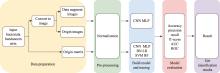
Hereditas(Beijing) ›› 2024, Vol. 46 ›› Issue (10): 886-896.doi: 10.16288/j.yczz.24-086
• Technique and Method • Previous Articles
Gut metagenome-derived image augmentation and deep learning improve prediction accuracy of metabolic disease classification
Huiyi Zheng( ), Huaxuan Wu(
), Huaxuan Wu( ), Zhiqiang Du(
), Zhiqiang Du( )
)
- College of Animal Science and Technology, Yangtze University, Jingzhou 434025, China
-
Received:2024-06-15Revised:2024-08-01Online:2024-08-08Published:2024-08-08 -
Contact:Zhiqiang Du E-mail:z15616428557@163.com;2021710855@yangtzeu.edu.cn;zhqdu@yangtzeu.edu.cn -
Supported by:Joint Research on Improved Livestock and Poultry Breeds in Anhui Province(2021-2025)
Cite this article
Huiyi Zheng, Huaxuan Wu, Zhiqiang Du. Gut metagenome-derived image augmentation and deep learning improve prediction accuracy of metabolic disease classification[J]. Hereditas(Beijing), 2024, 46(10): 886-896.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
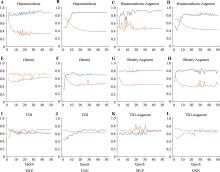
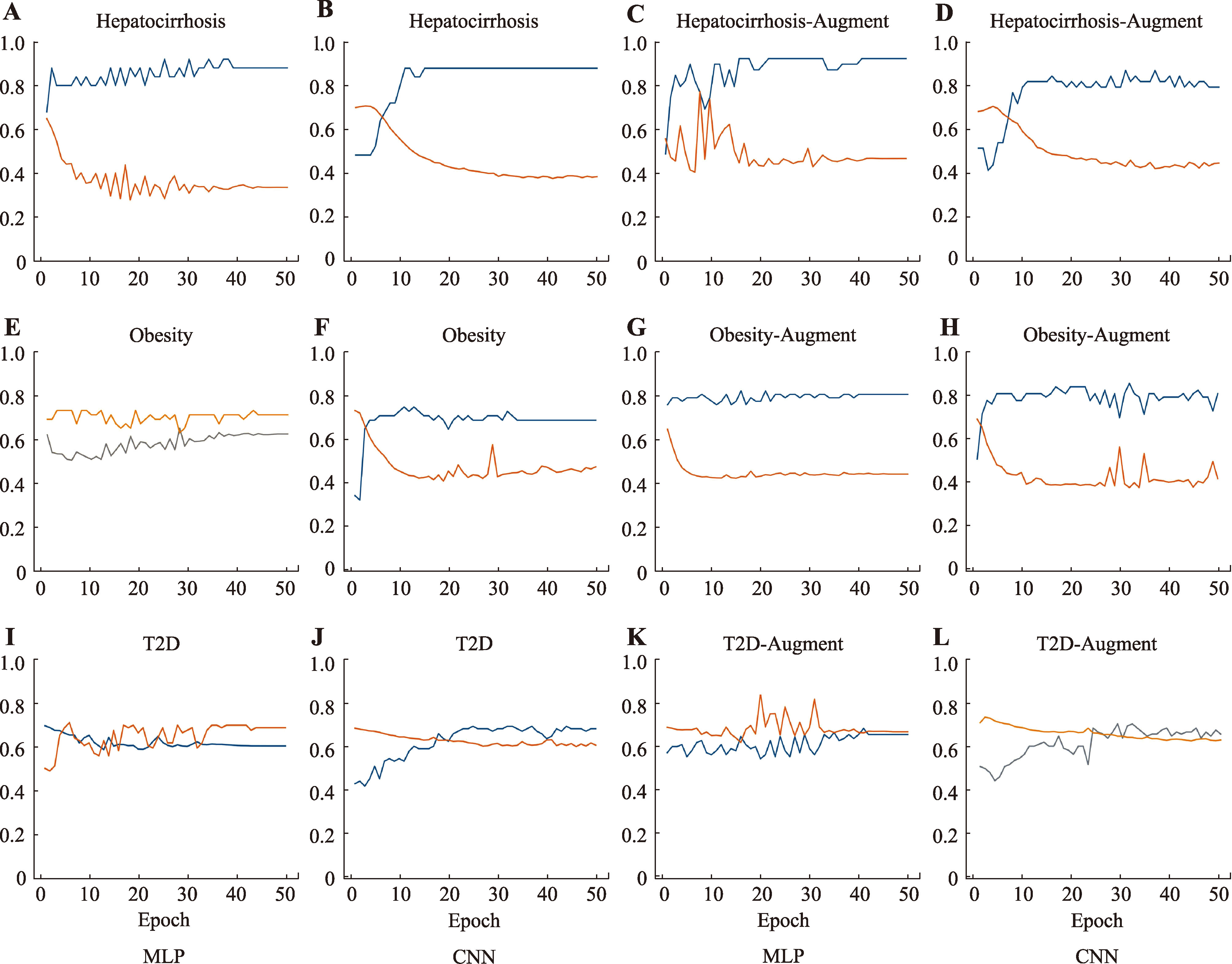
Table 2
Precision, recall and F1-score of machine learning and deep learning models"
| 数据集 | 指标 | MLP | CNN | BN | LR | SVM | RF | PopPhy- CNN | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Image Augmented | Image | Original | Image Augmented | Image | Original | |||||||
| Hepatocirrhosis | P | 1.00 | 1.00 | 0.85 | 0.90 | 0.86 | 0.85 | 0.75 | 0.73 | 0.73 | 0.85 | |
| R | 0.85 | 0.85 | 0.79 | 0.90 | 0.92 | 0.79 | 0.86 | 0.79 | 0.71 | 0.79 | ||
| F1 | 0.89 | 0.88 | 0.80 | 0.90 | 0.90 | 0.80 | 0.81 | 0.76 | 0.72 | 0.81 | ||
| Obesity | P | 0.81 | 0.74 | 0.77 | 0.79 | 0.86 | 0.67 | 0.54 | 0.71 | 0.65 | 0.63 | |
| R | 0.91 | 0.91 | 0.73 | 0.94 | 0.78 | 0.97 | 0.45 | 0.73 | 1.00 | 0.94 | ||
| F1 | 0.87 | 0.81 | 0.71 | 0.89 | 0.78 | 0.79 | 0.42 | 0.67 | 0.79 | 0.74 | 0.587 | |
| T2D | P | 0.66 | 0.69 | 0.66 | 0.69 | 0.65 | 0.82 | 0.68 | 0.60 | 0.61 | 0.59 | |
| R | 0.74 | 0.80 | 0.60 | 0.74 | 0.89 | 0.40 | 0.60 | 0.53 | 0.67 | 0.60 | ||
| F1 | 0.71 | 0.75 | 0.62 | 0.72 | 0.78 | 0.49 | 0.57 | 0.56 | 0.64 | 0.59 | 0.611 | |
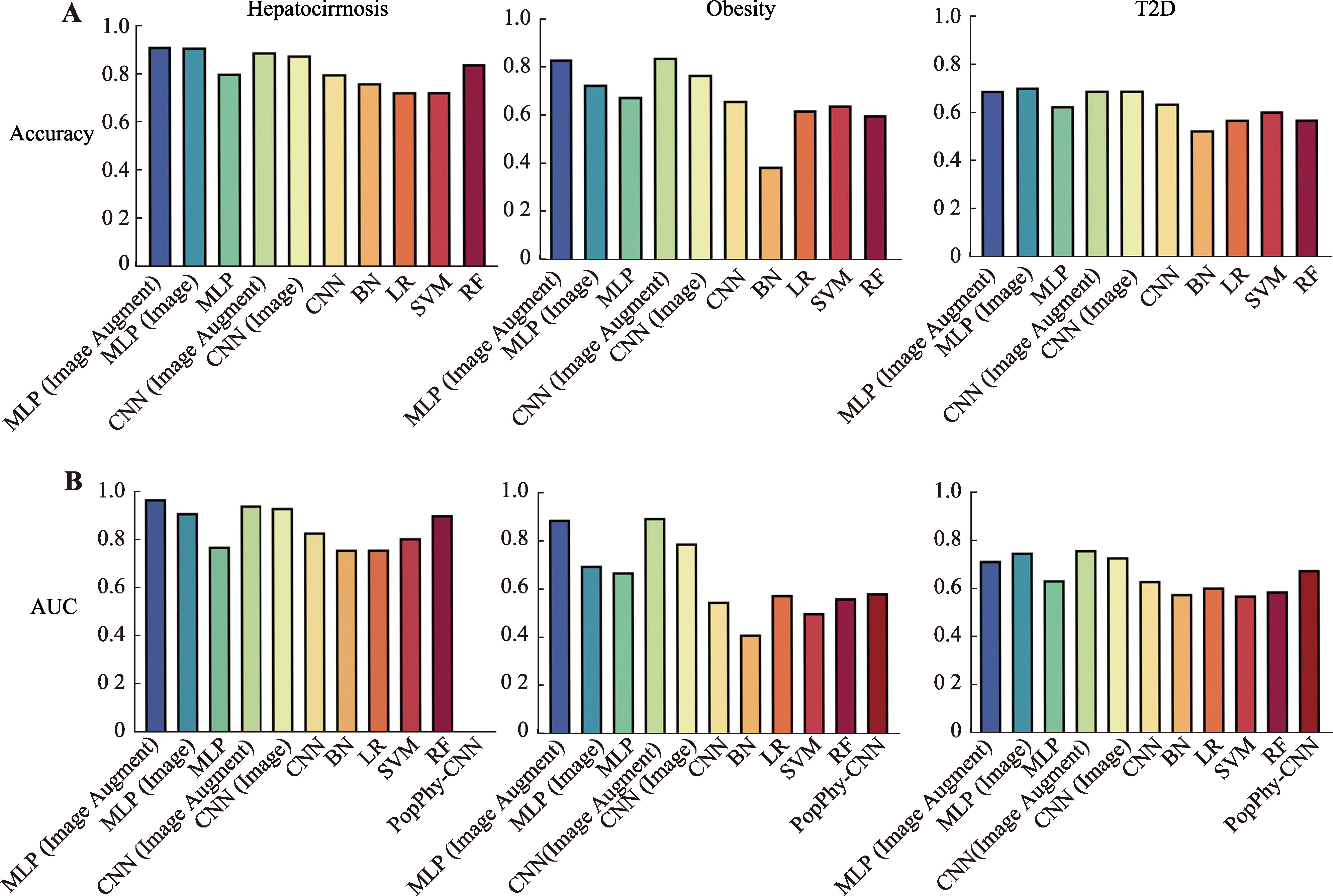
Table 3
Accuracy of cross-validation for machine learning and deep learning models"
| 数据集 | MLP | CNN | SVM | RF | BN | LR | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Image Augmented | Image | Original | Image Augmented | Image | Original | |||||
| Hepatocirrhosis | 0.921±0.021 | 0.908±0.039 | 0.835±0.042 | 0.890±0.043 | 0.888±0.047 | 0.808±0.034 | 0.738±0.080 | 0.688±0.032 | 0.862±0.055 | 0.765±0.047 |
| Obesity | 0.867±0.050 | 0.720±0.026 | 0.676±0.016 | 0.841±0.044 | 0.773±0.054 | 0.678±0.028 | 0.561±0.037 | 0.655±0.016 | 0.639±0.032 | 0.573±0.054 |
| T2D | 0.686±0.010 | 0.682±0.054 | 0.657±0.039 | 0.708±0.010 | 0.708±0.029 | 0.656±0.034 | 0.576±0.043 | 0.620±0.043 | 0.652±0.057 | 0.618±0.035 |
| [1] |
Bäckhed F, Ding H, Wang T, Hooper LV, Koh GY, Nagy A, Semenkovich CF, Gordon JI. The gut microbiota as an environmental factor that regulates fat storage. Proc Natl Acad Sci USA, 2004, 101(44): 15718-15723.
doi: 10.1073/pnas.0407076101 pmid: 15505215 |
| [2] | Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature, 2006, 444(7122): 1027-1031. |
| [3] | Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: human gut microbes associated with obesity. Nature, 2006, 444(7122): 1022-1023. |
| [4] | Tremaroli V, Bäckhed F. Functional interactions between the gut microbiota and host metabolism. Nature, 2012, 489(7415): 242-249. |
| [5] | Fan Y, Pedersen O. Gut microbiota in human metabolic health and disease. Nat Rev Microbiol, 2021, 19(1): 55-71. |
| [6] | Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, Affourtit JP, Egholm M, Henrissat B, Heath AC, Knight R, Gordon JI. A core gut microbiome in obese and lean twins. Nature, 2009, 457(7228): 480-484. |
| [7] | Ridaura VK, Faith JJ, Rey FE, Cheng JY, Duncan AE, Kau AL, Griffin NW, Lombard V, Henrissat B, Bain JR, Muehlbauer MJ, Ilkayeva O, Semenkovich CF, Funai K, Hayashi DK, Lyle BJ, Martini MC, Ursell LK, Clemente JC, Van Treuren W, Walters WA, Knight R, Newgard CB, Heath AC, Gordon JI. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science, 2013, 341(6150): 1241214. |
| [8] | Zeevi D, Korem T, Godneva A, Bar N, Kurilshikov A, Lotan-Pompan M, Weinberger A, Fu JY, Wijmenga C, Zhernakova A, Segal E. Structural variation in the gut microbiome associates with host health. Nature, 2019, 568(7750): 43-48. |
| [9] |
Kurilshikov A, Medina-Gomez C, Bacigalupe R, Radjabzadeh D, Wang J, Demirkan A, Le Roy CI, Raygoza Garay JA, Finnicum CT, Liu XR, Zhernakova DV, Bonder MJ, Hansen TH, Frost F, Rühlemann MC, Turpin W, Moon JY, Kim HN, Lüll K, Barkan E, Shah SA, Fornage M, Szopinska-Tokov J, Wallen ZD, Borisevich D, Agreus L, Andreasson A, Bang C, Bedrani L, Bell JT, Bisgaard H, Boehnke M, Boomsma DI, Burk RD, Claringbould A, Croitoru K, Davies GE, van Duijn CM, Duijts L, Falony G, Fu JY, van der Graaf A, Hansen T, Homuth G, Hughes DA, Ijzerman RG, Jackson MA, Jaddoe VWV, Joossens M, Jørgensen T, Keszthelyi D, Knight R, Laakso M, Laudes M, Launer LJ, Lieb W, Lusis AJ, Masclee AAM, Moll HA, Mujagic Z, Qibin Q, Rothschild D, Shin H, Sørensen SJ, Steves CJ, Thorsen J, Timpson NJ, Tito RY, Vieira-Silva S, Völker U, Völzke H, Võsa U, Wade KH, Walter S, Watanabe K, Weiss S, Weiss FU, Weissbrod O, Westra HJ, Willemsen G, Payami H, Jonkers DMAE, Arias Vasquez A, de Geus EJC, Meyer KA, Stokholm J, Segal E, Org E, Wijmenga C, Kim HL, Kaplan RC, Spector TD, Uitterlinden AG, Rivadeneira F, Franke A, Lerch MM, Franke L, Sanna S, D'Amato M, Pedersen O, Paterson AD, Kraaij R, Raes J, Zhernakova A. Large-scale association analyses identify host factors influencing human gut microbiome composition. Nat Genet, 2021, 53(2): 156-165.
doi: 10.1038/s41588-020-00763-1 pmid: 33462485 |
| [10] |
Lopera-Maya EA, Kurilshikov A, van der Graaf A, Hu SX, Andreu-Sánchez S, Chen LM, Vila AV, Gacesa R, Sinha T, Collij V, Klaassen MAY, Bolte LA, Gois MFB, Neerincx PBT, Swertz MA, LifeLines Cohort Study, Harmsen HJM, Wijmenga C, Fu JY, Weersma RK, Zhernakova A, Sanna S. Effect of host genetics on the gut microbiome in 7,738 participants of the Dutch Microbiome Project. Nat Genet, 2022, 54(2): 143-151.
doi: 10.1038/s41588-021-00992-y pmid: 35115690 |
| [11] |
Chen LM, Zhernakova DV, Kurilshikov A, Andreu- Sánchez S, Wang DM, Augustijn HE, Vich Vila A, Lifelines Cohort Study, Weersma RK, Medema MH, Netea MG, Kuipers F, Wijmenga C, Zhernakova A, Fu JY. Influence of the microbiome, diet and genetics on inter-individual variation in the human plasma metabolome. Nat Med, 2022, 28(11): 2333-2343.
doi: 10.1038/s41591-022-02014-8 pmid: 36216932 |
| [12] |
Sanna S, Kurilshikov A, van der Graaf A, Fu JY, Zhernakova A. Challenges and future directions for studying effects of host genetics on the gut microbiome. Nat Genet, 2022, 54(2): 100-106.
doi: 10.1038/s41588-021-00983-z pmid: 35115688 |
| [13] | Kim G, Yoon Y, Park JH, Park JW, Noh MG, Kim H, Park C, Kwon H, Park JH, Kim Y, Sohn J, Park S, Kim H, Im SK, Kim Y, Chung HY, Nam MH, Kwon JY, Kim IY, Kim YJ, Baek JH, Kim HS, Weinstock GM, Cho B, Lee C, Fang S, Park H, Seong JK. Bifidobacterial carbohydrate/ nucleoside metabolism enhances oxidative phosphorylation in white adipose tissue to protect against diet-induced obesity. Microbiome, 2022, 10(1): 188. |
| [14] |
Khan S, Hauptman R, Kelly L. Engineering the microbiome to prevent adverse events: challenges and opportunities. Annu Rev Pharmacol Toxicol, 2021, 61: 159-179.
doi: 10.1146/annurev-pharmtox-031620-031509 pmid: 33049161 |
| [15] | Giliberti R, Cavaliere S, Mauriello IE, Ercolini D, Pasolli E. Host phenotype classification from human microbiome data is mainly driven by the presence of microbial taxa. PLoS Comput Biol, 2022, 18(4): e1010066. |
| [16] |
LaPierre N, Ju CJT, Zhou GY, Wang W. MetaPheno: a critical evaluation of deep learning and machine learning in metagenome-based disease prediction. Methods, 2019, 166: 74-82.
doi: S1046-2023(18)30362-1 pmid: 30885720 |
| [17] | Aryal S, Alimadadi A, Manandhar I, Joe B, Cheng X. Machine learning strategy for gut microbiome-based diagnostic screening of cardiovascular disease. Hypertension, 2020, 76(5): 1555-1562. |
| [18] | Marcos-Zambrano LJ, Karaduzovic-Hadziabdic K, Loncar Turukalo T, Przymus P, Trajkovik V, Aasmets O, Berland M, Gruca A, Hasic J, Hron K, Klammsteiner T, Kolev M, Lahti L, Lopes MB, Moreno V, Naskinova I, Org E, Paciência I, Papoutsoglou G, Shigdel R, Stres B, Vilne B, Yousef M, Zdravevski E, Tsamardinos I, Carrillo de Santa Pau E, Claesson MJ, Moreno-Indias I, Truu J. Applications of machine learning in human microbiome studies: a review on feature selection, biomarker identification, disease prediction and treatment. Front Microbiol, 2021, 12: 634511. |
| [19] | Holmes S. Successful strategies for human microbiome data generation, storage and analyses. J Biosci, 2019, 44(5): 111. |
| [20] | Galloway-Peña J, Hanson B. Tools for analysis of the microbiome. Dig Dis Sci, 2020, 65(3): 674-685. |
| [21] | Clausen DS, Willis AD. Evaluating replicability in microbiome data. Biostatistics, 2022, 23(4): 1099-1114. |
| [22] | Datta S, Guha S. Statistical Analysis of Microbiome Data. Springer International Publishing, 2021. |
| [23] |
Zhang XY, Mallick H, Tang ZX, Zhang L, Cui XQ, Benson AK, Yi NJ. Negative binomial mixed models for analyzing microbiome count data. BMC Bioinformatics, 2017, 18(1): 4.
doi: 10.1186/s12859-016-1441-7 pmid: 28049409 |
| [24] |
Zhang XY, Yi NJ. NBZIMM: negative binomial and zero-inflated mixed models, with application to microbiome/metagenomics data analysis. BMC Bioinformatics, 2020, 21(1): 488.
doi: 10.1186/s12859-020-03803-z pmid: 33126862 |
| [25] | Liu TT, Xu PR, Du YY, Lu H, Zhao HY, Wang T. MZINBVA: variational approximation for multilevel zero-inflated negative-binomial models for association analysis in microbiome surveys. Brief Bioinform, 2022, 23(1): bbab443. |
| [26] |
Curry KD, Nute MG, Treangen TJ. It takes guts to learn: machine learning techniques for disease detection from the gut microbiome. Emerg Top Life Sci, 2021, 5(6): 815-827.
doi: 10.1042/ETLS20210213 pmid: 34779841 |
| [27] | Vilne B, Ķibilds J, Siksna I, Lazda I, Valciņa O, Krūmiņa A. Could artificial intelligence/machine learning and inclusion of diet-gut microbiome interactions improve disease risk prediction? case study: coronary artery disease. Front Microbiol, 2022, 13: 627892. |
| [28] |
Li Y, Xu ZL, Han WK, Cao HL, Umarov R, Yan AX, Fan M, Chen H, Duarte CM, Li LH, Ho PL, Gao X. HMD-ARG: hierarchical multi-task deep learning for annotating antibiotic resistance genes. Microbiome, 2021, 9(1): 40.
doi: 10.1186/s40168-021-01002-3 pmid: 33557954 |
| [29] |
Cai Y, Gu H, Kenney T. Learning microbial community structures with supervised and unsupervised non-negative matrix factorization. Microbiome, 2017, 5(1): 110.
doi: 10.1186/s40168-017-0323-1 pmid: 28859695 |
| [30] |
Chen D, Wang SJ, Zhao ZJ, Ji X, Shen Q, Yu Y, Cui SD, Wang JG, Chen ZY, Wang JY, Guo ZY, Wu PX, Tang GQ. Genomic prediction of pig growth traits based on machine learning. Hereditas(Beijing), 2023, 45(10): 922-932.
doi: 10.16288/j.yczz.23-120 pmid: 37872114 |
| 陈栋, 王书杰, 赵真坚, 姬祥, 申琦, 余杨, 崔晟頔, 王俊戈, 陈子旸, 王金勇, 郭宗义, 吴平先, 唐国庆. 基于机器学习的猪生长性状基因组预测. 遗传, 2023, 45(10): 922-932. | |
| [31] |
Liu B, Sträuber H, Saraiva J, Harms H, Silva SG, Kasmanas JC, Kleinsteuber S, Nunes da Rocha U. Machine learning-assisted identification of bioindicators predicts medium-chain carboxylate production performance of an anaerobic mixed culture. Microbiome, 2022, 10(1): 48.
doi: 10.1186/s40168-021-01219-2 pmid: 35331330 |
| [32] | Vangay P, Hillmann BM, Knights D. Microbiome Learning Repo (ML Repo): a public repository of microbiome regression and classification tasks. Gigascience, 2019, 8(5): giz042. |
| [33] | Reiman D, Metwally AA, Sun J, Dai Y. PopPhy-CNN: a phylogenetic tree embedded architecture for convolutional neural networks to predict host phenotype from metagenomic data. IEEE J Biomed Health Inform, 2020, 24(10): 2993-3001. |
| [34] |
Sharma A, Vans E, Shigemizu D, Boroevich KA, Tsunoda T. DeepInsight: a methodology to transform a non-image data to an image for convolution neural network architecture. Sci Rep, 2019, 9(1): 11399.
doi: 10.1038/s41598-019-47765-6 pmid: 31388036 |
| [35] | Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, Blondel M, Prettenhofer P, Weiss R, Dubourg V, Vanderplas J, Passos A, Cournapeau D, Brucher M, Perrot M, Duchesnay E. Scikit-learn: machine learning in Python. J Mach Learn Res, 2011, 12(85): 2825-2830. |
| [36] | Paszke A, Gross S, Massa F, Lerer A, Bradbury J, Chanan G, Killeen T, Lin ZM, Gimelshein N, Antiga L, Desmaison A, Köpf A, Yang E, DeVito Z, Raison M, Tejani A, Chilamkurthy S, Steiner B, Fang L, Bai JJ, Chintala S. Pytorch: an imperative style, high-performance deep learning library. Advances in neural information processing systems, 2019, 32: 8024-8035. |
| [37] | Zhang XH, Zou YX, Shi W. Dilated convolution neural network with LeakyReLU for environmental sound classification. The 22nd International Conference on Digital Signal Processing (DSP), 2017, 1-5. |
| [38] | Wen CL, Sun CJ, Yang N. The concepts and research progress: from heritability to microbiability. Hereditas(Beijing), 2019, 41(11): 1023-1040. |
| 文超良, 孙从佼, 杨宁. 从遗传力到肠菌力:概念及研究进展. 遗传, 2019, 41(11): 1023-1040. | |
| [39] | Szegedy C, Liu W, Jia YQ, Sermanet P, Reed S, Anguelov D, Erhan D, Vanhoucke V, Rabinovich A. Going deeper with convolutions. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), 2015, 1-9. |
| [40] | He KM, Zhang XY, Ren SQ, Sun J. Deep residual learning for image recognition. Proceedings of the IEEE conference on computer vision and pattern recognition, 2016, 770-778. |
| [41] | Krizhevsky A, Sutskever I, Hinton GE. Imagenet classification with deep convolutional neural networks. Commun ACM, 2017, 60(6): 84-90. |
| [42] | Shorten C, Khoshgoftaar TM. A survey on image data augmentation for deep learning. J Big Data, 2019, 6(1): 1-48. |
| [43] | Goodfellow I, Pouget-Abadie J, Mirza M, Xu B, Warde-Farley D, Ozair S, Courville A, Bengio Y. Generative adversarial networks. Commun ACM, 2020, 63(11): 139-144. |
| [44] |
Chen XN, Chen DG, Zhao ZM, Balko JM, Chen JC. Artificial image objects for classification of breast cancer biomarkers with transcriptome sequencing data and convolutional neural network algorithms. Breast Cancer Res, 2021, 23(1): 96.
doi: 10.1186/s13058-021-01474-z pmid: 34629099 |
| [45] | Chen XN, Chen DG, Zhao ZM, Zhan J, Ji CR, Chen JC. Artificial image objects for classification of schizophrenia with GWAS-selected SNVs and convolutional neural network. Patterns (NY), 2021, 2(8): 100303. |
| [46] | Tang H, Yu XT, Liu R, Zeng T. Vec2image: an explainable artificial intelligence model for the feature representation and classification of high-dimensional biological data by vector-to-image conversion. Brief Bioinform, 2022, 23(2): bbab584. |
| [47] | He T, Zhang Z, Zhang H, Zhang ZY, Xie JY, Li M. Bag of tricks for image classification with convolutional neural networks. Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, 2019, 558-567. |
| [48] | Aasmets O, Lüll K, Lang JM, Pan C, Kuusisto J, Fischer K, Laakso M, Lusis AJ, Org E. Machine learning reveals time-varying microbial predictors with complex effects on glucose regulation. mSystems, 2021, 6(1): e01191-20. |
| [49] | Kodikara S, Ellul S, Lê Cao KA. Statistical challenges in longitudinal microbiome data analysis. Brief Bioinform, 2022, 23(4): bbac273. |
| [50] |
Wang C, Segal LN, Hu JY, Zhou BY, Hayes RB, Ahn J, Li HL. Microbial risk score for capturing microbial characteristics, integrating multi-omics data, and predicting disease risk. Microbiome, 2022, 10(1): 121.
doi: 10.1186/s40168-022-01310-2 pmid: 35932029 |
| [51] | Corander J, Hanage WP, Pensar J. Causal discovery for the microbiome. Lancet Microbe, 2022, 3(11): e881-e887. |
| [52] |
Xu YW, Nash K, Acharjee A, Gkoutos GV. CACONET: a novel classification framework for microbial correlation networks. Bioinformatics, 2022, 38(6): 1639-1647.
doi: 10.1093/bioinformatics/btab879 pmid: 34983063 |
| [1] | Yanchun Bao, Caixia Shi, Chuanqiang Zhang, Mingjuan Gu, Lin Zhu, Zaixia Liu, Le Zhou, Fengying Ma, Risu Na, Wenguang Zhang. Progress on deep learning in genomics [J]. Hereditas(Beijing), 2024, 46(9): 701-715. |
| [2] | Fan Yang, Qiaoling Han, Wendi Zhao, Yue Zhao. EC number prediction of protein sequences based on combination of hierarchical and global features [J]. Hereditas(Beijing), 2024, 46(8): 661-669. |
| [3] | Hui Liang, Xue Wang, Jingfang Si, Yi Zhang. Classification accuracy of machine learning algorithms for Chinese local cattle breeds using genomic markers [J]. Hereditas(Beijing), 2024, 46(7): 530-539. |
| [4] | Ziyi Zhang, Qilin Wang, Junyou Zhang, Yingying Duan, Jiaxin Liu, Zhaoshuo Liu, Chunyan Li. Machine learning applications in breast cancer survival and therapeutic outcome prediction based on multi-omic analysis [J]. Hereditas(Beijing), 2024, 46(10): 820-832. |
| [5] | Dong Chen, Shujie Wang, Zhenjian Zhao, Xiang Ji, Qi Shen, Yang Yu, Shengdi Cui, Junge Wang, Ziyang Chen, Jinyong Wang, Zongyi Guo, Pingxian Wu, Guoqing Tang. Genomic prediction of pig growth traits based on machine learning [J]. Hereditas(Beijing), 2023, 45(10): 922-932. |
| [6] | Yongqiang Kong, Jinkai Liu, Jiaqi Gu, Jingyi Xu, Yunuo Zheng, Yiliang Wei, Shaoyuan Wu. Optimization scheme of machine learning model for genetic division between northern Han, southern Han, Korean and Japanese [J]. Hereditas(Beijing), 2022, 44(11): 1028-1043. |
| [7] | Yali Hu, Rui Dai, Yongxin Liu, Jingying Zhang, Bin Hu, Chengcai Chu, Huaibo Yuan, Yang Bai. Analysis of rice root bacterial microbiota of Nipponbare and IR24 [J]. Hereditas(Beijing), 2020, 42(5): 506-518. |
| [8] | Weipeng Hu, Youping Li, Xiuqing Zhang. MHC-I epitope presentation prediction based on transfer learning [J]. Hereditas(Beijing), 2019, 41(11): 1041-1049. |
| [9] | Zhao Xuetong, Yang Yadong, Qu Hongzhu, Fang Xiangdong. Applications of machine learning in clinical decision support in the omic era [J]. Hereditas(Beijing), 2018, 40(9): 693-703. |
| [10] | Zhang Guishan, Yang Yong, Zhang Lingmin, Dai Xianhua. Application of machine learning in the CRISPR/Cas9 system [J]. Hereditas(Beijing), 2018, 40(9): 704-723. |
| [11] | Zhe-ye Peng,Zi-jun Tang,Min-zhu Xie. Research progress in machine learning methods for gene-gene interaction detection [J]. Hereditas(Beijing), 2018, 40(3): 218-226. |
| [12] | HOU Yan-Yan, YING Xiao-Min, LI Wu-Ju . Computational approaches to microRNA discovery [J]. HEREDITAS, 2008, 30(6): 687-696. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||