Hereditas(Beijing) ›› 2025, Vol. 47 ›› Issue (3): 382-392.doi: 10.16288/j.yczz.24-213
• Technique and Method • Previous Articles
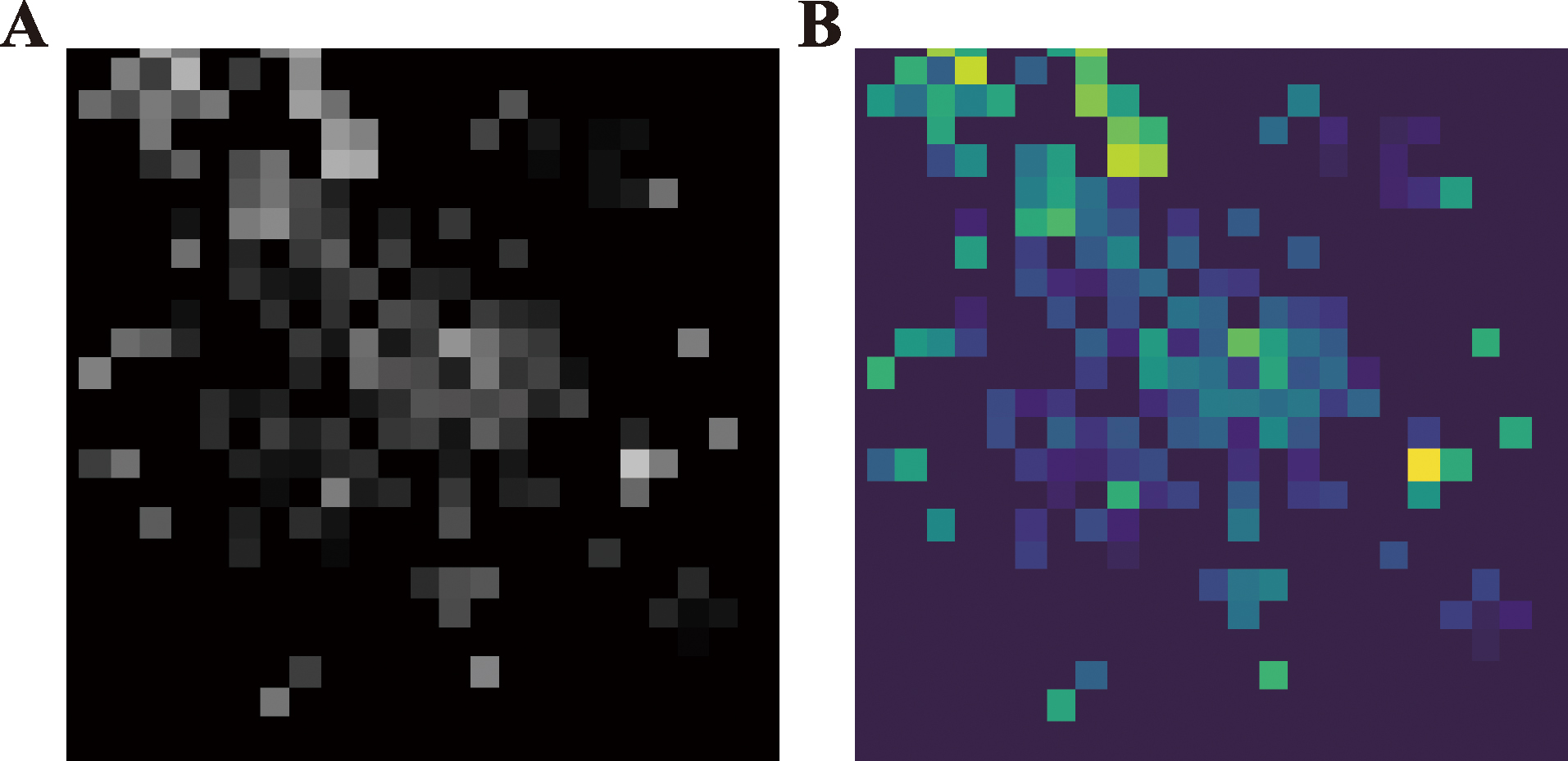
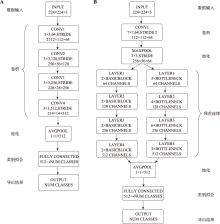
Enhancing single-cell classification accuracy using image conversion and deep learning
Bingxi Gao( ), Huaxuan Wu, Zhiqiang Du(
), Huaxuan Wu, Zhiqiang Du( )
)
- College of Animal Science and Technology, Yangtze University, Jingzhou 434025, China
-
Received:2024-07-19Revised:2024-09-23Online:2025-03-20Published:2024-09-26 -
Contact:Zhiqiang Du E-mail:gbx15020771250@163.com;zhqdu@yangtzeu.edu.cn -
Supported by:Joint Research on Improved Livestock and Poultry Breeds in Anhui Province(2021~2025)
Cite this article
Bingxi Gao, Huaxuan Wu, Zhiqiang Du. Enhancing single-cell classification accuracy using image conversion and deep learning[J]. Hereditas(Beijing), 2025, 47(3): 382-392.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
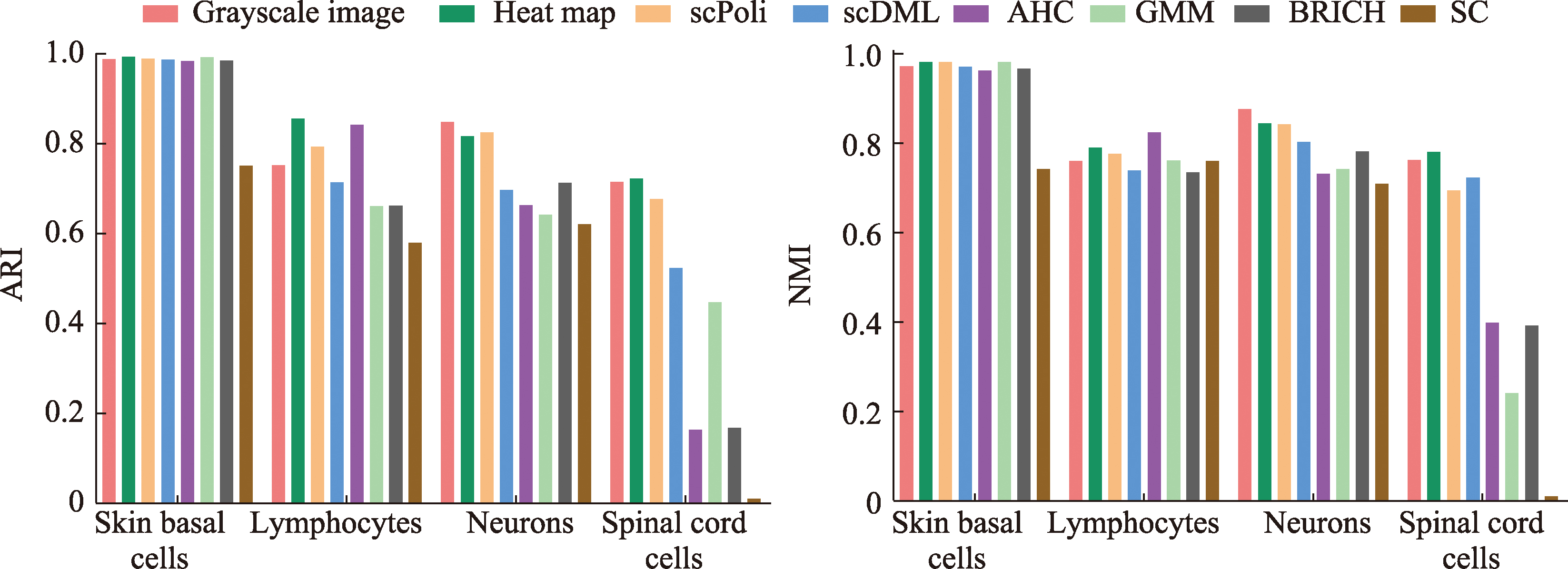
Table 2
Performance metrics for seven clustering methods"
| 数据集 | 指标 | 聚类方法 | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Grayscale image | Heat map | scPoli | scDML | AHC | GMM | BRICH | SC | ||
| 小鼠皮肤基底细胞 | ARI | 0.988 | 0.993 | 0.989 | 0.987 | 0.984 | 0.992 | 0.985 | 0.751 |
| NMI | 0.972 | 0.982 | 0.982 | 0.971 | 0.962 | 0.982 | 0.967 | 0.743 | |
| 小鼠淋巴细胞 | ARI | 0.752 | 0.856 | 0.793 | 0.714 | 0.842 | 0.661 | 0.662 | 0.58 |
| NMI | 0.76 | 0.79 | 0.776 | 0.740 | 0.825 | 0.761 | 0.735 | 0.76 | |
| 人类神经元细胞 | ARI | 0.848 | 0.816 | 0.825 | 0.697 | 0.663 | 0.642 | 0.713 | 0.62 |
| NMI | 0.876 | 0.845 | 0.843 | 0.803 | 0.732 | 0.743 | 0.782 | 0.71 | |
| 小鼠脊髓细胞 | ARI | 0.715 | 0.723 | 0.677 | 0.524 | 0.164 | 0.447 | 0.168 | 0.01 |
| NMI | 0.763 | 0.781 | 0.695 | 0.723 | 0.399 | 0.241 | 0.392 | 0.01 | |
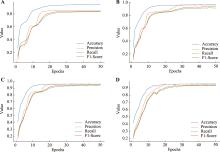
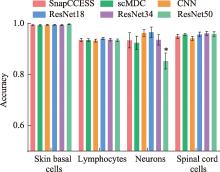
Table 3
Cell classification model evaluation metrics"
| 数据集 | 指标 | 细胞分类模型 | |||||
|---|---|---|---|---|---|---|---|
| SnapCCESS | scMDC | CNN | ResNet18 | ResNet34 | ResNet50 | ||
| 小鼠皮肤基底细胞 | ACC | 0.995(0.002) | 0.994(0.003) | 0.995 (0.002) | 0.995 (0.002) | 0.994 (0.003) | 0.998 (0.001) |
| P | 0.995(0.003) | 0.995(0.003) | 0.995 (0.002) | 0.995 (0.002) | 0.994 (0.002) | 0.997 (0.001) | |
| R | 0.995(0.003) | 0.996(0.003) | 0.995 (0.003) | 0.995 (0.003) | 0.994 (0.003) | 0.998 (0.002) | |
| F1 | 0.994(0.002) | 0.996(0.002) | 0.995 (0.002) | 0.995 (0.003) | 0.994 (0.003) | 0.998 (0.001) | |
| Loss | 0.040 (0.005) | 0.020(0.005) | 0.030 (0.005) | 0.010 (0.005) | 0.020 (0.006) | 0.010 (0.003) | |
| 小鼠淋巴细胞 | ACC | 0.935(0.006) | 0.935(0.005) | 0.934 (0.005) | 0.941 (0.003) | 0.938 (0.004) | 0.934 (0.005) |
| P | 0.934(0.002) | 0.937(0.004) | 0.907 (0.004) | 0.929 (0.003) | 0.912 (0.003) | 0.920 (0.004) | |
| R | 0.932(0.005) | 0.936(0.004) | 0.856 (0.006) | 0.900 (0.004) | 0.896 (0.004) | 0.891 (0.006) | |
| F1 | 0.941(0.005) | 0.937(0.004) | 0.874 (0.005) | 0.905 (0.003) | 0.902 (0.004) | 0.896 (0.005) | |
| Loss | 0.310 (0.005) | 0.230(0.005) | 0.210 (0.010) | 0.190 (0.006) | 0.220 (0.008) | 0.210 (0.001) | |
| 人类神经元细胞 | ACC | 0.933(0.029) | 0.926(0.026) | 0.967 (0.015) | 0.961 (0.018) | 0.925 (0.025) | 0.849 (0.031) |
| P | 0.937(0.031) | 0.932(0.023) | 0.963 (0.012) | 0.959 (0.014) | 0.932 (0.021) | 0.819 (0.028) | |
| R | 0.947(0.027) | 0.947(0.030) | 0.958 (0.018) | 0.945 (0.021) | 0.891 (0.030) | 0.787 (0.042) | |
| F1 | 0.947(0.023) | 0.947(0.030) | 0.953 (0.015) | 0.946 (0.018) | 0.890 (0.025) | 0.776 (0.035) | |
| Loss | 0.130 (0.051) | 0.142(0.042) | 0.320 (0.030) | 0.150 (0.036) | 0.390 (0.050) | 0.600 (0.070) | |
| 小鼠脊髓细胞 | ACC | 0.952(0.007) | 0.957(0.003) | 0.944 (0.008) | 0.958 (0.008) | 0.960 (0.007) | 0.958 (0.009) |
| P | 0.942(0.003) | 0.945(0.005) | 0.855 (0.008) | 0.959 (0.006) | 0.961 (0.006) | 0.957 (0.007) | |
| R | 0.932(0.007) | 0.928(0.012) | 0.835 (0.012) | 0.912 (0.009) | 0.939 (0.008) | 0.934 (0.009) | |
| F1 | 0.925(0.008) | 0.934(0.013) | 0.844 (0.010) | 0.929 (0.008) | 0.947 (0.007) | 0.942 (0.009) | |
| Loss | 0.130 (0.005) | 0.210(0.006) | 0.200 (0.020) | 0.130 (0.016) | 0.130 (0.014) | 0.140 (0.018) | |
| [1] |
Giladi A, Amit I. Single-cell genomics: a stepping stone for future immunology discoveries. Cell, 2018, 172(1-2): 14-21.
doi: S0092-8674(17)31320-X pmid: 29328909 |
| [2] | Han H, Luo FC. Application of single-cell RNA sequencing in probing oligodendroglia heterogeneity and neurological disorders. Hereditas (Beijing), 2023, 45(3): 198-211. |
| 韩熙, 罗富成. 单细胞转录组测序在少突胶质谱系细胞异质性与神经系统疾病中的应用. 遗传, 2023, 45(3): 198-211. | |
| [3] | Qu L, Li S, Qiu HJ. Applications of single-cell RNA sequencing in virology. Hereditas (Beijing), 2020, 42(3): 269-277. |
| 屈亮, 李素, 仇华吉. 单细胞RNA测序技术在病毒研究中的应用. 遗传, 2020, 42(3): 269-277. | |
| [4] | Zhang CQ, Geng Y, Han ZB, Liu YQ, Fu HZ, Hu QH. Autoencoder in autoencoder networks. IEEE Trans Neural Netw Learn Syst, 2024, 35(2): 2263-2275. |
| [5] | Li ZW, Liu F, Yang WJ, Peng SH, Zhou J. A survey of convolutional neural networks: analysis, applications, and prospects. IEEE Trans Neural Netw Learn Syst, 2022, 33(12): 6999-7019. |
| [6] | Arafa A, El-Fishawy N, Badawy M, Radad M. RNAutoencoder: reduced noise autoencoder for classifying imbalanced cancer genomic data. J Biol Eng, 2023, 17(1): 7. |
| [7] | Zhao JP, Wang N, Wang HY, Zheng CH, Su YS. SCDRHA: a scrna-seq data dimensionality reduction algorithm based on hierarchical autoencoder. Front Genet, 2021, 12: 733906. |
| [8] | Zhu YJ, Chen FX. Research progress of object recognition methods based on machine vision. Sci. Technol. Soc. 2023, 21(21): 21-24. |
| 朱亚军, 陈砆兴. 基于机器视觉的目标识别方法的研究进展. 科技资讯, 2023, 21(21): 21-24. | |
| [9] | Eltager M, Abdelaal T, Mahfouz A, Reinders MJT. scMoC: single-cell multi-omics clustering. Bioinform Adv, 2022, 2(1): vbac011. |
| [10] |
Song Q, Wang JT, Bar-Joseph Z. scSTEM: clustering pseudotime ordered single-cell data. Genome Biol, 2022, 23(1): 150.
doi: 10.1186/s13059-022-02716-9 pmid: 35799304 |
| [11] |
Islam MT, Xing L. Cartography of genomic interactions enables deep analysis of single-cell expression data. Nat Commun, 2023, 14(1): 679.
doi: 10.1038/s41467-023-36383-6 pmid: 36755047 |
| [12] | Jia SR, Lysenko A, Boroevich KA, Sharma A, Tsunoda T. scDeepInsight: a supervised cell-type identification method for scRNA-seq data with deep learning. Brief Bioinform, 2023, 24(5): bbad266. |
| [13] | Du ZH, Hu WL, Li JQ, Shang XQ, You ZH, Chen ZZ, Huang YA. scPML: pathway-based multi-view learning for cell type annotation from single-cell RNA-seq data. Commun Biol, 2023, 6(1): 1268. |
| [14] |
Derry A, Krzywinski M, Altman N. Convolutional neural networks. Nat Methods, 2023, 20(9): 1269-1270.
doi: 10.1038/s41592-023-01973-1 pmid: 37580560 |
| [15] | He FX, Liu TL, Tao DC. Why resnet works? Residuals generalize. IEEE Trans Neural Netw Learn Syst, 2020, 31(12): 5349-5362. |
| [16] |
Yu XK, Xu XY, Zhang JX, Li XJ. Batch alignment of single-cell transcriptomics data using deep metric learning. Nat Commun, 2023, 14(1): 960.
doi: 10.1038/s41467-023-36635-5 pmid: 36810607 |
| [17] |
Sharma A, Vans E, Shigemizu D, Boroevich KA, Tsunoda T. DeepInsight: a methodology to transform a non-image data to an image for convolution neural network architecture. Sci Rep, 2019, 9(1): 11399.
doi: 10.1038/s41598-019-47765-6 pmid: 31388036 |
| [18] |
Kang MJ, Lee S, Lee D, Kim S. Learning cell-type-specific gene regulation mechanisms by multi-attention based deep learning with regulatory latent space. Front Genet, 2020, 11: 869.
doi: 10.3389/fgene.2020.00869 pmid: 33133123 |
| [19] | Bao YC, Shi CX, Zhang CQ, Gu MJ, Zhu L, Liu ZX, Zhou L, Ma FY, Na RS, Zhang WG. Progress on deep learning in genomics. Hereditas (Beijing), 2024, 46(9): 701-715. |
| 鲍艳春, 石彩霞, 张传强, 谷明娟, 朱琳, 刘在霞, 周乐, 马凤英, 娜日苏, 张文广. 深度学习在基因组学中的研究进展. 遗传, 2024, 46(9): 701-715. | |
| [20] | Zheng HY, Wu HX, Du ZQ. Gut macrogenomic image enhancement and deep learning improve metabolic disease classification prediction accuracy. Hereditas (Beijing), 2024, 46(10): 886-896. |
| 郑慧怡, 吴华煊, 杜志强. 肠道宏基因组图像增强和深度学习改善代谢性疾病分类预测精度. 遗传, 2024, 46(10): 886-896. | |
| [21] |
Padovani F, Mairhörmann B, Falter-Braun P, Lengefeld J, Schmoller KM. Segmentation, tracking and cell cycle analysis of live-cell imaging data with Cell-ACDC. BMC Biol, 2022, 20(1): 174.
doi: 10.1186/s12915-022-01372-6 pmid: 35932043 |
| [22] | Salvatore M, Horlacher M, Marsico A, Winther O, Andersson R. Transfer learning identifies sequence determinants of cell-type specific regulatory element accessibility. NAR Genom Bioinform, 2023, 5(2): lqad026. |
| [23] | Lou RD, Chen JB, Hou HH, Liu YL, Tian Z, Zhang PC, Gui ZG. A new method of cell classification based on deep convolution neural networks. Journal of Test and Measurement Technology, 2019, 33(6): 509-515. |
| 娄润东, 陈俊彪, 侯宏花, 刘艳莉, 田珠, 张鹏程, 桂志国. 基于深度卷积神经网络的细胞分类新方法. 测试技术学报, 2019, 33(6): 509-515. | |
| [24] | Alaeddine H, Jihene M. Deep residual network in network. Comput Intell Neurosci, 2021, 2021: 6659083. |
| [25] |
De Donno C, Hediyeh-Zadeh S, Moinfar AA, Wagenstetter M, Zappia L, Lotfollahi M, Theis FJ. Population-level integration of single-cell datasets enables multi-scale analysis across samples. Nat Methods, 2023, 20(11): 1683-1692.
doi: 10.1038/s41592-023-02035-2 pmid: 37813989 |
| [26] | Yu LJ, Liu CL, Yang JYH, Yang PY. Ensemble deep learning of embeddings for clustering multimodal single- cell omics data. Bioinformatics, 2023, 39(6): btad382. |
| [27] |
Lin X, Tian T, Wei Z, Hakonarson H. Clustering of single-cell multi-omics data with a multimodal deep learning method. Nat Commun, 2022, 13(1): 7705.
doi: 10.1038/s41467-022-35031-9 pmid: 36513636 |
| [28] | Amin MR, Hasan M, Arnab SP, DeGiorgio M. Tensor decomposition-based feature extraction and classification to detect natural selection from genomic data. Mol Biol Evol. 2023 Oct 4; 40(10): msad216. |
| [29] |
Wu TC, Yu JX, Huang XF, Chen S, Wang YX, Pu YM. Preliminary study on deep learning picture classification model for identification and classification of invasion pattern of oral squamous cell carcinoma. J Oral Sci Res, 2023, 39(10): 917-922.
doi: 10.13701/j.cnki.kqyxyj.2023.10.013 |
|
吴天赐, 郁佳鑫, 黄晓峰, 陈盛, 王育新, 蒲玉梅. 深度学习图片分类模型ResNet-18用于判定口腔鳞状细胞癌浸润方式的初步研究. 口腔医学研究, 2023, 39(10): 917-922.
doi: 10.13701/j.cnki.kqyxyj.2023.10.013 |
|
| [30] | Qiao L, Wu WN, Lu ZT. Radiomics-based cerebrospinal fluid cell classification. Chinese Journal of Medical Physics, 2023, 40(2): 244-250. |
| 乔琳, 吴文娜, 卢振泰. 基于影像组学的脑脊液细胞分类方法. 中国医学物理学杂志, 2023, 40(2): 244-250. | |
| [31] | Liu M, Zhou L. Classification of cervical cells based on transfer learning and label smoothing strategy. Modern Computer, 2022, 28(19): 1-9+32. |
| 刘美, 周龙. 基于迁移学习与标签平滑策略的宫颈细胞分类方法. 现代计算机, 2022, 28(19): 1-9+32. | |
| [32] | Wu FQ, Lv LL, Lv D, Feng CB, Shi T, Wang W, Cui HH, Zhou Y. Deep learning model for automatic recognition of erythroid cells and granulocyte cells in bone marrow. Journal of Jilin University(lnformation Science Edition). 2020, 38(6): 729-736. |
| 吴汾奇, 吕丽丽, 吕迪, 冯辰彬, 施恬, 王维, 崔红花, 周柚. 骨髓红粒细胞自动识别的深度学习模型. 吉林大学学报(信息科学版), 2020, 38(6): 729-736. | |
| [33] | Sun K, Yao XF, Ma FL, Zhao WS, Huang G. Blood cell classification based on machine learning. Chinese Journal of Medical Physics, 2020, 37(1): 127-132. |
| 孙凯, 姚旭峰, 马风玲, 赵文硕, 黄钢. 基于机器学习的血细胞分类研究进展. 中国医学物理学杂志, 2020, 37(01): 127-132. | |
| [34] |
Frank SM, Qi A, Ravasio D, Sasaki Y, Rosen EL, Watanabe T. Supervised learning occurs in visual perceptual learning of complex natural images. Curr Biol, 2020, 30(15): 2995-3000.e3.
doi: S0960-9822(20)30737-5 pmid: 32502415 |
| [35] | Ju W, Luo X, Ma ZY, Yang JW, Deng MH, Zhang M. GHNN: graph harmonic neural networks for semi-supervised graph-level classification. Neural Netw, 2022, 151: 70-79. |
| [36] |
Kiselev VY, Andrews TS, Hemberg M. Challenges in unsupervised clustering of single-cell rna-seq data. Nat Rev Genet, 2019, 20(5): 273-282.
doi: 10.1038/s41576-018-0088-9 pmid: 30617341 |
| [37] | Stahlschmidt SR, Ulfenborg B, Synnergren J. Multimodal deep learning for biomedical data fusion: a review. Brief Bioinform, 2022, 23(2): bbab569. |
| [38] | Felix MA, Wagner A. Robustness and evolution: concepts, insights and challenges from a developmental model system. Heredity (Edinb), 2008, 100(2): 132-140. |
| [1] | Yanchun Bao, Caixia Shi, Chuanqiang Zhang, Mingjuan Gu, Lin Zhu, Zaixia Liu, Le Zhou, Fengying Ma, Risu Na, Wenguang Zhang. Progress on deep learning in genomics [J]. Hereditas(Beijing), 2024, 46(9): 701-715. |
| [2] | Fan Yang, Qiaoling Han, Wendi Zhao, Yue Zhao. EC number prediction of protein sequences based on combination of hierarchical and global features [J]. Hereditas(Beijing), 2024, 46(8): 661-669. |
| [3] | Huiyi Zheng, Huaxuan Wu, Zhiqiang Du. Gut metagenome-derived image augmentation and deep learning improve prediction accuracy of metabolic disease classification [J]. Hereditas(Beijing), 2024, 46(10): 886-896. |
| [4] | Weipeng Hu, Youping Li, Xiuqing Zhang. MHC-I epitope presentation prediction based on transfer learning [J]. Hereditas(Beijing), 2019, 41(11): 1041-1049. |
| [5] | Lan Kang, Jiayu Chen, Shaorong Gao. Historical review of reprogramming and pluripotent stem cell research in China [J]. Hereditas(Beijing), 2018, 40(10): 825-840. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||