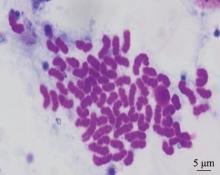
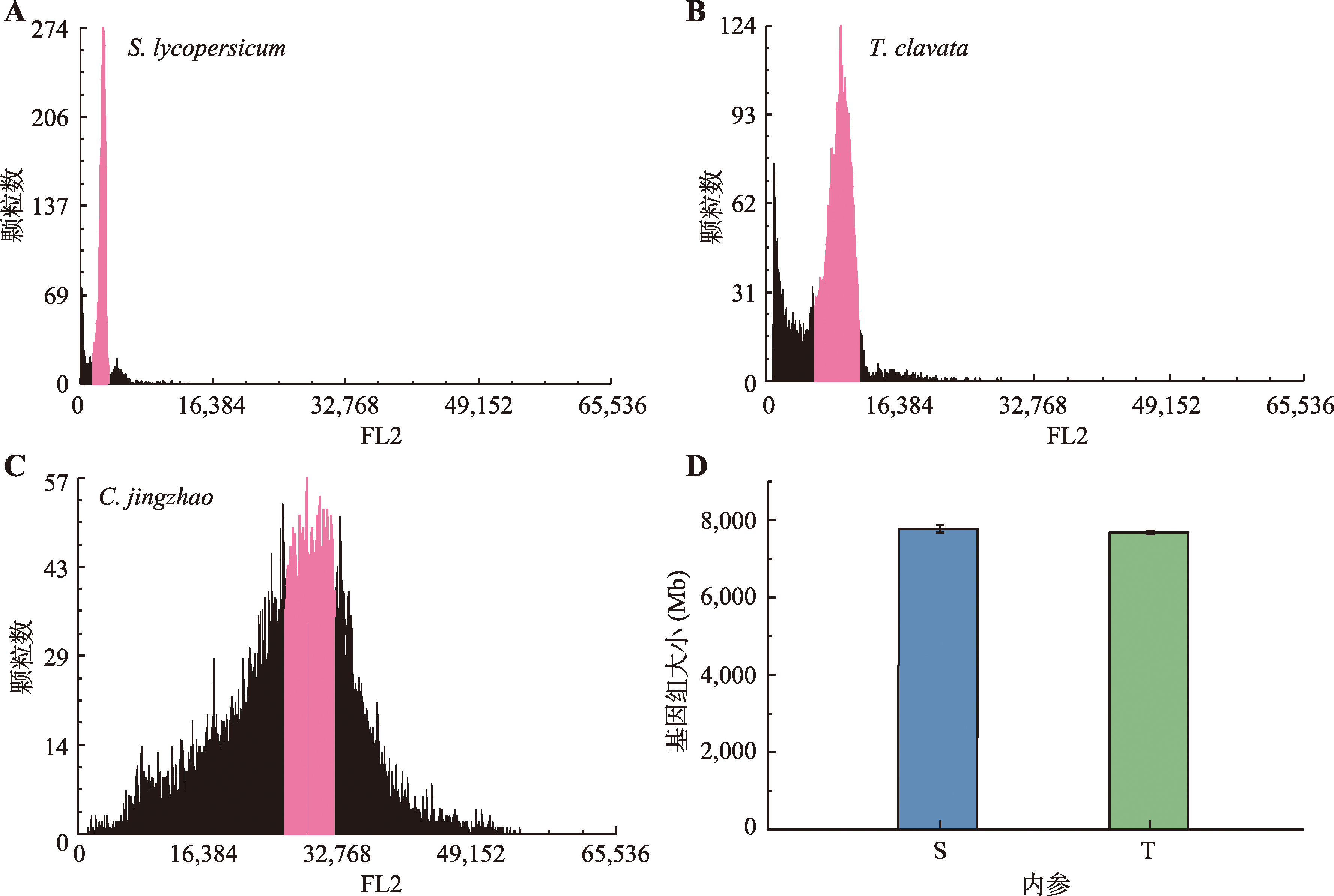
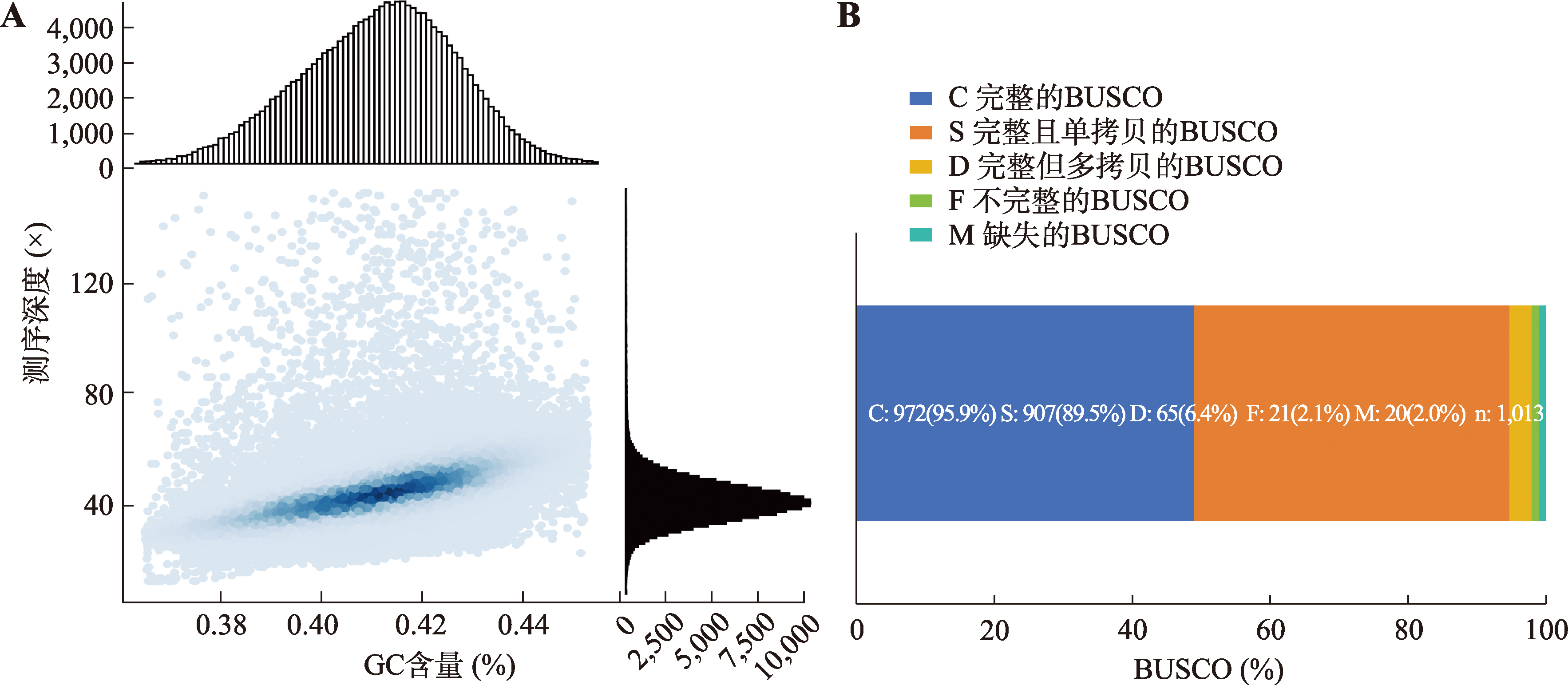
Hereditas(Beijing) ›› 2025, Vol. 47 ›› Issue (12): 1351-1364.doi: 10.16288/j.yczz.25-026
• Research Article • Previous Articles Next Articles
Karyotype and genome characterization analysis of Chilobrachys jingzhao (Theraphosidae: Chilobrachys)
Yuxuan Zhang1,2( ), Mengying Zhang3, Hanting Yang1,2, Chi Song1,2(
), Mengying Zhang3, Hanting Yang1,2, Chi Song1,2( ), Zizhong Yang3(
), Zizhong Yang3( ), Shilin Chen1,2(
), Shilin Chen1,2( )
)
1. School of Pharmacy ,Chengdu University of Traditional Chinese Medicine Chengdu 611137, China 2. Institute of Herbgenomics ,Chengdu University of Traditional Chinese Medicine Chengdu 611137, China 3. Yunnan Provincial Key Laboratory of Entomological Biopharmaceutical R&D ,College of Pharmacy, Dali University Dali 671000, China
-
Received:2025-01-21Revised:2025-03-23Online:2025-05-21Published:2025-05-21 -
Contact:Chi Song, Zizhong Yang, Shilin Chen E-mail:18744989564@163.com;songchi@cdutcm.edu.cn;yangzzh69@163.com;slchen@cdutcm.edu.cn -
Supported by:Chengdu University of Traditional Chinese Medicine Talent Introduction Research Startup Fund(030040015);Chengdu University of Traditional Chinese Medicine Talent Introduction Research Startup Fund(030040017)
Cite this article
Yuxuan Zhang, Mengying Zhang, Hanting Yang, Chi Song, Zizhong Yang, Shilin Chen. Karyotype and genome characterization analysis of Chilobrachys jingzhao (Theraphosidae: Chilobrachys)[J]. Hereditas(Beijing), 2025, 47(12): 1351-1364.
share this article
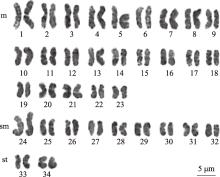
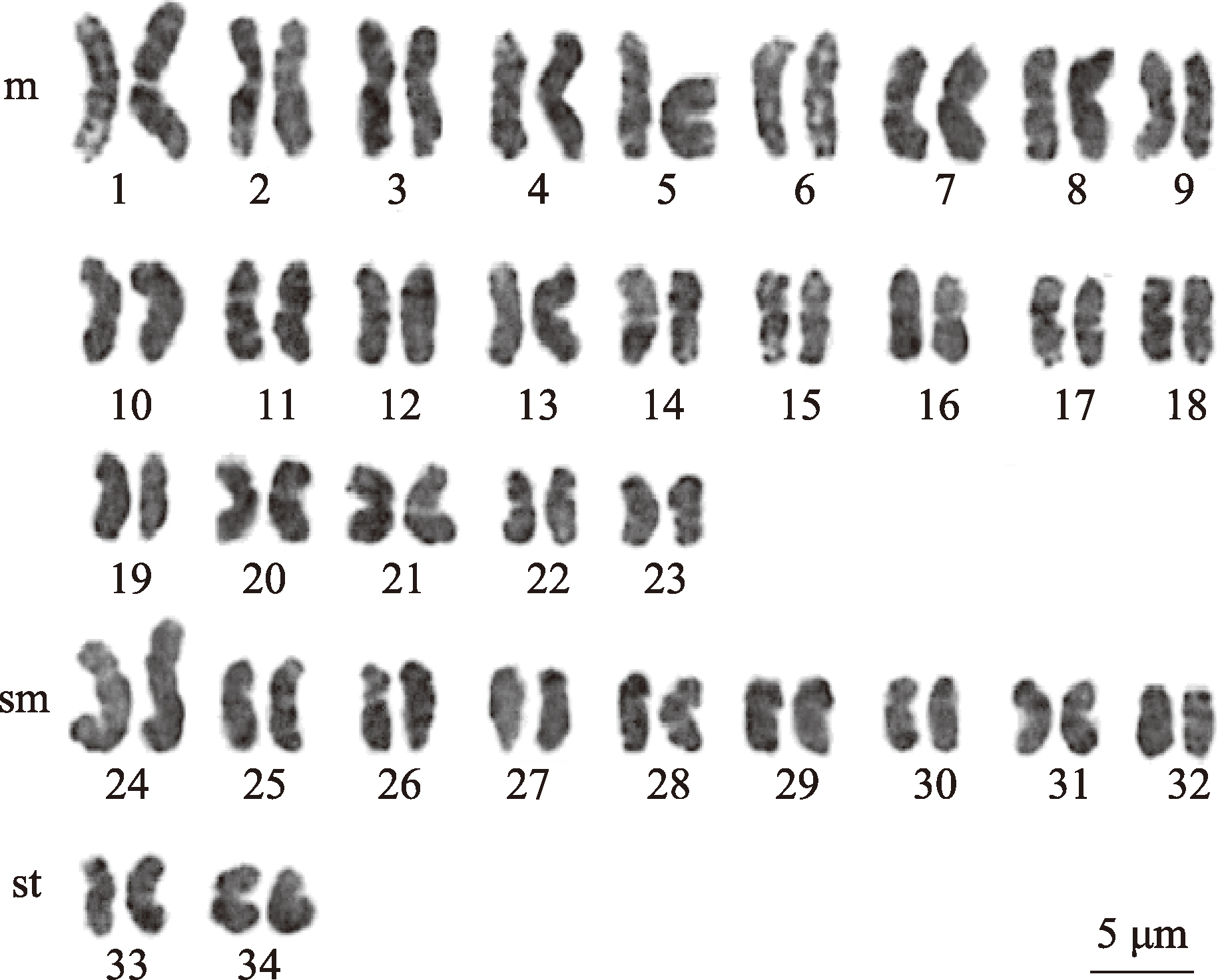
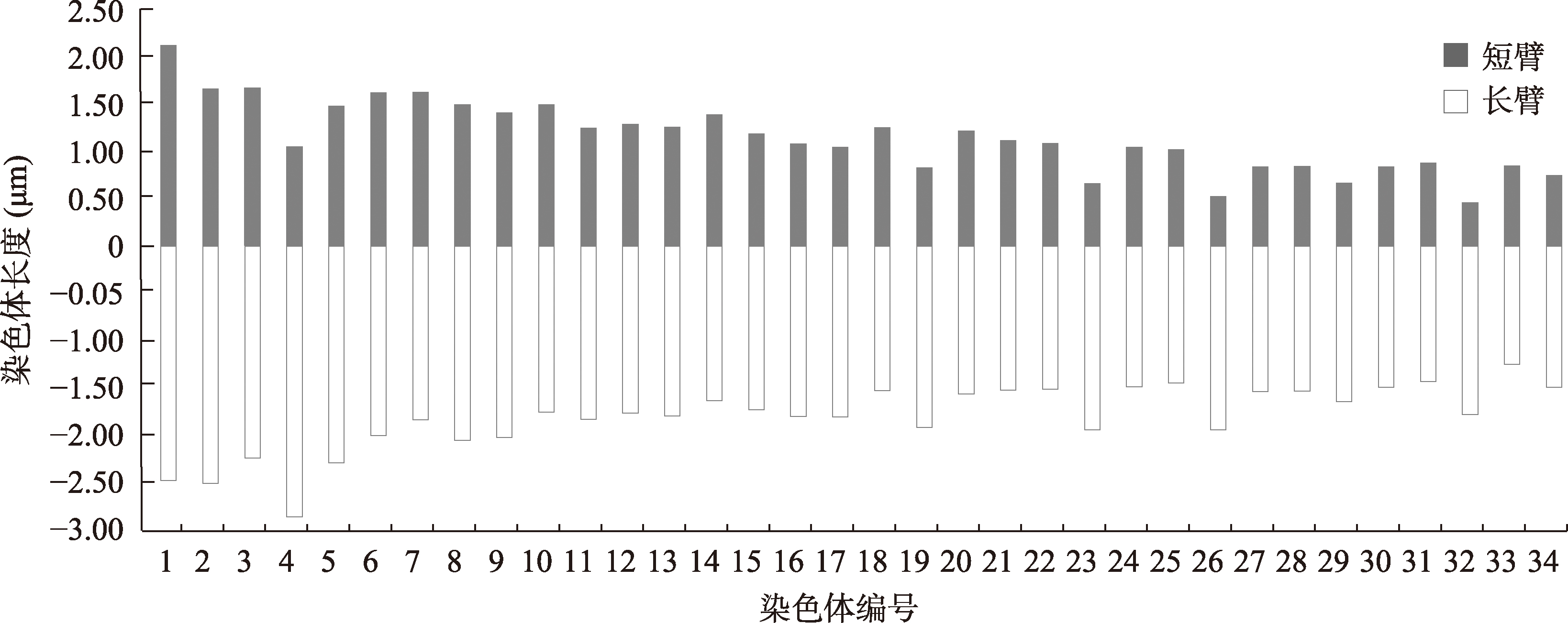
Table 1
Parameters of the chromosome karyotype of C. jingzhao"
| 编号 | 染色体相对长度(%)±标准差 | 臂比±标准差 | 相对长度系数 | 染色体类型 | |
|---|---|---|---|---|---|
| Levan分类标准 | Kuo分类标准 | ||||
| 1 | 4.59±0.14 | 1.17±0.02 | 1.56 | m | L |
| 2 | 4.17±0.16 | 1.52±0.02 | 1.41 | m | L |
| 3 | 3.91±0.05 | 1.35±0.03 | 1.33 | m | L |
| 4 | 3.77±0.03 | 1.56±0.02 | 1.33 | m | L |
| 5 | 3.62±0.02 | 1.24±0.04 | 1.28 | m | L |
| 6 | 3.54±0.05 | 1.38±0.02 | 1.23 | m | M2 |
| 7 | 3.46±0.02 | 1.13±0.02 | 1.20 | m | M2 |
| 8 | 3.43±0.02 | 1.44±0.02 | 1.17 | m | M2 |
| 9 | 3.24±0.03 | 1.18±0.01 | 1.16 | m | M2 |
| 10 | 3.07±0.04 | 1.48±0.03 | 1.10 | m | M2 |
| 11 | 3.04±0.03 | 1.38±0.02 | 1.04 | m | M2 |
| 12 | 3.04±0.03 | 1.44±0.02 | 1.03 | m | M2 |
| 13 | 3.01±0.03 | 1.18±0.01 | 1.03 | m | M2 |
| 14 | 2.91±0.03 | 1.47±0.02 | 1.02 | m | M2 |
| 15 | 2.88±0.03 | 1.68±0.03 | 0.99 | m | M1 |
| 16 | 2.78±0.03 | 1.23±0.01 | 0.98 | m | M1 |
| 17 | 2.78±0.03 | 1.29±0.01 | 0.97 | m | M1 |
| 18 | 2.63±0.03 | 1.37±0.02 | 0.94 | m | M1 |
| 19 | 2.59±0.03 | 1.41±0.03 | 0.94 | m | M1 |
| 20 | 2.52±0.03 | 1.43±0.01 | 0.93 | m | M1 |
| 21 | 2.46±0.03 | 1.43±0.02 | 0.89 | m | M1 |
| 22 | 2.30±0.02 | 1.64±0.02 | 0.88 | m | M1 |
| 23 | 2.09±0.02 | 1.49±0.02 | 0.88 | m | M1 |
| 24 | 3.91±0.04 | 2.74±0.02 | 0.86 | sm | M1 |
| 25 | 2.85±0.03 | 1.74±0.02 | 0.83 | sm | M1 |
| 26 | 2.74±0.03 | 2.34±0.04 | 0.83 | sm | M1 |
| 27 | 2.59±0.03 | 2.97±0.03 | 0.80 | sm | M1 |
| 28 | 2.37±0.03 | 1.85±0.02 | 0.80 | sm | M1 |
| 29 | 2.37±0.03 | 1.84±0.02 | 0.79 | sm | M1 |
| 30 | 2.33±0.02 | 1.79±0.02 | 0.78 | sm | M1 |
| 31 | 2.30±0.02 | 2.50±0.03 | 0.78 | sm | M1 |
| 32 | 2.23±0.02 | 2.02±0.03 | 0.76 | sm | M1 |
| 33 | 2.46±0.03 | 3.77±0.06 | 0.76 | st | M1 |
| 34 | 2.23±0.02 | 3.94±0.12 | 0.71 | st | S |
Table 3
Chromosome karyotype statistics in Theraphosidae"
| 物种 | 2n | 性染色体系统 | 核型公式 | 参考文献 |
|---|---|---|---|---|
| Acanthoscurria gomesiana | 43~46 | m, sm, st, t | [ | |
| Aphonopelma hentzi | 44 | X1X2O | [ | |
| Avicularia minatrix | 78 | X1X2X3X4O | 52m/sm+4st+18t+X1sm+X2m+X3m+X4t | [ |
| Chaetopelma olivaceum | (16) | [ | ||
| Grammostola rosea | 72 | X1X2O | 64m+4sm+2st+X1m+X2sm | [ |
| Grammostola aff. rosea | 72 | X1X2O+2CSCP | X1m+X2t+CSCP1m+CSCP2sm | [ |
| Holothele cf. longipes | 73 | X1X2X3O | 66m/sm+4t/T+X1X2X3m/sm | [ |
| Idiothele mira | 25 | XO | 18m+2sm+2st+2t+Xm | [ |
| Ischnocolus jickelii | 85 | X1X2X3O | 70m+12sm+X1X2X3m/sm | [ |
| Pelinobius muticus | 67 | X1X2X3O | 46t+18m/sm+X1m+X2st+X3t | [ |
| Poecilotheria formosa | 110 | X1X2X3X4O | 30m+18sm+18st+40t+X1X2X3m+X4sm | [ |
| Psalmopoeus cambridgei | 84 | X1X2O | 64m/sm+18t+X1X2m | [ |
| Pterinochilus lugardi | 23 | XO+CSCP | sm, m | [ |
| Pterinochilus murinus | 43 | XO | 36m+6sm+Xm | [ |
| Pterinopelma roseum | 48 | st, t | [ | |
| Tekoapora wacketi | 48 | st, t | [ | |
| Tliltocatl albopilosus | 74 | X1X2X3X4O | 64m/sm+6st+X1X2X3X4m | [ |
| Vitalius dubius | 74~81 | m, sm, st, t | [ | |
| Vitalius sorocabae | 48 | st, t | [ | |
| Vitalius sp. | 48 | m, sm, st, t | [ |
| [1] | 田建平, 胡远艳, 杨卫丽, 潘坤, 张俊清, 曾渝. 海南黎族药用动物大型蜘蛛资源调查及其传统习用研究. 中药材, 2017, 40(8): 1811-1814. |
| [2] | Tian JP, Hu YY, Zhang MN, Chen GL. History and application characteristics of domestic spider used for medicine and edible resources. Chin J Exp Tradit Med Formulae, 2018, 24(3): 214-218. |
| 田建平, 胡远艳, 张名楠, 陈国良. 国产蜘蛛药用及食用的历史沿革与应用特点. 中国实验方剂学杂志, 2018, 24(3): 214-218. | |
| [3] |
Pineda SS, Chaumeil PA, Kunert A, Kaas Q, Thang MWC, Le L, Nuhn M, Herzig V, Saez NJ, Cristofori-Armstrong B, Anangi R, Senff S, Gorse D, King GF. ArachnoServer 3.0: an online resource for automated discovery, analysis and annotation of spider toxins. Bioinformatics, 2018, 34(6): 1074-1076.
pmid: 29069336 |
| [4] |
Nicolas S, Zoukimian C, Bosmans F, Montnach J, Diochot S, Cuypers E, De Waard S, Béroud R, Mebs D, Craik D, Boturyn D, Lazdunski M, Tytgat J, De Waard M. Chemical synthesis, proper folding, Nav channel selectivity profile and analgesic properties of the spider peptide Phlotoxin 1. Toxins (Basel), 2019, 11(6): 367.
pmid: 31234412 |
| [5] | Liu XY, Qian F. The inhibitory effect and mechanism of the JZ-VF3 fragment from the venom toxin JZTX-V of Chilobrachys jingzhao on macrophage inflammatory response. J Shihezi Univ (Nat Sci), 2024, 42(5): 614-620. |
| 刘欣悦, 钱峰. 敬钊缨毛蛛肽类毒素JZTX-V中JZ-VF3片段对巨噬细胞炎症反应的抑制作用及机制研究. 石河子大学学报(自然科学版), 2024, 42(5): 614-620. | |
| [6] |
Wu WF, Yin Y, Feng PH, Chen G, Pan LY, Gu PY, Zhou SQ, Lin FL, Ji SY, Zheng CB, Deng MC. Spider venom-derived peptide JZTX-14 prevents migration and invasion of breast cancer cells via inhibition of sodium channels. Front Pharmacol, 2023, 14: 1067665.
pmid: 37033662 |
| [7] |
Chen JJ, Zhao LQ, Jiang LP, Meng E, Zhang YQ, Xiong X, Liang SP. Transcriptome analysis revealed novel possible venom components and cellular processes of the tarantula Chilobrachys jingzhao venom gland. Toxicon, 2008, 52(7): 794-806.
pmid: 18778726 |
| [8] |
Liao Z, Cao J, Li SM, Yan XJ, Hu WJ, He QY, Chen JJ, Tang JZ, Xie JY, Liang SP. Proteomic and peptidomic analysis of the venom from Chinese tarantula Chilobrachys jingzhao. Proteomics, 2007, 7(11): 1892-1907.
pmid: 17476710 |
| [9] | Shi JJ, Xu RJ, Xu H, Ma S, Jia QR, Gao ZM. Karyotype and genome size analyses of Daemonorops jenkinsiana. J Trop Subtrop Bot, 2019, 27(3): 315-322. |
| 史晶晶, 徐瑞晶, 徐浩, 马霜, 贾秋蕊, 高志民. 黄藤染色体核型及基因组大小分析. 热带亚热带植物学报, 2019, 27(3): 315-322. | |
| [10] | Vimala Y, Lavania S, Lavania UC. Chromosome change and karyotype differentiation-implications in speciation and plant systematics. Nucleus, 2021, 64(1): 33-54. |
| [11] |
Graphodatsky AS, Trifonov VA, Stanyon R. The genome diversity and karyotype evolution of mammals. Mol Cytogenet, 2011, 4(1): 22.
pmid: 21992653 |
| [12] |
Bennett MD, Leitch IJ. Nuclear DNA amounts in angiosperms: progress, problems and prospects. Ann Bot, 2005, 95(1): 45-90.
pmid: 15596457 |
| [13] |
Kim JH, Roh JY, Kwon DH, Kim YH, Yoon KA, Yoo S, Noh SJ, Park J, Shin EH, Park MY, Lee SH. Estimation of the genome sizes of the chigger mites Leptotrombidium pallidum and Leptotrombidium scutellare based on quantitative PCR and k-mer analysis. Parasit Vectors, 2014, 7(1): 279.
pmid: 24947244 |
| [14] |
Zhou YT, Xiao SJ, Lin G, Chen D, Cen W, Xue T, Liu ZY, Zhong JX, Chen YT, Xiao YJ, Chen JH, Guo YH, Chen YQ, Zhang YD, Hu XF, Huang Z. Chromosome genome assembly and annotation of the yellowbelly pufferfish with PacBio and Hi-C sequencing data. Sci Data, 2019, 6(1): 267.
pmid: 31704938 |
| [15] |
Liu JF, Qin KW, Wu CL, Fu KF, Yu XJ, Zhou LJ. De novo sequencing provides insights into the pathogenicity of Foodborne Vibrio parahaemolyticus. Front Cell Infect Microbiol, 2021, 11: 652957.
pmid: 34055666 |
| [16] |
Král J, Forman M, Kořínková T, Lerma ACR, Haddad CR, Musilová J, Řezáč M, Herrera IMÁ, Thakur S, Dippenaar- Schoeman AS, Marec F, Horová L, Bureš P. Insights into the karyotype and genome evolution of haplogyne spiders indicate a polyploid origin of lineage with holokinetic chromosomes. Sci Rep, 2019, 9(1): 3001.
pmid: 30816146 |
| [17] |
Yang RD, Nelson AC, Henzler C, Thyagarajan B, Silverstein KAT. ScanIndel: a hybrid framework for indel detection via gapped alignment, split reads and de novo assembly. Genome Med, 2015, 7: 127.
pmid: 26643039 |
| [18] |
Deng N, Hou C, Ma FF, Liu CX, Tian YX. Single- molecule long-read sequencing reveals the diversity of full-length transcripts in leaves of Gnetum (Gnetales). Int J Mol Sci, 2019, 20(24): 6350.
pmid: 31861078 |
| [19] | Akan Z, Varol I, Özaslan M. A cytotaxonomical investigation on spiders (Arachnida: Araneae). Biotechnol Biotechnol Equip, 2005, 19(2): 101-104. |
| [20] |
Král J, Kořínková T, Forman M, Krkavcová L. Insights into the meiotic behavior and evolution of multiple sex chromosome systems in spiders. Cytogenet Genome Res, 2011, 133(1): 43-66.
pmid: 21282941 |
| [21] |
Sanggaard KW, Bechsgaard JS, Fang XD, Duan JJ, Dyrlund TF, Gupta V, Jiang XT, Cheng L, Fan DD, Feng Y, Han LJ, Huang ZY, Wu ZZ, Liao L, Settepani V, Thøgersen IB, Vanthournout B, Wang T, Zhu YB, Funch P, Enghild JJ, Schauser L, Andersen SU, Villesen P, Schierup MH, Bilde T, Wang J. Spider genomes provide insight into composition and evolution of venom and silk. Nat Commun, 2014, 5(1): 3765.
pmid: 24801114 |
| [22] | Salim MMR, Rashid MH, Hossain MM, Zakaria M. Morphological characterization of tomato (Solanum lycopersicum L.) genotypes. J Saudi Soc Agric Sci, 2020, 19(3): 233-240. |
| [23] |
Hu WB, Jia AQ, Ma SY, Zhang GQ, Wei ZY, Lu F, Luo YJ, Zhang ZS, Sun JH, Yang TF, Xia TT, Li QH, Yao T, Zheng JY, Jiang ZJ, Xu ZH, Xia QY, Wang Y. A molecular atlas reveals the tri-sectional spinning mechanism of spider dragline silk. Nat Commun, 2023, 14(1): 837.
pmid: 36792670 |
| [24] | Ma Q, Jiang C, Zhou LQ, Sun T, Liu SF, Zhuang ZM. Karyotype characteristics of white trevally (Pseudocaranx dentex). J Fish Sci China, 2021, 28(5): 561-568. |
| 马青, 姜晨, 周丽青, 孙涛, 柳淑芳, 庄志猛. 黄带拟鲹染色体核型特征分析. 中国水产科学, 2021, 28(5): 561-568. | |
| [25] | Levan A, Fredga K, Sandberg AA. Nomenclature for centromeric position on chromosomes. Hereditas, 1964, 52(2): 201-220. |
| [26] | Kuo SR, Wang TT, Huang TC. Karyotype analysis of some Formosan gymnosperms. Taiwania, 1972, 17(1): 66-80. |
| [27] | Dong L. Predict of genome size and analysis of karyotype in Calycanthaceae[Dissertation]. Huazhong Agricultural University, 2017. |
| 董雷. 蜡梅科植物基因组大小预测与核型分析[学位论文]. 华中农业大学, 2017. | |
| [28] | Shi XM, Zhang ZZ, Ye ZX, Luo XY, Chang K, Zhang YH, Li C, You L, Zheng LL. Genome size analysis of Hemiboea subcapitata by flow cytometry and genome survey. Mol Plant Breed, 2024, 1-10. |
| 石宪铭, 张泽志, 叶峥秀, 罗湘胤, 常堃, 张勇洪, 李琛, 游磊, 郑兰兰. 联合流式细胞术和K-mer分析法测定半蒴苣苔基因组大小. 分子植物育种, 2024, 1-10. | |
| [29] | Aboul-Maaty NAF, Oraby HAS. Extraction of high- quality genomic DNA from different plant orders applying a modified CTAB-based method. Bull Natl Res Cent, 2019, 43(1): 25. |
| [30] |
Marçais G, Kingsford C. A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics, 2011, 27(6): 764-770.
pmid: 21217122 |
| [31] |
Luo DD, Zeng ZY, Wu ZQ, Chen CJ, Zhao TT, Du HZ, Miao YH, Liu DH. Intraspecific variation in genome size in Artemisia argyi determined using flow cytometry and a genome survey. 3 Biotech, 2023, 13(2): 57.
pmid: 36698769 |
| [32] |
Li NN, Li W, Feng JX, Zhang WW, Zhang R, Du SH, Liu SY, Xue GH, Yan C, Cui JH, Zhao HQ, Feng YL, Gan L, Zhang Q, Chen C, Liu D, Yuan J. High alcohol-producing Klebsiella pneumoniae causes fatty liver disease through 2,3-butanediol fermentation pathway in vivo. Gut Microbes, 2021, 13(1): 1979883.
pmid: 34632939 |
| [33] |
Qi WH, Lim YW, Patrignani A, Schläpfer P, Bratus-Neuenschwander A, Grüter S, Chanez C, Rodde N, Prat E, Vautrin S, Fustier MA, Pratas D, Schlapbach R, Gruissem W. The haplotype-resolved chromosome pairs of a heterozygous diploid African cassava cultivar reveal novel pan-genome and allele-specific transcriptome features. GigaScience, 2022, 11: giac028.
pmid: 35333302 |
| [34] |
Xu ZZ, Chen JD, Meng S, Xu P, Zhai CJ, Huang F, Guo Q, Zhao L, Quan YG, Shangguan YX, Meng Z, Wen T, Zhang Y, Zhang XG, Zhao J, Xu JW, Liu JG, Gao J, Ni WC, Chen XL, Ji W, Wang NY, Lu XX, Wang SH, Wang K, Zhang TZ, Shen XL. Genome sequence of Gossypium anomalum facilitates interspecific introgression breeding. Plant Commun, 2022, 3(5): 100350.
pmid: 35733334 |
| [35] |
Tang D, Jia YX, Zhang JZ, Li HB, Cheng L, Wang P, Bao ZG, Liu ZH, Feng SS, Zhu XJ, Li DW, Zhu GT, Wang HR, Zhou Y, Zhou YF, Bryan GJ, Buell CR, Zhang CZ, Huang SW. Genome evolution and diversity of wild and cultivated potatoes. Nature, 2022, 606(7914): 535-541.
pmid: 35676481 |
| [36] |
Sang YP, Long ZQ, Dan XM, Feng JJ, Shi TT, Jia CF, Zhang XX, Lai Q, Yang GL, Zhang HY, Xu XT, Liu HH, Jiang YZ, Ingvarsson PK, Liu JQ, Mao KS, Wang J. Genomic insights into local adaptation and future climate-induced vulnerability of a keystone forest tree in East Asia. Nat Commun, 2022, 13(1): 6541.
pmid: 36319648 |
| [37] |
Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics, 2015, 31(19): 3210-3212.
pmid: 26059717 |
| [38] |
Liu QP, Chen YY, Yu YY, An P, Xing YZ, Yang HX, Zhang YJ, Rahman K, Zhang L, Luan X, Zhang H. Bie-Jia-Ruan- Mai-Tang, a Chinese medicine formula, inhibits retinal neovascularization in diabetic mice through inducing the apoptosis of retinal vascular endothelial cells. Front Cardiovasc Med, 2022, 9: 959298.
pmid: 35903668 |
| [39] | Sato S, Tabata S, Hirakawa H, Asamizu E, Shirasawa K, Isobe S, Kaneko T, Nakamura Y, Shibata D, Aoki K, Egholm M, Knight J, Bogden R, Li CB, Shuang Y, Xu X, Pan SK, Cheng SF, Liu X, Ren YY, Wang J, Albiero A, Dal Pero F, Todesco S, Van Eck J, Buels RM, Bombarely A, Gosselin JR, Huang MY, Leto JA, Menda N, Strickler S, Mao LY, Gao S, Tecle IY, York T, Zheng Y, Vrebalov JT, Lee J, Zhong SL, Mueller LA, Stiekema WJ, Ribeca P, Alioto T, Yang WC, Huang SW, Du YC, Zhang ZH, Gao JC, Guo YM, Wang XX, Li Y, He J, Li CY, Cheng ZK, Zuo JR, Ren JF, Zhao JH, Yan LH, Jiang HL, Wang B, Li HS, Li ZJ, Fu FY, Chen BT, Han B, Feng Q, Fan DL, Wang Y, Ling HQ, Xue YB, Ware D, Richard McCombie W, Lippman ZB, Chia JM, Jiang K, Pasternak S, Gelley L, Kramer M, Anderson LK, Chang SB, Royer SM, Shearer LA, Stack SM, Rose JKC, Xu YM, Eannetta N, Matas AJ, McQuinn R, Tanksley SD, Camara F, Guigó R, Rombauts S, Fawcett J, Van de Peer Y, Zamir D, Liang CB, Spannagl M, Gundlach H, Bruggmann R, Mayer K, Jia ZQ, Zhang JH, Ye ZB, Bishop GJ, Butcher S, Lopez-Cobollo R, Buchan D, Filippis I, Abbott J, Dixit R, Singh M, Singh A, Kumar Pal J, Pandit A, Kumar Singh P, Kumar Mahato A, Dogra V, Gaikwad K, Raj Sharma T, Mohapatra T, Kumar Singh N, Causse M, Rothan C, Schiex T, Noirot C, Bellec A, Klopp C, Delalande C, Berges H, Mariette J, Frasse P, Vautrin S, Zouine M, Latché A, Rousseau C, Regad F, Pech JC, Philippot M, Bouzayen M, Pericard P, Osorio S, Fernandez del Carmen A, Monforte A, Granell A, Fernandez-Muñoz R, Conte M, Lichtenstein G, Carrari F, De Bellis G, Fuligni F, Peano C, Grandillo S, Termolino P, Pietrella M, Fantini E, Falcone G, Fiore A, Giuliano G, Lopez L, Facella P, Perrotta G, Daddiego L, Bryan G, Orozco M, Pastor X, Torrents D, van Schriek MGM, Feron RMC, van Oeveren J, de Heer P, daPonte L, Jacobs-Oomen S, Cariaso M, Prins M, van Eijk MJT, Janssen A, van Haaren MJJ, Jo SH, Kim J, Kwon SY, Kim S, Koo DH, Lee S, Hur CG, Clouser C, Rico A, Hallab A, Gebhardt C, Klee K, Jöcker A, Warfsmann J, Göbel U, Kawamura S, Yano K, Sherman JD, Fukuoka H, Negoro S, Bhutty S, Chowdhury P, Chattopadhyay D, Datema E, Smit S, Schijlen EGWM, van de Belt J, van Haarst JC, Peters SA, van Staveren MJ, Henkens MHC, Mooyman PJW, Hesselink T, van Ham RCHJ, Jiang GY, Droege M, Choi D, Kang BC, Dong Kim B, Park M, Kim S, Yeom SI, Lee YH, Choi YD, Li GC, Gao JW, Liu YS, Huang SX, Fernandez-Pedrosa V, Collado C, Zuñiga S, Wang GP, Cade R, Dietrich RA, Rogers J, Knapp S, Fei ZJ, White RA, Thannhauser TW, Giovannoni JJ, Angel Botella M, Gilbert L, Gonzalez R. The tomato genome sequence provides insights into fleshy fruit evolution. Nature, 2012, 485(7400): 635-641. |
| [40] |
Yan N, Yang T, Yu XT, Shang LG, Guo DP, Zhang Y, Meng L, Qi QQ, Li YL, Du YM, Liu XM, Yuan XL, Qin P, Qiu J, Qian Q, Zhang ZF. Chromosome-level genome assembly of Zizania latifolia provides insights into its seed shattering and phytocassane biosynthesis. Commun Biol, 2022, 5(1): 36.
pmid: 35017643 |
| [41] |
Rezác M, Král J, Musilová J, Pekár S. Unusual karyotype diversity in the European spiders of the genus Atypus (Araneae: Atypidae). Hereditas, 2006, 143(2006): 123-129.
pmid: 17362345 |
| [42] | Painter TS. Spermatogenesis in spiders. Zool Jahrb Abt Anat Ontog Tiere, 1914, 38: 509-576. |
| [43] | Kořínková T, Král J. Karyotypes, sex chromosomes, and meiotic division in spiders. In: Nentwig W, eds. Spider Ecophysiology. Springer, Berlin, Heidelberg, 2013, 159-171. |
| [44] | Král J, Kořínková T, Krkavcová L, Musilová J, Forman M, Herrera IMÁ, Haddad CR, Vítková M, Henriques S, Vargas JGP, Hedin M. Evolution of karyotype, sex chromosomes, and meiosis in mygalomorph spiders (Araneae: Mygalomorphae). Biol J Linn Soc, 2013, 109(2): 377-408. |
| [45] |
Sember A, Pappová M, Forman M, Nguyen P, Marec F, Dalíková M, Divišová K, Doležálková-Kaštánková M, Zrzavá M, Sadílek D, Hrubá B, Král J. Patterns of sex chromosome differentiation in spiders: insights from comparative genomic Hybridisation. Genes (Basel), 2020, 11(8): 849.
pmid: 32722348 |
| [46] |
Cavenagh AF, Rincão MP, Dias FC, Brescovit AD, Dias AL. Chromosomal diversity in three species of Lycosa Latreille, 1804 (Araneae, Lycosidae): inferences on diversification of diploid number and sexual chromosome systems in Lycosinae. Genet Mol Biol, 2022, 45(1): e20200440.
pmid: 35098965 |
| [47] | Xian M, Chen Y, Liang YH, Song YX, Niu H, Zhang WG, Xia JW, Zhang TL, Xu SZ, Zhang LP, Gao HJ, Li JY, Gao X. Study on genome size of Bos frontalis by flow cytometry. Genomics Appl Biol, 2018, 37(12): 5214-5219. |
| 贤明, 陈燕, 梁永虎, 宋禹昕, 牛红, 张文刚, 夏江威, 张天留, 许尚忠, 张路培, 高会江, 李俊雅, 高雪. 利用流式细胞术测定大额牛基因组大小的研究. 基因组学与应用生物学, 2018, 37(12): 5214-5219. | |
| [48] |
Qin ZK, Li XY, Liu DW, Wang Q, Lu L, Zhang ZF. Analysis of chromosome karyotype and genome size in echiuran Urechisunicinctus Drasche, 1880 (Polychaeta, Urechidae). Comp Cytogenet, 2019, 13(1): 75-85.
pmid: 30918599 |
| [49] |
Shi LL, Yi SK, Li YH. Genome survey sequencing of red swamp crayfish Procambarus clarkii. Mol Biol Rep, 2018, 45(5): 799-806.
pmid: 29931535 |
| [50] | Gao SH, Yu HY, Wu SY, Wang S, Geng JN, Luo YF, Hu SN. Advances of sequencing and assembling technologies for complex genomes. Hereditas(Beijing), 2018, 40(11): 944-963. |
| 高胜寒, 禹海英, 吴双阳, 王森, 耿佳宁, 骆迎峰, 胡松年. 复杂基因组测序技术研究进展. 遗传, 2018, 40(11): 944-963. | |
| [51] |
Mgwatyu Y, Stander AA, Ferreira S, Williams W, Hesse U. Rooibos (Aspalathus linearis) genome size estimation using flow cytometry and K-mer analyses. Plants (Basel), 2020, 9(2): 270.
pmid: 32085566 |
| [52] |
He K, Lin KJ, Wang GR, Li F. Genome sizes of nine insect species determined by flow cytometry and k-mer analysis. Front Physiol, 2016, 7: 569.
pmid: 27932995 |
| [53] | Huang SW, Li RQ, Zhang ZH, Li L, Gu XF, Fan W, Lucas WJ, Wang XW, Xie BY, Ni PX, Ren YY, Zhu HM, Li J, Lin K, Jin WW, Fei ZJ, Li GC, Staub J, Kilian A, Van Der Vossen EAG, Wu Y, Guo J, He J, Jia ZQ, Ren Y, Tian G, Lu Y, Ruan J, Qian WB, Wang MW, Huang QF, Li B, Xuan ZL, Cao JJ, Asan, Wu ZG, Zhang JB, Cai QL, Bai YQ, Zhao BW, Han YH, Li Y, Li XF, Wang SH, Shi QX, Liu SQ, Cho WK, Kim JY, Xu Y, Heller-Uszynska K, Miao H, Cheng ZC, Zhang SP, Wu J, Yang YH, Kang HX, Li M, Liang HQ, Ren XL, Shi ZB, Wen M, Jian M, Yang HL, Zhang GJ, Yang ZT, Chen R, Liu SF, Li JW, Ma LJ, Liu H, Zhou Y, Zhao J, Fang XD, Li GQ, Fang L, Li YR, Liu DY, Zheng HK, Zhang Y, Qin N, Li Z, Yang GH, Yang S, Bolund L, Kristiansen K, Zheng HC, Li SC, Zhang XQ, Yang HM, Wang J, Sun RF, Zhang BX, Jiang SZ, Wang J, Du YC, Li SG. The genome of the cucumber, Cucumis sativus L.. Nat Genet, 2009, 41(12): 1275-1281. |
| [54] |
Simbolo M, Gottardi M, Corbo V, Fassan M, Mafficini A, Malpeli G, Lawlor RT, Scarpa A. DNA qualification workflow for next generation sequencing of histopathological samples. PLoS One, 2013, 8(6): e62692.
pmid: 23762227 |
| [55] |
Sheffer MM, Hoppe A, Krehenwinkel H, Uhl G, Kuss AW, Jensen L, Jensen C, Gillespie RG, Hoff KJ, Prost S. Chromosome-level reference genome of the European wasp spider Argiope bruennichi: a resource for studies on range expansion and evolutionary adaptation. GigaScience, 2021, 10(1): giaa148.
pmid: 33410470 |
| [56] |
Cerca J, Armstrong EE, Vizueta J, Fernández R, Dimitrov D, Petersen B, Prost S, Rozas J, Petrov D, Gillespie RG. The Tetragnatha kauaiensis genome sheds light on the origins of genomic novelty in spiders. Genome Biol Evol, 2021, 13(12): evab262.
pmid: 34849853 |
| [57] |
Yahav T, Privman E. A comparative analysis of methods for de novo assembly of hymenopteran genomes using either haploid or diploid samples. Sci Rep, 2019, 9(1): 6480.
pmid: 31019201 |
| [58] |
Zhang L, Fang XD, Liao HR, Zhang ZM, Zhou X, Han LJ, Chen Y, Qiu QW, Li SC. A comprehensive investigation of metagenome assembly by linked-read sequencing. Microbiome, 2020, 8(1): 156.
pmid: 33176883 |
| [59] |
Espinosa E, Bautista R, Fernandez I, Larrosa R, Zapata EL, Plata O. Comparing assembly strategies for third- generation sequencing technologies across different genomes. Genomics, 2023, 115(5): 110700.
pmid: 37598732 |
| [60] |
Kajitani R, Toshimoto K, Noguchi H, Toyoda A, Ogura Y, Okuno M, Yabana M, Harada M, Nagayasu E, Maruyama H, Kohara Y, Fujiyama A, Hayashi T, Itoh T. Efficient de novo assembly of highly heterozygous genomes from whole-genome shotgun short reads. Genome Res, 2014, 24(8): 1384-1395.
pmid: 24755901 |
| [1] | CHEN Jian-Jun, WANG Ying. Recent progress in plant genome size evolution [J]. HEREDITAS, 2009, 31(5): 464-464―470. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||