (Amst), 2013, 12(9): 699-706. |
| [28] |
Alt FW, Zhang Y, Meng FL, Guo CG, Schwer B. Mechanisms of programmed DNA lesions and genomic instability in the immune system. Cell, 2013, 152(3): 417-429.
doi: 10.1016/j.cell.2013.01.007 pmid: 23374339 |
| [29] | Komor AC, Kim YB, Packer MS, Zuris JA, Liu DR. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature, 2016, 533(7603): 420-424. |
| [30] |
Ransom M, Bryan DS, Hesselberth JR. High-resolution mapping of modified DNA nucleobases using excision repair enzymes. Methods Mol Biol, 2018, 1672: 63-76.
doi: 10.1007/978-1-4939-7306-4_6 pmid: 29043617 |
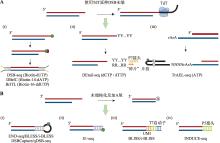
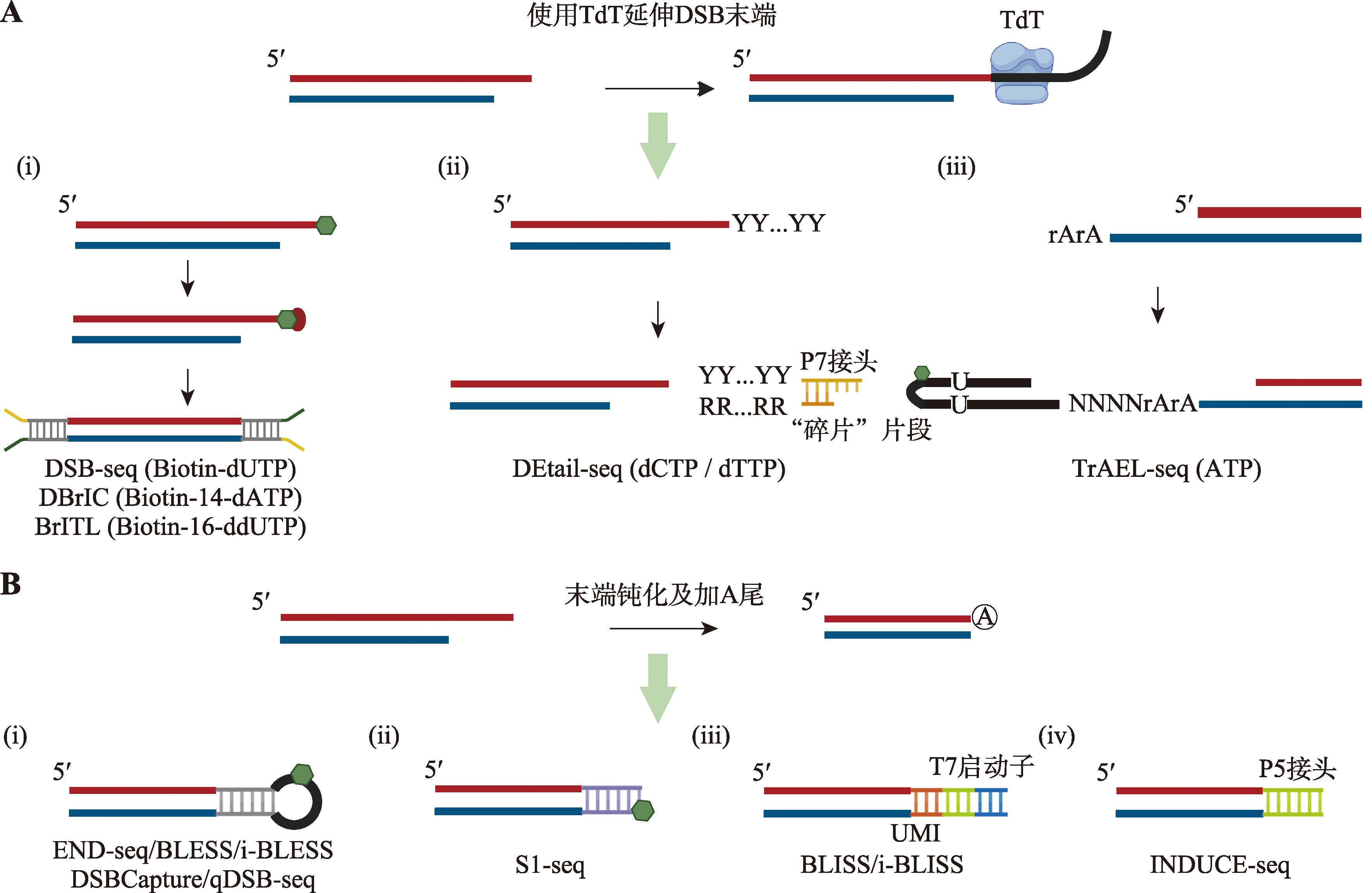
| [31] |
Sakhtemani R, Senevirathne V, Stewart J, Perera MLW, Pique-Regi R, Lawrence MS, Bhagwat AS. Genome- wide mapping of regions preferentially targeted by the human DNA-cytosine deaminase APOBEC3A using uracil-DNA pulldown and sequencing. J Biol Chem, 2019, 294(41): 15037-15051.
doi: 10.1074/jbc.RA119.008053 pmid: 31431505 |
| [32] |
Shu XT, Liu MH, Lu ZK, Zhu CX, Meng HW, Huang SH, Zhang XX, Yi CQ. Genome-wide mapping reveals that deoxyuridine is enriched in the human centromeric DNA. Nat Chem Biol, 2018, 14(7): 680-687.
doi: 10.1038/s41589-018-0065-9 pmid: 29785056 |
| [33] | Pálinkás H L, Békési A, Róna G, Pongor L, Papp G, Tihanyi G, Holub E, Póti Á, Gemma C, Ali S, Morten MJ, Rothenberg E, Pagano M, Szüts D, Györffy B, Vértessy BG. Genome-wide alterations of uracil distribution patterns in human DNA upon chemotherapeutic treatments. Elife, 2020, 9: e60498. |
| [34] |
Jiang LD, Yin JY, Qian MX, Rong SQ, Zhang SQ, Chen KJ, Zhao CC, Tan YQ, Guo JY, Chen H, Gao SY, Liu TT, Liu Y, Shen B, Yang J, Zhang Y, Meng FL, Hu JC, Ma HH, Chen YH. Udgx-mediated uracil sequencing at single-nucleotide resolution. J Am Chem Soc, 2022, 144(3): 1323-1331.
doi: 10.1021/jacs.1c11269 pmid: 35037455 |
| [35] |
Sang PB, Srinath T, Patil AG, Woo EJ, Varshney U. A unique uracil-DNA binding protein of the uracil DNA glycosylase superfamily. Nucleic Acids Res, 2015, 43(17): 8452-8463.
doi: 10.1093/nar/gkv854 pmid: 26304551 |
| [36] | Wang YF, Zhang X, Han SQ, Yang W, Chen ZG, Wu F, Liu JZ, Weng XC, Zhou X. Base-resolution analysis of deoxyuridine at the genome scale based on the artificial incorporation modified nucleobase. ACS Cent Sci, 2021, 7(6): 973-979. |
| [37] | Williams JS, Kunkel TA. Ribonucleotides in DNA: origins, repair and consequences. DNA Repair (Amst), 2014, 19: 27-37. |
| [38] | Huang SN, Williams JS, Arana ME, Kunkel TA, Pommier Y. Topoisomerase I-mediated cleavage at unrepaired ribonucleotides generates DNA double-strand breaks. EMBO J, 2017, 36(3): 361-373. |
| [39] |
Clausen AR, Murray MS, Passer AR, Pedersen LC, Kunkel TA. Structure-function analysis of ribonucleotide bypass by B family DNA replicases. Proc Natl Acad Sci USA, 2013, 110(42): 16802-16807.
doi: 10.1073/pnas.1309119110 pmid: 24082122 |
| [40] |
Andres SN, Schellenberg MJ, Wallace BD, Tumbale P, Williams RS. Recognition and repair of chemically heterogeneous structures at DNA ends. Environ Mol Mutagen, 2015, 56(1): 1-21.
doi: 10.1002/em.21892 pmid: 25111769 |
| [41] |
Sparks JL, Chon H, Cerritelli SM, Kunkel TA, Johansson E, Crouch RJ, Burgers PM. RNase H2-initiated ribonucleotide excision repair. Mol Cell, 2012, 47(6): 980-986.
doi: 10.1016/j.molcel.2012.06.035 pmid: 22864116 |
| [42] |
Crow YJ, Leitch A, Hayward BE, Garner A, Parmar R, Griffith E, Ali M, Semple C, Aicardi J, Babul-Hirji R, Baumann C, Baxter P, Bertini E, Chandler KE, Chitayat D, Cau D, Déry C, Fazzi E, Goizet C, King MD, Klepper J, Lacombe D, Lanzi G, Lyall H, Martínez-Frías ML, Mathieu M, McKeown C, Monier A, Oade Y, Quarrell OW, Rittey CD, Rogers RC, Sanchis A, Stephenson JB, Tacke U, Till M, Tolmie JL, Tomlin P, Voit T, Weschke B, Woods CG, Lebon P, Bonthron DT, Ponting CP, Jackson AP. Mutations in genes encoding ribonuclease H2 subunits cause Aicardi-Goutières syndrome and mimic congenital viral brain infection. Nat Genet, 2006, 38(8): 910-916.
doi: 10.1038/ng1842 pmid: 16845400 |
| [43] |
Koh KD, Balachander S, Hesselberth JR, Storici F. Ribose-seq: global mapping of ribonucleotides embedded in genomic DNA. Nat Methods, 2015, 12(3): 251-257.
doi: 10.1038/nmeth.3259 pmid: 25622106 |
| [44] | Clausen AR, Lujan SA, Burkholder AB, Orebaugh CD, Williams JS, Clausen MF, Malc EP, Mieczkowski PA, Fargo DC, Smith DJ, Kunkel TA. Tracking replication enzymology in vivo by genome-wide mapping of ribonucleotide incorporation. Nat Struct Mol Biol, 2015, 22(3): 185-191. |
| [45] |
Keszthelyi A, Daigaku Y, Ptasińska K, Miyabe I, Carr AM. Mapping ribonucleotides in genomic DNA and exploring replication dynamics by polymerase usage sequencing (Pu-seq). Nat Protoc, 2015, 10(11): 1786-1801.
doi: 10.1038/nprot.2015.116 pmid: 26492137 |
| [46] |
Lindahl T, Nyberg B. Rate of depurination of native deoxyribonucleic acid. Biochemistry, 1972, 11(19): 3610-3618.
doi: 10.1021/bi00769a018 pmid: 4626532 |
| [47] | Kohli RM, Zhang Y. TET enzymes, TDG and the dynamics of DNA demethylation. Nature, 2013, 502(7472): 472-479. |
| [48] | Boiteux S, Guillet M. Abasic sites in DNA: repair and biological consequences in Saccharomyces cerevisiae. DNA Repair (Amst), 2004, 3(1): 1-12. |
| [49] |
Liu ZJ, Martínez Cuesta S, van Delft P, Balasubramanian S. Sequencing abasic sites in DNA at single-nucleotide resolution. Nat Chem, 2019, 11(7): 629-637.
doi: 10.1038/s41557-019-0279-9 pmid: 31209299 |
| [50] |
Cai Y, Cao HF, Wang F, Zhang YF, Kapranov P. Complex genomic patterns of abasic sites in mammalian DNA revealed by a high-resolution SSiNGLe-AP method. Nat Commun, 2022, 13(1): 5868.
doi: 10.1038/s41467-022-33594-1 pmid: 36198706 |
| [51] |
Cao HF, Salazar-García L, Gao F, Wahlestedt T, Wu CL, Han XE, Cai Y, Xu DY, Wang F, Tang L, Ricciardi N, Cai DD, Wang HF, Chin MPS, Timmons JA, Wahlestedt C, Kapranov P. Novel approach reveals genomic landscapes of single-strand DNA breaks with nucleotide resolution in human cells. Nat Commun, 2019, 10(1): 5799.
doi: 10.1038/s41467-019-13602-7 pmid: 31862872 |
| [52] | Krokan HE, Bjørås M. Base excision repair. Cold Spring Harb Perspect Biol, 2013, 5(4): a012583. |
| [53] |
Mao P, Smerdon MJ, Roberts SA, Wyrick JJ. Chromosomal landscape of UV damage formation and repair at single-nucleotide resolution. Proc Natl Acad Sci USA, 2016, 113(32): 9057-9062.
doi: 10.1073/pnas.1606667113 pmid: 27457959 |
| [54] | Pfeifer GP, Besaratinia A. UV wavelength-dependent DNA damage and human non-melanoma and melanoma skin cancer. Photochem Photobiol Sci, 2012, 11(1): 90-97. |
| [55] |
Eastman A. Reevaluation of interaction of cis-dichloro(ethylenediamine)platinum(II) with DNA. Biochemistry, 1986, 25(13): 3912-3915.
pmid: 3741840 |
| [56] |
Siddik ZH. Cisplatin: mode of cytotoxic action and molecular basis of resistance. Oncogene, 2003, 22(47): 7265-7279.
doi: 10.1038/sj.onc.1206933 pmid: 14576837 |
| [57] |
Hu JC, Li WT, Adebali O, Yang YY, Oztas O, Selby CP, Sancar A. Genome-wide mapping of nucleotide excision repair with XR-seq. Nat Protoc, 2019, 14(1): 248-282.
doi: 10.1038/s41596-018-0093-7 pmid: 30552409 |
| [58] | Reardon JT, Sancar A. Nucleotide excision repair. Prog Nucleic Acid Res Mol Biol, 2005, 79: 183-235. |
| [59] |
Li WT, Hu JC, Adebali O, Adar S, Yang YY, Chiou YY, Sancar A. Human genome-wide repair map of DNA damage caused by the cigarette smoke carcinogen benzo[a]pyrene. Proc Natl Acad Sci USA, 2017, 114(26): 6752-6757.
doi: 10.1073/pnas.1706021114 pmid: 28607059 |
| [60] | Wu SZ, Huang YC, Selby CP, Gao M, Sancar A, Hu JC. A new technique for genome-wide mapping of nucleotide excision repair without immunopurification of damaged DNA. J Biol Chem, 2022, 298(5): 101863. |
| [61] |
Hu JC, Lieb JD, Sancar A, Adar S. Cisplatin DNA damage and repair maps of the human genome at single-nucleotide resolution. Proc Natl Acad Sci USA, 2016, 113(41): 11507-11512.
pmid: 27688757 |
| [62] | Shu XT, Xiong XS, Song JH, He C, Yi CQ. Base-resolution analysis of cisplatin-DNA adducts at the genome scale. Angew Chem Int Ed Engl, 2016, 55(46): 14246-14249. |
| [63] |
Hu JC, Adebali O, Adar S, Sancar A. Dynamic maps of UV damage formation and repair for the human genome. Proc Natl Acad Sci USA, 2017, 114(26): 6758-6763.
doi: 10.1073/pnas.1706522114 pmid: 28607063 |
| [64] | Zhu YC, Tan YQ, Li L, Xiang YN, Huang YC, Zhang XP, Yin JY, Li J, Lan F, Qian MX, Hu JC. Genome-wide mapping of protein-DNA damage interaction by PADD-seq. Nucleic Acids Res, 2023, 51(6): e32. |
| [65] | Alhegaili AS, Ji Y, Sylvius N, Blades MJ, Karbaschi M, Tempest HG, Jones GDD, Cooke MS. Genome-wide adductomics analysis reveals heterogeneity in the induction and loss of cyclobutane thymine dimers across both the nuclear and mitochondrial genomes. Int J Mol Sci, 2019, 20(20): 5112. |
| [66] |
Fineev NE, Grebennikov BV. Experimental study of the technical light characteristics of endoscopic optical systems. Nov Med Tekh, 1976, (2): 58-62.
pmid: 1032541 |
| [67] | Laughery MF, Brown AJ, Bohm KA, Sivapragasam S, Morris HS, Tchmola M, Washington AD, Mitchell D, Mather S, Malc EP, Mieczkowski PA, Roberts SA, Wyrick JJ. Atypical UV photoproducts induce non-canonical mutation classes associated with driver mutations in melanoma. Cell Rep, 2020, 33(7): 108401. |
| [68] |
Cafardi JA, Elmets CA. T4 endonuclease V: review and application to dermatology. Expert Opin Biol Ther, 2008, 8(6): 829-838.
doi: 10.1517/14712598.8.6.829 pmid: 18476794 |
| [69] |
Premi S, Han L, Mehta S, Knight J, Zhao DJ, Palmatier MA, Kornacker K, Brash DE. Genomic sites hypersensitive to ultraviolet radiation. Proc Natl Acad Sci USA, 2019, 116(48): 24196-24205.
doi: 10.1073/pnas.1907860116 pmid: 31723047 |
| [70] | Jin SG, Pettinga D, Johnson J, Li PP, Pfeifer GP. The major mechanism of melanoma mutations is based on deamination of cytosine in pyrimidine dimers as determined by circle damage sequencing. Sci Adv, 2021, 7(31): eabi6508. |
| [71] |
Avery AM, Kaur B, Taylor JS, Mello JA, Essigmann JM, Doetsch PW. Substrate specificity of ultraviolet DNA endonuclease (UVDE/Uve1p) from Schizosaccharomyces pombe. Nucleic Acids Res, 1999, 27(11): 2256-2264.
pmid: 10325412 |
| [72] | Kara N, Krueger F, Rugg-Gunn P, Houseley J. Genome-wide analysis of DNA replication and DNA double-strand breaks using TrAEL-seq. PLoS Biol, 2021, 19(3): e3000886. |
| [73] | Shastri N, Tsai YC, Hile S, Jordan D, Powell B, Chen J, Maloney D, Dose M, Lo Y, Anastassiadis T, Rivera O, Kim T, Shah S, Borole P, Asija K, Wang X, Smith KD, Finn D, Schug J, Casellas R, Yatsunyk LA, Eckert KA, Brown EJ. Genome-wide identification of structure- forming repeats as principal sites of fork collapse upon ATR inhibition. Mol Cell, 2018, 72(2): 222-238.e11. |
| [74] |
Baranello L, Kouzine F, Wojtowicz D, Cui KR, Przytycka TM, Zhao KJ, Levens D. DNA break mapping reveals topoisomerase II activity genome-wide. Int J Mol Sci, 2014, 15(7): 13111-13122.
doi: 10.3390/ijms150713111 pmid: 25056547 |
| [75] | Gregoire MC, Massonneau J, Leduc F, Arguin M, Brazeau MA, Boissonneault G. Quantification and genome-wide mapping of DNA double-strand breaks. DNA Repair (Amst), 2016, 48: 63-68. |
| [76] |
Xu W, Liu C, Zhang Z, Sun CB, Li Q, Li K, Jiang H, Li W, Sun QW. DEtail-seq is an ultra-efficient and convenient method for meiotic DNA break profiling in multiple organisms. Sci China Life Sci, 2023, 66(6): 1392-1407.
doi: 10.1007/s11427-022-2277-y pmid: 36723795 |
| [77] |
Canela A, Sridharan S, Sciascia N, Tubbs A, Meltzer P, Sleckman BP, Nussenzweig A. DNA breaks and end resection measured genome-wide by end sequencing. Mol Cell, 2016, 63(5): 898-911.
doi: 10.1016/j.molcel.2016.06.034 pmid: 27477910 |
| [78] |
Crosetto N, Mitra A, Silva MJ, Bienko M, Dojer N, Wang Q, Karaca E, Chiarle R, Skrzypczak M, Ginalski K, Pasero P, Rowicka M, Dikic I. Nucleotide-resolution DNA double-strand break mapping by next-generation sequencing. Nat Methods, 2013, 10(4): 361-365.
doi: 10.1038/nmeth.2408 pmid: 23503052 |
| [79] |
Lensing SV, Marsico G, Hänsel-Hertsch R, Lam EY, Tannahill D, Balasubramanian S. DSBCapture: in situ capture and sequencing of DNA breaks. Nat Methods, 2016, 13(10): 855-857.
doi: 10.1038/nmeth.3960 |
| [80] |
Zhu YJ, Biernacka A, Pardo B, Dojer N, Forey R, Skrzypczak M, Fongang B, Nde J, Yousefi R, Pasero P, Ginalski K, Rowicka M. qDSB-seq is a general method for genome-wide quantification of DNA double-strand breaks using sequencing. Nat Commun, 2019, 10(1): 2313.
doi: 10.1038/s41467-019-10332-8 pmid: 31127121 |
| [81] |
Biernacka A, Zhu YJ, Skrzypczak M, Forey R, Pardo B, Grzelak M, Nde J, Mitra A, Kudlicki A, Crosetto N, Pasero P, Rowicka M, Ginalski K. i-BLESS is an ultra-sensitive method for detection of DNA double-strand breaks. Commun Biol, 2018, 1: 181.
doi: 10.1038/s42003-018-0165-9 pmid: 30393778 |
| [82] | Mimitou EP, Yamada S, Keeney S. A global view of meiotic double-strand break end resection. Science, 2017, 355(6320): 40-45. |
| [83] |
Hoffman EA, McCulley A, Haarer B, Arnak R, Feng WY. Break-seq reveals hydroxyurea-induced chromosome fragility as a result of unscheduled conflict between DNA replication and transcription. Genome Res, 2015, 25(3): 402-412.
doi: 10.1101/gr.180497.114 pmid: 25609572 |
| [84] |
Bouwman BAM, Agostini F, Garnerone S, Petrosino G, Gothe HJ, Sayols S, Moor AE, Itzkovitz S, Bienko M, Roukos V, Crosetto N. Genome-wide detection of DNA double-strand breaks by in-suspension BLISS. Nat Protoc, 2020, 15(12): 3894-3941.
doi: 10.1038/s41596-020-0397-2 pmid: 33139954 |
| [85] |
Motea EA, Berdis AJ. Terminal deoxynucleotidyl transferase: the story of a misguided DNA polymerase. Biochim Biophys Acta, 2010, 1804(5): 1151-1166.
doi: 10.1016/j.bbapap.2009.06.030 pmid: 19596089 |
| [86] |
Dobbs FM, van Eijk P, Fellows MD, Loiacono L, Nitsch R, Reed SH. Precision digital mapping of endogenous and induced genomic DNA breaks by INDUCE-seq. Nat Commun, 2022, 13(1): 3989.
doi: 10.1038/s41467-022-31702-9 pmid: 35810156 |
| [87] |
Cejka P, Symington LS. DNA end resection: mechanism and control. Annu Rev Genet, 2021, 55: 285-307.
doi: 10.1146/annurev-genet-071719-020312 pmid: 34813349 |
| [88] |
Baudat F, Imai Y, de Massy B. Meiotic recombination in mammals: localization and regulation. Nat Rev Genet, 2013, 14(11): 794-806.
doi: 10.1038/nrg3573 pmid: 24136506 |
| [89] |
Khil PP, Smagulova F, Brick KM, Camerini-Otero RD, Petukhova GV. Sensitive mapping of recombination hotspots using sequencing-based detection of ssDNA. Genome Res, 2012, 22(5): 957-965.
doi: 10.1101/gr.130583.111 pmid: 22367190 |
| [90] |
Pan J, Sasaki M, Kniewel R, Murakami H, Blitzblau HG, Tischfield SE, Zhu X, Neale MJ, Jasin M, Socci ND, Hochwagen A, Keeney S. A hierarchical combination of factors shapes the genome-wide topography of yeast meiotic recombination initiation. Cell, 2011, 144(5): 719-731.
doi: 10.1016/j.cell.2011.02.009 pmid: 21376234 |
| [91] |
Gittens WH, Johnson DJ, Allison RM, Cooper TJ, Thomas H, Neale MJ. A nucleotide resolution map of Top2-linked DNA breaks in the yeast and human genome. Nat Commun, 2019, 10(1): 4846.
doi: 10.1038/s41467-019-12802-5 pmid: 31649282 |
| [92] |
Chiarle R, Zhang Y, Frock RL, Lewis SM, Molinie B, Ho YJ, Myers DR, Choi VW, Compagno M, Malkin DJ, Neuberg D, Monti S, Giallourakis CC, Gostissa M, Alt FW. Genome-wide translocation sequencing reveals mechanisms of chromosome breaks and rearrangements in B cells. Cell, 2011, 147(1): 107-119.
doi: 10.1016/j.cell.2011.07.049 pmid: 21962511 |
| [93] |
Oliveira TY, Resch W, Jankovic M, Casellas R, Nussenzweig MC, Klein IA. Translocation capture sequencing: A method for high throughput mapping of chromosomal rearrangements. J Immunol Methods, 2012, 375(1-2): 176-181.
doi: 10.1016/j.jim.2011.10.007 pmid: 22033343 |
| [94] | Delaney JR, La Spada AR. Protocol for mapping double-stranded DNA break sites across the genome with translocation capture sequencing. STAR Protoc, 2023, 4(2): 102205. |
| [95] |
Gabriel R, Lombardo A, Arens A, Miller JC, Genovese P, Kaeppel C, Nowrouzi A, Bartholomae CC, Wang JB, Friedman G, Holmes MC, Gregory PD, Glimm H, Schmidt M, Naldini L, von Kalle C. An unbiased genome-wide analysis of zinc-finger nuclease specificity. Nat Biotechnol, 2011, 29(9): 816-823.
doi: 10.1038/nbt.1948 pmid: 21822255 |
| [96] | Liu Y, Yin JH, Gan TT, Liu MZ, Xin CC, Zhang WW, Hu JZ. PEM-seq comprehensively quantifies DNA repair outcomes during gene-editing and DSB repair. STAR Protoc, 2022, 3(1): 101088. |
| [97] | Zhang XF, Zhang Y, Ba ZQ, Kyritsis N, Casellas R, Alt FW. Fundamental roles of chromatin loop extrusion in antibody class switching. Nature, 2019, 575(7782): 385-389. |
| [98] |
Hu JZ, Meyers RM, Dong JC, Panchakshari RA, Alt FW, Frock RL. Detecting DNA double-stranded breaks in mammalian genomes by linear amplification-mediated high-throughput genome-wide translocation sequencing. Nat Protoc, 2016, 11(5): 853-871.
doi: 10.1038/nprot.2016.043 pmid: 27031497 |
| [99] | Dai HQ, Hu HL, Lou JM, Ye AY, Ba ZQ, Zhang XF, Zhang YW, Zhao LJ, Yoon HS, Chapdelaine-Williams AM, Kyritsis N, Chen H, Johnson K, Lin S, Conte A, Casellas R, Lee CS, Alt FW. Loop extrusion mediates physiological Igh locus contraction for RAG scanning. Nature, 2021, 590(7845): 338-343. |
| [100] | Fowler FC, Chen BR, Zolnerowich N, Wu W, Pavani R, Paiano J, Peart C, Chen ZL, Nussenzweig A, Sleckman BP, Tyler JK. DNA-PK promotes DNA end resection at DNA double strand breaks in G0 cells. Elife, 2022, 11: e74700. |
| [101] |
Paiano J, Wu W, Yamada S, Sciascia N, Callen E, Paola Cotrim A, Deshpande RA, Maman Y, Day A, Paull TT, Nussenzweig A. ATM and PRDM9 regulate SPO11-bound recombination intermediates during meiosis. Nat Commun, 2020, 11(1): 857.
doi: 10.1038/s41467-020-14654-w pmid: 32051414 |
| [102] |
Lin SG, Ba ZQ, Du Z, Zhang Y, Hu JZ, Alt FW. Highly sensitive and unbiased approach for elucidating antibody repertoires. Proc Natl Acad Sci USA, 2016, 113(28): 7846-7851.
doi: 10.1073/pnas.1608649113 pmid: 27354528 |
| [103] |
Wang YY, Zhang SX, Yang XR, Hwang JK, Zhan CZ, Lian CY, Wang C, Gui TT, Wang BB, Xie X, Dai PF, Zhang L, Tian Y, Zhang HZ, Han C, Cai YN, Hao Q, Ye XF, Liu XJ, Liu JQ, Cao ZW, Huang SH, Song J, Pan-Hammarström Q, Zhao YF, Alt FW, Zheng XQ, Da LT, Yeap LS, Meng FL. Mesoscale DNA feature in antibody-coding sequence facilitates somatic hypermutation. Cell, 2023, 186(10): 2193-2207.e19.
doi: 10.1016/j.cell.2023.03.030 pmid: 37098343 |
| [104] |
Matos-Rodrigues G, van Wietmarschen N, Wu W, Tripathi V, Koussa NC, Pavani R, Nathan WJ, Callen E, Belinky F, Mohammed A, Napierala M, Usdin K, Ansari AZ, Mirkin SM, Nussenzweig A. S1-END-seq reveals DNA secondary structures in human cells. Mol Cell, 2022, 82(19): 3538-3552.e5.
doi: 10.1016/j.molcel.2022.08.007 pmid: 36075220 |
| [105] |
Wang DP, Wu W, Callen E, Pavani R, Zolnerowich N, Kodali S, Zong DL, Wong N, Noriega S, Nathan WJ, Matos-Rodrigues G, Chari R, Kruhlak MJ, Livak F, Ward M, Caldecott K, Di Stefano B, Nussenzweig A. Active DNA demethylation promotes cell fate specification and the DNA damage response. Science, 2022, 378(6623): 983-989.
doi: 10.1126/science.add9838 pmid: 36454826 |
| [106] | Wu SH, Liang Y, Wu W. High throughput sequencing technologies for DNA double-strand break. Chinese Journal of Cell Biology, 2024, 46(8): 1488-1496. |
| 吴树恒, 梁羽, 吴薇. DNA双链断裂高通量测序技术的研究进展. 中国细胞生物学学报, 2024, 46(8): 1488-1496. | |
| [107] |
Caldecott KW. Single-strand break repair and genetic disease. Nat Rev Genet, 2008, 9(8): 619-931.
doi: 10.1038/nrg2380 pmid: 18626472 |
| [108] |
Caldecott KW. DNA single-strand break repair and human genetic disease. Trends Cell Biol, 2022, 32(9): 733-745.
doi: 10.1016/j.tcb.2022.04.010 pmid: 35643889 |
| [109] |
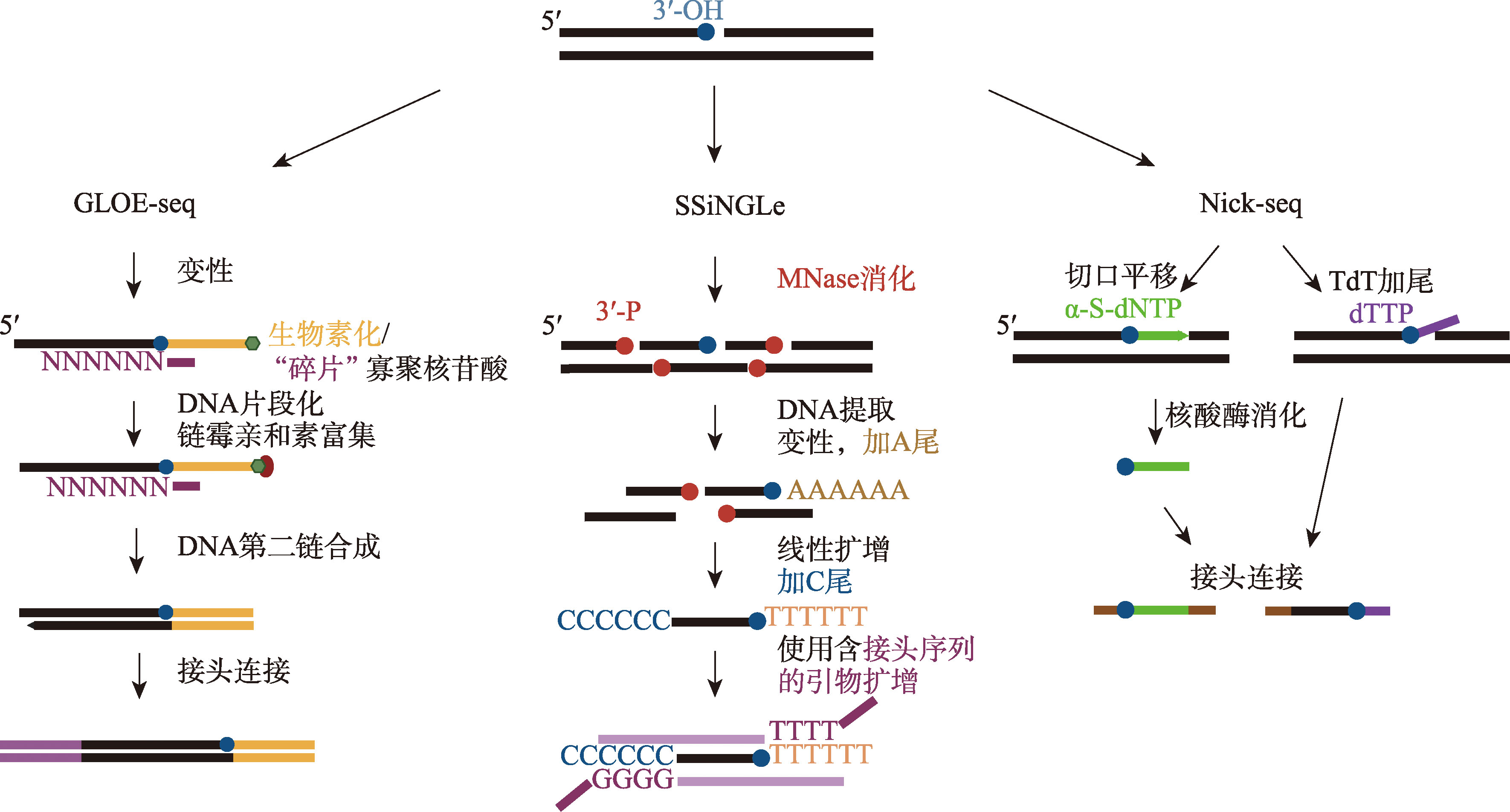
Sriramachandran AM, Petrosino G, Méndez-Lago M, Schäfer AJ, Batista-Nascimento LS, Zilio N, Ulrich HD. Genome-wide nucleotide-resolution mapping of DNA replication patterns, single-strand breaks, and lesions by GLOE-seq. Mol Cell, 2020, 78(5): 975-985.e7.
doi: S1097-2765(20)30195-7 pmid: 32320643 |
| [110] |
Elacqua JJ, Ranu N, DiIorio SE, Blainey PC. DENT-seq for genome-wide strand-specific identification of DNA single-strand break sites with single-nucleotide resolution. Genome Res, 2021, 31(1): 75-87.
doi: 10.1101/gr.265223.120 pmid: 33355294 |
| [111] |
Cao B, Wu XL, Zhou JL, Wu H, Liu LL, Zhang QH, DeMott MS, Gu C, Wang LR, You DL, Dedon PC. Nick-seq for single-nucleotide resolution genomic maps of DNA modifications and damage. Nucleic Acids Res, 2020, 48(12): 6715-6725.
doi: 10.1093/nar/gkaa473 pmid: 32484547 |
| [112] | Wu W, Hill SE, Nathan WJ, Paiano J, Callen E, Wang DP, Shinoda K, van Wietmarschen N, Colón-Mercado JM, Zong DL, De Pace R, Shih HY, Coon S, Parsadanian M, Pavani R, Hanzlikova H, Park S, Jung SK, McHugh PJ, Canela A, Chen C, Casellas R, Caldecott KW, Ward ME, Nussenzweig A. Neuronal enhancers are hotspots for DNA single-strand break repair. Nature, 2021, 593(7859): 440-444. |
| [113] |
Zhang JY, Wang Y, Wang YQ, Zhang PK, Chen HY, Huang S. Discrimination between different DNA lesions by monitoring single-molecule polymerase stalling kinetics during nanopore sequencing. Nano Lett, 2022, 22(13): 5561-5569.
doi: 10.1021/acs.nanolett.2c01833 pmid: 35713465 |
| [114] | Wang Y, Patil KM, Yan SH, Zhang PK, Guo WM, Wang YQ, Chen HY, Gillingham D, Huang S. Nanopore sequencing accurately identifies the mutagenic DNA lesion o6-carboxymethyl guanine and reveals its behavior in replication. Angew Chem Int Ed Engl, 2019, 58(25): 8432-8436. |
| [115] |
An N, Fleming AM, White HS, Burrows CJ. Nanopore detection of 8-oxoguanine in the human telomere repeat sequence. ACS Nano, 2015, 9(4): 4296-4307.
doi: 10.1021/acsnano.5b00722 pmid: 25768204 |
| [116] | Ma FB, Yan SH, Zhang JY, Wang Y, Wang LY, Wang YQ, Zhang SY, Du XY, Zhang PK, Chen HY, Huang S. Nanopore sequencing accurately identifies the cisplatin adduct on DNA. ACS Sens, 2021, 6(8): 3082-3092. |
| [117] |
Wang Z, Shen XH, Shi QH. Advances in single-cell whole genome sequencing technology and its application in biomedicine. Hereditas(Beijing), 2021, 43(2): 108-117.
doi: 10.16288/j.yczz.20-363 pmid: 33724214 |
| 王卓, 申笑涵, 施奇惠. 单细胞基因组测序技术新进展及其在生物医学中的应用. 遗传, 2021, 43(2): 108-117. |
| [1] | Daiyuan Liu, Zhaohui Zhang, Xianjiang Kang. Research progress on the effect of sperm chromatin integrity on function and its detection methods [J]. Hereditas(Beijing), 2024, 46(7): 511-529. |
| [2] | Nan Zhang, Jue Zhang, Ge Lin. Advances in the study of DNA damage and repair in mammalian oocytes [J]. Hereditas(Beijing), 2023, 45(5): 379-394. |
| [3] |
Zhuo Wang, Xiaohan Shen, Qihui Shi.
|
| [4] | Qingyi Zhang, Yingzi Zhang, Kai Shen, Shuyu Zhang, Jianping Cao. Histone ubiquitylation and its roles in DNA damage response [J]. Hereditas(Beijing), 2019, 41(1): 29-40. |
| [5] | Zhen Liu, Jianhong Xu. The application of the high throughput sequencing technology in the transposable elements [J]. HEREDITAS(Beijing), 2015, 37(9): 885-898. |
| [6] | Yuchen Guo, Bingkun Lei, Xiaolong Deng, Yao Yu, Hong LV. Large scale screening of genetic interaction with sgf73+ in fission yeast [J]. HEREDITAS(Beijing), 2014, 36(7): 723-731. |
| [7] | Deling Lin, Ying Luo, Yi Song. The post-transcriptional regulation of the DNA damage response [J]. HEREDITAS, 2014, 36(4): 309-318. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||