Hereditas(Beijing) ›› 2019, Vol. 41 ›› Issue (11): 979-993.doi: 10.16288/j.yczz.19-227
• Review • Next Articles
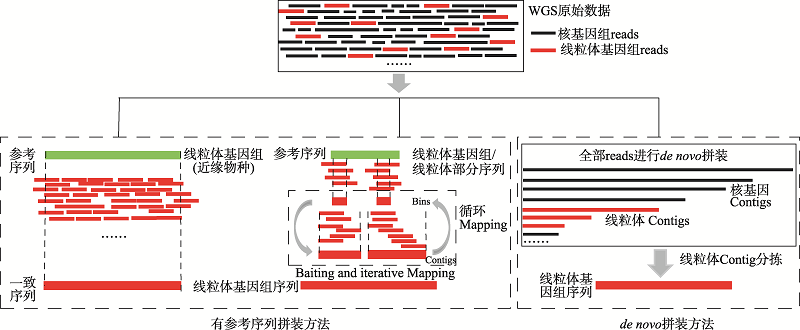
Mitogenome assembly strategies and software applications in the genome era
- State Key Laboratory for Conservation and Utilization of Bio-Resource in Yunnan, School of Life Sciences, Yunnan University, Kunming 650091, China
-
Received:2019-08-07Revised:2019-09-25Online:2019-11-20Published:2019-10-15 -
Contact:Yu Li E-mail:yuli@ynu.edu.cn -
Supported by:Supported by the National Natural Science Foundation of China No(31872213);Industrialization Cultivation Project of Scientific Research Fund of Yunnan Education Department No(2016CYH02);and the Academic Graduate Students Foundation of Yunnan Province
Cite this article
Weimin Kuang, Li Yu. Mitogenome assembly strategies and software applications in the genome era[J]. Hereditas(Beijing), 2019, 41(11): 979-993.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
Table 1
Mitogenome assembly software"
| 软件名称 | 是否需要参考序列/ 参考序列类型 | 适用 物种 | 输入文件格式、 类型 | 变异 注释 | 结构可 视化 | 运行 环境 | 编程 语言 | 软件网址 |
|---|---|---|---|---|---|---|---|---|
| MIA | 是/自定义参考序列 | 任意 物种 | Fastq、SE reads和 PE reads | × | × | CUI | C/C++ | https://github.com/mpieva/mapping-iterative-assembler |
| MitoBamAnnotator | 是/ rCRS | 人 | Bam | √ | √ | Web | Java | http://bioinfo.bgu.ac.il/bsu/software/MITO-BAM |
| MitoSeek | 是/rCRS和hg19 | 人 | Bam | √ | × | GUI | Perl | |
| mtDNA- profiler | 是/rCRS | 人 | Fasta | × | √ | Web | Java | http://mtprofiler.yonsei.ac.kr |
| MITObim | 是/自定义参考序列 | 任意 物种 | Bam | × | × | CUI | Perl | |
| Mit-o-matic | 是/rCRS | 人 | Fastq、SE reads和 PE reads | √ | √ | Web/GUI | Java | |
| MToolBox | 是/rCRS和RSRS | 人 | Fastq/Bam/Sam、 SE reads和PE reads | √ | × | Web/CUI | Python | https://sourceforge.net/projects/mtoolbox |
| ARC | 是/自定义参考序列 | 任意 物种 | Fastq、SE reads和 PE reads | × | × | Web/CUI | Python | |
| Phy-Mer | 是/自定义参考序列 | 任意 物种 | Fasta/fastq/Bam、 SE reads和PE reads | × | √ | CUI | Python | https://github.com/danielnavarrogomez/phy-mer |
| mtDNA- Server | 是/rCRS和RSRS | 人 | Fastq/Bam/VCF、 SE reads和PE reads | √ | √ | Web | Java | |
| IOGA | 是/自定义参考序列 | 任意 物种 | Fastq、SE reads和 PE reads | × | × | CUI | Python | |
| NOVOPlasty | 是/自定义参考序列 | 任意 物种 | Fastq/fasta、SE reads 和PE reads | × | × | Web/CUI | Perl | https://github.com/ndierckx/NOVOPlasty |
| Norgal | 否 | 任意 物种 | Fastq、SE reads和 PE reads | × | × | CUI | Python/ Java | https://bitbucket.org/kosaidtu/norgal |
| Organelle- PBA | 是/自定义参考序列 | 任意 物种 | PacBio reads | × | × | CUI | Perl | https://github.com/aubombarely/Organelle_PBA |
| MitoSuite | 是/rCRS, RSRS, hg19, GRCh37和38 | 人 | Bam/Sam | √ | √ | GUI | Python | |
| ORG.Asm | 是/自定义参考序列 | 任意 物种 | Fastq、SE reads和 PE reads | × | × | CUI | Python | |
| MitoZ | 否 | 任意 物种 | Fastq、SE reads和 PE reads | √ | √ | CUI | Python | |
| GetOrganelle | 是/自定义参考序列 | 任意 物种 | Fastq、SE reads和 PE reads | × | × | CUI | Python | |
| Trimitomics | 是/自定义参考序列 | 任意 物种 | RNA-seq reads、 PE reads | × | × | Unknown | Unknown | Unknown |
Table 1
| [1] |
Brown WM, George M Jr., Wilson AC . Rapid evolution of animal mitochondrial DNA . Proc Natl Acad Sci USA, 1979,76(4):1967-1971.
doi: 10.1073/pnas.76.4.1967 pmid: 109836 |
| [2] |
Lei R, Frasier CL, Hawkins MT, Engberg SE, Bailey CA, Johnson SE, Mclain AT, Groves CP, Perry GH, Nash SD, Mittermeier RA, Louis EE . Phylogenomic reconstruction of Sportive Lemurs (genus Lepilemur) recovered from mitogenomes with inferences for madagascar biogeography . J Hered, 2017,108(2):107-119.
doi: 10.1093/jhered/esw072 pmid: 28173059 |
| [3] |
Mueller RL, Macey JR, Jaekel M, Wake DB, Boore JL . Morphological homoplasy, life history evolution, and historical biogeography of plethodontid salamanders inferred from complete mitochondrial genomes . Proc Natl Acad Sci USA, 2004,101(38):13820-13825.
doi: 10.1073/pnas.0405785101 pmid: 15365171 |
| [4] |
Zhang P, Chen YQ, Zhou H, Liu YF, Wang XL, Papenfuss TJ, Wake DB, Qu LH . Phylogeny, evolution, and biogeography of Asiatic Salamanders (Hynobiidae) . Proc Natl Acad Sci USA, 2006,103(19):7360-7365.
doi: 10.1073/pnas.0602325103 pmid: 16648252 |
| [5] |
Zhang P, Papenfuss TJ, Wake MH, Qu LH, Wake DB . Phylogeny and biogeography of the family Salamandridae (Amphibia: Caudata) inferred from complete mitochondrial genomes . Mol Phylogenet Evol, 2008,49(2):586-597.
doi: 10.1016/j.ympev.2008.08.020 |
| [6] |
Cerný V, Fernandes V, Costa MD, Hájek M, Mulligan CJ, Pereira L . Migration of Chadic speaking pastoralists within Africa based on population structure of Chad Basin and phylogeography of mitochondrial L3f haplogroup . BMC Evol Biol, 2009,9:63.
doi: 10.1186/1471-2148-9-63 pmid: 19309521 |
| [7] |
Klimova A, Phillips CD, Fietz K, Olsen MT, Harwood J, Amos W, Hoffman JI . Global population structure and demographic history of the grey seal . Mol Ecol, 2014,23(16):3999-4017.
doi: 10.1111/mec.12850 |
| [8] |
Lin LH, Ji X, Diong CH, Du Y, Lin CX . Phylogeography and population structure of the Reevese's Butterfly Lizard (Leiolepis reevesii) inferred from mitochondrial DNA sequences . Mol Phylogenet Evol, 2010,56(2):601-607.
doi: 10.1016/j.ympev.2010.04.032 pmid: 20433932 |
| [9] |
Miller W, Hayes VM, Ratan A, Petersen DC, Wittekindt NE, Miller J, Walenz B, Knight J, Qi J, Zhao F, Wang Q, Bedoya-Reina OC, Katiyar N, Tomsho LP, Kasson LM, Hardie RA, Woodbridge P, Tindall EA, Bertelsen MF, Dixon D, Pyecroft S, Helgen KM, Lesk AM, Pringle TH, Patterson N, Zhang Y, Kreiss A, Woods GM, Jones ME, Schuster SC . Genetic diversity and population structure of the endangered marsupial Sarcophilus harrisii (Tasmanian devil) . Proc Natl Acad Sci USA, 2011,108(30):12348-12353.
doi: 10.1073/pnas.1102838108 pmid: 21709235 |
| [10] |
Roslin T . Spatial population structure in a patchily distributed beetle . Mol Ecol, 2001,10(4):823-837.
doi: 10.1046/j.1365-294x.2001.01235.x pmid: 11348492 |
| [11] |
Teacher AG, André C, Merilä J, Wheat CW . Whole mitochondrial genome scan for population structure and selection in the Atlantic herring . BMC Evol Biol, 2012,12:248.
doi: 10.1186/1471-2148-12-248 pmid: 23259908 |
| [12] |
Uren C, Kim M, Martin AR, Bobo D, Gignoux CR, Van Helden PD, Möller M, Hoal EG, Henn BM . Fine-scale human population structure in southern Africa reflects ecogeographic boundaries . Genetics, 2016,204(1):303-314.
doi: 10.1534/genetics.116.187369 pmid: 27474727 |
| [13] |
Kuang WM, Ming C, Li HP, Wu H, Frantz L, Roos C, Zhang YP, Zhang CL, Jia T, Yang JY, Yu L . The origin and population history of the endangered golden snub- nosed monkey (Rhinopithecus roxellana) . Mol Biol Evol, 2019,36(3):487-499.
doi: 10.1093/molbev/msy220 pmid: 30481341 |
| [14] |
Haberman Y, Karns R, Dexheimer PJ, Schirmer M, Somekh J, Jurickova I, Braun T, Novak E, Bauman L, Collins MH, Mo A, Rosen MJ, Bonkowski E, Gotman N, Marquis A, Nistel M, Rufo PA, Baker SS, Sauer CG, Markowitz J, Pfefferkorn MD, Rosh JR, Boyle BM, Mack DR, Baldassano RN, Shah S, Leleiko NS, Heyman MB, Grifiths AM, Patel AS, Noe JD, Aronow BJ, Kugathasan S, Walters TD, Gibson G, Thomas SD, Mollen K, Shen-Orr S, Huttenhower C, Xavier RJ, Hyams JS, Denson LA . Ulcerative colitis mucosal transcriptomes reveal mitochondriopathy and personalized mechanisms underlying disease severity and treatment response . Nat Commun, 2019,10(1):38.
doi: 10.1038/s41467-018-07841-3 pmid: 30604764 |
| [15] |
Inak G, Lorenz C, Lisowski P, Zink A, Mlody B, Prigione A . Concise review: induced pluripotent stem cell-based drug discovery for mitochondrial disease . Stem Cells, 2017,35(7):1655-1662.
doi: 10.1002/stem.2637 pmid: 28544378 |
| [16] |
Suomalainen A . Mitochondrial DNA and disease . Ann Med, 1997,29(3):235-246.
doi: 10.3109/07853899708999341 pmid: 9240629 |
| [17] |
Toda T . Molecular genetics of Parkinson's disease . Brain Nerve, 2007,59(8):815-823.
pmid: 17713117 |
| [18] |
Armbrust EV, Berges JA, Bowler C, Green BR, Martinez D, Putnam NH, Zhou S, Allen AE, Apt KE, Bechner M, Brzezinski MA, Chaal BK, Chiovitti A, Davis AK, Demarest MS, Detter JC, Glavina T, Goodstein D, Hadi MZ, Hellsten U, Hildebrand M, Jenkins BD, Jurka J, Kapitonov VV, Kröger N, Lau WW, Lane TW, Larimer FW, Lippmeier JC, Lucas S, Medina M, Montsant A, Obornik M, Parker MS, Palenik B, Pazour GJ, Richardson PM, Rynearson TA, Saito MA, Schwartz DC, Thamatrakoln K, Valentin K, Vardi A, Wilkerson FP, Rokhsar DS . The genome of the diatom Thalassiosira pseudonana: ecology, evolution, and metabolism . Science, 2004,306(5693):79-86.
doi: 10.1126/science.1101156 pmid: 15459382 |
| [19] |
Janzen DH, Burns JM, Cong Q, Hallwachs W, Dapkey T, Manjunath R, Hajibabaei M, Hebert PDN, Grishin NV . Nuclear genomes distinguish cryptic species suggested by their DNA barcodes and ecology . Proc Natl Acad Sci USA, 2017,114(31):8313-8318.
doi: 10.1073/pnas.1621504114 pmid: 28716927 |
| [20] |
Zarowiecki MZ, Huyse T, Littlewood DT . Making the most of mitochondrial genomes--markers for phylogeny, molecular ecology and barcodes in Schistosoma (Platyhelminthes: Digenea) . Int J Parasitol, 2007,37(12):1401-1418.
doi: 10.1016/j.ijpara.2007.04.014 |
| [21] |
Hu M, Jex AR, Campbell BE, Gasser RB . Long PCR amplification of the entire mitochondrial genome from individual helminths for direct sequencing . Nat Protoc, 2007,2(10):2339-2344.
doi: 10.1038/nprot.2007.358 pmid: 17947975 |
| [22] |
Nabholz B, Jarvis ED, Ellegren H . Obtaining mtDNA genomes from next-generation transcriptome sequencing: a case study on the basal Passerida (Aves: Passeriformes) phylogeny . Mol Phylogenet Evol, 2010,57(1):466-470.
doi: 10.1016/j.ympev.2010.06.009 pmid: 20601014 |
| [23] |
Timmermans MJ, Dodsworth S, Culverwell CL, Bocak L, Ahrens D, Littlewood DT, Pons J, Vogler AP . Why barcode? High-throughput multiplex sequencing of mitochondrial genomes for molecular systematics . Nucleic Acids Res, 2010,38(21):e197.
doi: 10.1093/nar/gkq807 pmid: 20876691 |
| [24] |
Metzker ML . Sequencing technologies - the next generation . Nat Rev Genet, 2010,11(1):31-46.
doi: 10.1038/nrg2626 pmid: 19997069 |
| [25] |
Lounsberry ZT, Brown SK, Collins PW, Henry RW, Newsome SD, Sacks BN . Next-generation sequencing workflow for assembly of nonmodel mitogenomes exemplified with North Pacific albatrosses (Phoebastria spp.) . Mol Ecol Resour, 2015,15(4):893-902.
doi: 10.1111/1755-0998.12365 pmid: 25545584 |
| [26] |
Shearman JR, Sonthirod C, Naktang C, Pootakham W, Yoocha T, Sangsrakru D, Jomchai N, Tragoonrung S, Tangphatsornruang S . The two chromosomes of the mitochondrial genome of a sugarcane cultivar: assembly and recombination analysis using long PacBio reads . Sci Rep, 2016,6:31533.
doi: 10.1038/srep31533 pmid: 27530092 |
| [27] |
Kovar L, Nageswara-Rao M, Ortega-Rodriguez S, Dugas DV, Straub S, Cronn R, Strickler SR, Hughes CE, Hanley KA, Rodriguez DN, Langhorst BW, Dimalanta ET, Bailey CD . PacBio-based mitochondrial genome assembly of Leucaena trichandra (Leguminosae) and an intrageneric assessment of mitochondrial RNA editing . Genome Biol Evol, 2018,10(9):2501-2517.
doi: 10.1093/gbe/evy179 pmid: 30137422 |
| [28] |
Wang SB, Song QW, Li SS, Hu ZG, Dong GQ, Song C, Huang HW, Liu YF . Assembly of a complete mitogenome of chrysanthemum nankingense using Oxford Nanopore long reads and the diversity and evolution of Asteraceae mitogenomes . Genes, 2018,9(11):547.
doi: 10.3390/genes9110547 pmid: 30424578 |
| [29] |
Gan HM, Linton SM, Austin CM . Two reads to rule them all: Nanopore long read-guided assembly of the iconic Christmas Island red crab, Gecarcoidea natalis (Pocock, 1888), mitochondrial genome and the challenges of AT-rich mitogenomes . Mar Genom, 2019,45:64-71.
doi: 10.1016/j.margen.2019.02.002 pmid: 30928201 |
| [30] |
Maughan PJ, Chaney L, Lightfoot DJ, Cox BJ, Tester M, Jellen EN, Jarvis DE . Mitochondrial and chloroplast genomes provide insights into the evolutionary origins of quinoa (Chenopodium quinoa Willd.) . Sci Rep, 2019,9(1):185.
doi: 10.1038/s41598-018-36693-6 pmid: 30655548 |
| [31] |
Mofiz E, Seemann T, Bahlo M, Holt D, Currie BJ, Fischer K, Papenfuss AT . Mitochondrial genome sequence of the Scabies Mite provides insight into the genetic diversity of individual scabies infections . PLoS Negl Trop Dis, 2016,10(2):e0004384.
doi: 10.1371/journal.pntd.0004384 pmid: 26872064 |
| [32] |
Ni P, Bhuiyan AA, Chen JH, Li J, Zhang C, Zhao S, Du X, Li H, Yu H, Liu X, Li K . De novo assembly of mitochondrial genomes provides insights into genetic diversity and molecular evolution in wild boars and domestic pigs . Genetica, 2018,146(3):277-285.
doi: 10.1007/s10709-018-0018-y pmid: 29748765 |
| [33] |
Niu WT, Yu SG, Tian P, Xiao JG . Complete mitochondrial genome of Echinophyllia aspera (Scleractinia, Lobophylliidae): mitogenome characterization and phylogenetic positioning . Zookeys, 2018,793:1-14.
doi: 10.3897/zookeys.793.28977 pmid: 30405308 |
| [34] |
Sahoo PK, Singh L, Sharma L, Kumar R, Singh VK, Ali S, Singh AK, Barat A . The complete mitogenome of brown trout (Salmo trutta fario) and its phylogeny . Mitochondrial DNA A DNA Mapp Seq Anal, 2016,27(6):4563-4565.
doi: 10.3109/19401736.2015.1101565 pmid: 26641940 |
| [35] |
Shi YC, Liu Y, Zhang SZ, Zou R, Tang JM, Mu WX, Peng Y, Dong SS . Assembly and comparative analysis of the complete mitochondrial genome sequence of Sophora japonica 'JinhuaiJ2' . PLoS One, 2018,13(8):e0202485.
doi: 10.1371/journal.pone.0202485 pmid: 30114217 |
| [36] |
Al-Nakeeb K, Petersen TN, Sicheritz-Pontén T . Norgal: extraction and de novo assembly of mitochondrial DNA from whole-genome sequencing data . BMC Bioinformatics, 2017,18(1):510.
doi: 10.1186/s12859-017-1927-y pmid: 29162031 |
| [37] |
Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA . SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing . J Comput Biol, 2012,19(5):455-477.
doi: 10.1089/cmb.2012.0021 |
| [38] |
Dierckxsens N, Mardulyn P, Smits G . NOVOPlasty: de novo assembly of organelle genomes from whole genome data . Nucleic Acids Res, 2017,45(4):e18.
doi: 10.1093/nar/gkw955 pmid: 28204566 |
| [39] |
Meng GL, Li YY, Yang CT, Liu SL . MitoZ: a toolkit for animal mitochondrial genome assembly, annotation and visualization . Nucleic Acids Res, 2019,47(11):63.
doi: 10.1093/nar/gkz173 pmid: 30864657 |
| [40] |
Bignell GR, Miller AR, Evans IH . Isolation of mitochondrial DNA . Methods Mol Biol, 1996,53:109-116.
doi: 10.1385/0-89603-319-8:109 pmid: 8924972 |
| [41] |
Li G, Davis BW, Eizirik E, Murphy WJ . Phylogenomic evidence for ancient hybridization in the genomes of living cats (Felidae) . Genome Res, 2016,26(1):1-11.
doi: 10.1101/gr.186668.114 pmid: 26518481 |
| [42] | Yang QQ, Li ZH, Liu LJ . Advance and application of mtDNA COⅠ barcodes on insects. Chin Bull Entomol, 2012,49(06):1687-1695. |
| 杨倩倩, 李志红, 伍祎, 柳丽君 . 线粒体COⅠ基因在昆虫DNA条形码中的研究与应用. 应用昆虫学报, 2012,49(06):1687-1695. | |
| [43] | Sha M, Lin LL, Li XJ, Huang Y . Strategy and methods for sequencing mitochondrial genome. Chin Bull Entomol, 2013,50(01):293-297. |
| 沙淼, 林立亮, 李雪娟, 黄原 . 线粒体基因组测序策略和方法. 应用昆虫学报, 2013,50(01):293-297. | |
| [44] | Li TJ, Cao YX, Zhao HC, Yu Y, Qiao J . Research progress of sequencing method for animal mitochondrial genome. Tianjin Med J, 2016,44(06):796-800. |
| 李天杰, 曹延祥, 赵红翠, 于洋, 乔杰 . 动物线粒体基因组测序方法的研究进展. 天津医药, 2016,44(06):796-800. | |
| [45] |
Groenenberg DSJ, Harl J, Duijm E, Gittenberger E . The complete mitogenome of Orcula dolium (Draparnaud, 1801); ultra-deep sequencing from a single long-range PCR using the Ion-Torrent PGM . Hereditas, 2017,154:7.
doi: 10.1186/s41065-017-0028-2 pmid: 28396619 |
| [46] |
King JL, Larue BL, Novroski NM, Stoljarova M, Seo SB, Zeng X, Warshauer DH, Davis CP, Parson W, Sajantila A, Budowle B . High-quality and high-throughput massively parallel sequencing of the human mitochondrial genome using the Illumina MiSeq . Forensic Sci Int Genet, 2014,12:128-135.
doi: 10.1016/j.fsigen.2014.06.001 pmid: 24973578 |
| [47] | Hunter SS, Lyon RT, Sarver BAJ, Hardwick K, Forney LJ, Settles ML . Assembly by Reduced Complexity (ARC): a hybrid approach for targeted assembly of homologous sequences . bioRxiv, 2015: 014662. |
| [48] |
Machado DJ, Lyra ML, Grant T . Mitogenome assembly from genomic multiplex libraries: comparison of strategies and novel mitogenomes for five species of frogs . Mol Ecol Resour, 2016,16(3):686-693.
doi: 10.1111/1755-0998.12492 pmid: 26607054 |
| [49] |
Li H, Durbin R . Fast and accurate short read alignment with Burrows-Wheeler transform . Bioinformatics, 2009,25(14):1754-1760.
doi: 10.1093/bioinformatics/btp324 pmid: 19451168 |
| [50] |
Min-Shan Ko A, Zhang YQ, Yang MA, Hu YB, Cao P, Feng XT, Zhang LZ, Wei FW, Fu QM . Mitochondrial genome of a 22,000-year-old giant panda from southern China reveals a new panda lineage . Curr Biol, 2018,28(12):R693-R694.
doi: 10.1016/j.cub.2018.05.008 pmid: 29920259 |
| [51] |
Taylor RW, Turnbull DM . Mitochondrial DNA mutations in human disease . Nat Rev Genet, 2005,6(5):389-402.
doi: 10.1038/nrg1606 pmid: 15861210 |
| [52] |
Torroni A, Achilli A, Macaulay V, Richards M, Bandelt HJ . Harvesting the fruit of the human mtDNA tree . Trends Genet, 2006,22(6):339-345.
doi: 10.1016/j.tig.2006.04.001 pmid: 16678300 |
| [53] |
Green RE, Malaspinas AS, Krause J, Briggs AW, Johnson PLF, Uhler C, Meyer M, Good JM, Maricic T, Stenzel U, Prüfer K, Siebauer M, Burbano HA, Ronan M, Rothberg JM, Egholm M, Rudan P, Brajković D, Kućan Z, Gušić I, Wikström M, Laakkonen L, Kelso J, Slatkin M, Pääbo S . A complete Neandertal mitochondrial genome sequence determined by high-throughput sequencing . Cell, 2008,134(3):416-426.
doi: 10.1016/j.cell.2008.06.021 pmid: 18692465 |
| [54] |
Zhidkov I, Nagar T, Mishmar D, Rubin E . MitoBamAnnotator: A web-based tool for detecting and annotating heteroplasmy in human mitochondrial DNA sequences . Mitochondrion, 2011,11(6):924-928.
doi: 10.1016/j.mito.2011.08.005 |
| [55] |
Guo Y, Li J, Li CI, Shyr Y, Samuels DC . MitoSeek: extracting mitochondria information and performing high-throughput mitochondria sequencing analysis . Bioinformatics, 2013,29(9):1210-1211.
doi: 10.1093/bioinformatics/btt118 |
| [56] |
Yang IS, Lee HY, Yang WI, Shin KJ. mtDNAprofiler: a Web application for the nomenclature and comparison of human mitochondrial DNA sequences . J Forensic Sci, 2013,58(4):972-980.
doi: 10.1111/1556-4029.12139 pmid: 23682804 |
| [57] |
Vellarikkal SK, Dhiman H, Joshi K, Hasija Y, Sivasubbu S, Scaria V. mit-o-matic: a comprehensive computational pipeline for clinical evaluation of mitochondrial variations from next-generation sequencing datasets . Hum Mutat, 2015,36(4):419-424.
doi: 10.1002/humu.22767 pmid: 25677119 |
| [58] |
Calabrese C, Simone D, Diroma MA, Santorsola M, Guttà C, Gasparre G, Picardi E, Pesole G, Attimonelli M . MToolBox: a highly automated pipeline for heteroplasmy annotation and prioritization analysis of human mitochondrial variants in high-throughput sequencing . Bioinformatics, 2014,30(21):3115-3117.
doi: 10.1093/bioinformatics/btu483 |
| [59] |
Navarro-Gomez D, Leipzig J, Shen L, Lott M, Stassen AP, Wallace DC, Wiggs JL, Falk MJ, Van Oven M, Gai X . Phy-Mer: a novel alignment-free and reference-independent mitochondrial haplogroup classifier . Bioinformatics, 2015,31(8):1310-1312.
doi: 10.1093/bioinformatics/btu825 pmid: 25505086 |
| [60] |
Weissensteiner H, Forer L, Fuchsberger C, Schöpf B, Kloss-Brandstätter A, Specht G, Kronenberg F , Schönherr S. mtDNA-Server: next-generation sequencing data analysis of human mitochondrial DNA in the cloud . Nucleic Acids Res, 2016,44(W1):W64-69.
doi: 10.1093/nar/gkw247 pmid: 27084948 |
| [61] |
Ishiya K, Ueda S . MitoSuite: a graphical tool for human mitochondrial genome profiling in massive parallel sequencing . PeerJ, 2017,5:e3406.
doi: 10.7717/peerj.3406 pmid: 28584729 |
| [62] |
Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA . Circos: an information aesthetic for comparative genomics . Genome Res, 2009,19(9):1639-1645.
doi: 10.1101/gr.092759.109 pmid: 19541911 |
| [63] |
Mckenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, Depristo MA . The Genome Analysis Toolkit: a MapReduce framework for analyzing next- generation DNA sequencing data . Genome Res, 2010,20(9):1297-1303.
doi: 10.1101/gr.107524.110 pmid: 20644199 |
| [64] |
Langmead B, Salzberg SL . Fast gapped-read alignment with Bowtie 2 . Nat Methods, 2012,9(4):357-359.
doi: 10.1038/NMETH.1923 |
| [65] |
Mardis ER . The impact of next-generation sequencing technology on genetics . Trends Genet, 2008,24(3):133-141.
doi: 10.1016/j.tig.2007.12.006 |
| [66] |
Hahn C, Bachmann L, Chevreux B . Reconstructing mitochondrial genomes directly from genomic next- generation sequencing reads--a baiting and iterative mapping approach . Nucleic Acids Res, 2013,41(13):e129.
doi: 10.1093/nar/gkt371 pmid: 23661685 |
| [67] |
Hahn C . Assembly of ancient mitochondrial genomes without a closely related reference sequence . Methods Mol Biol, 2019,1963:195-213.
doi: 10.1007/978-1-4939-9176-1_18 pmid: 30875055 |
| [68] |
Li R, Ren X, Bi Y, Ding Q, Ho VWS, Zhao Z . Comparative mitochondrial genomics reveals a possible role of a recent duplication of NADH dehydrogenase subunit 5 in gene regulation . DNA Res, 2018,25(6):577-586.
doi: 10.1093/dnares/dsy026 pmid: 30085012 |
| [69] |
Warren RL, Sutton GG, Jones SJ, Holt RA . Assembling millions of short DNA sequences using SSAKE . Bioinformatics, 2007,23(4):500-501.
doi: 10.1093/bioinformatics/btl629 pmid: 17158514 |
| [70] |
Jeck WR, Reinhardt JA, Baltrus DA, Hickenbotham MT, Magrini V, Mardis ER, Dangl JL, Jones CD . Extending assembly of short DNA sequences to handle error . Bioinformatics, 2007,23(21):2942-2944.
doi: 10.1093/bioinformatics/btm451 pmid: 17893086 |
| [71] | Bakker FT, Lei D, Yu JY, Mohammadin S, Wei Z, Van De Kerke S, Gravendeel B, Nieuwenhuis M, Staats M, Alquezar-Planas DE, Holmer R. Herbarium genomics: plastome sequence assembly from a range of herbarium specimens using an iterative organelle genome assembly pipeline . Biolo J Linn Soc, 2016,117(1):33-43. |
| [72] | Jin JJ, Yu WB, Yang JB, Song Y, dePamphilis CW, Yi TS, Li DZ, . GetOrganelle: a simple and fast pipeline for de novo assembly of a complete circular chloroplast genome using genome skimming data . bioRxiv, 2018: 256479. |
| [73] |
Coissac E, Hollingsworth PM, Lavergne S, Taberlet P . From barcodes to genomes: extending the concept of DNA barcoding . Mol Ecol, 2016,25(7):1423-1428.
doi: 10.1111/mec.13549 pmid: 26821259 |
| [74] |
Clark SC, Egan R, Frazier PI, Wang Z . ALE: a generic assembly likelihood evaluation framework for assessing the accuracy of genome and metagenome assemblies . Bioinformatics, 2013,29(4):435-443.
doi: 10.1093/bioinformatics/bts723 |
| [75] |
Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL . BLAST+: architecture and applications . BMC Bioinformatics, 2009,10:421.
doi: 10.1186/1471-2105-10-421 pmid: 20003500 |
| [76] |
Bayliss SC, Hunt VL, Yokoyama M, Thorpe HA, Feil EJ . The use of Oxford Nanopore native barcoding for complete genome assembly . Gigascience, 2017,6(3):1-6.
doi: 10.1093/gigascience/gix007 pmid: 28327976 |
| [77] |
Cao MD, Nguyen SH, Ganesamoorthy D, Elliott AG, Cooper MA, Coin LJ . Scaffolding and completing genome assemblies in real-time with nanopore sequencing . Nat Commun, 2017,8:14515.
doi: 10.1038/ncomms14515 pmid: 28218240 |
| [78] |
Deschamps S, Zhang Y, Llaca V, Ye L, Sanyal A, King M, May G, Lin HN . A chromosome-scale assembly of the sorghum genome using nanopore sequencing and optical mapping . Nat Commun, 2018,9(1):4844.
doi: 10.1038/s41467-018-07271-1 pmid: 30451840 |
| [79] |
Goodwin S, Gurtowski J, Ethe-Sayers S, Deshpande P, Schatz MC, Mccombie WR . Oxford Nanopore sequencing, hybrid error correction, and de novo assembly of a eukaryotic genome . Genome Res, 2015,25(11):1750-1756.
doi: 10.1101/gr.191395.115 pmid: 26447147 |
| [80] |
Lin MM, Qi XJ, Chen JY, Sun LM, Zhong YP, Fang JB, Hu CG . The complete chloroplast genome sequence of Actinidia arguta using the PacBio RS II platform . PLoS One, 2018,13(5):e0197393.
doi: 10.1371/journal.pone.0197393 pmid: 29795601 |
| [81] |
Chin CS, Alexander DH, Marks P, Klammer AA, Drake J, Heiner C, Clum A, Copeland A, Huddleston J, Eichler EE, Turner SW, Korlach J . Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data . Nat Methods, 2013,10(6):563-569.
doi: 10.1038/nmeth.2474 pmid: 23644548 |
| [82] |
Koren S, Walenz BP, Berlin K, Miller JR, Bergman NH, Phillippy AM . Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation . Genome Res, 2017,27(5):722-736.
doi: 10.1101/gr.215087.116 pmid: 28298431 |
| [83] |
Miyamoto M, Motooka D, Gotoh K, Imai T, Yoshitake K, Goto N, Iida T, Yasunaga T, Horii T, Arakawa K, Kasahara M, Nakamura S . Performance comparison of second- and third-generation sequencers using a bacterial genome with two chromosomes . BMC Genomics, 2014,15(1):699.
doi: 10.1186/1471-2164-15-699 pmid: 25142801 |
| [84] |
Soorni A, Haak D, Zaitlin D, Bombarely A . Organelle_PBA, a pipeline for assembling chloroplast and mitochondrial genomes from PacBio DNA sequencing data . BMC Genomics, 2017,18(1):49.
doi: 10.1186/s12864-016-3412-9 pmid: 28061749 |
| [85] |
Chaisson MJ, Tesler G . Mapping single molecule sequencing reads using basic local alignment with successive refinement (BLASR): application and theory . BMC Bioinformatics, 2012,13:238.
doi: 10.1186/1471-2105-13-238 pmid: 22988817 |
| [86] |
Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R , Genome Project Data Processing S. The sequence alignment/map format and SAMtools . Bioinformatics, 2009,25(16):2078-2079.
doi: 10.1093/bioinformatics/btp352 pmid: 19505943 |
| [87] |
Mcginnis S, Madden TL . BLAST: at the core of a powerful and diverse set of sequence analysis tools . Nucleic Acids Res, 2004,32(Web Server issue):W20-25.
doi: 10.1093/nar/gkh435 pmid: 15215342 |
| [88] |
Boetzer M, Pirovano W . SSPACE-LongRead: scaffolding bacterial draft genomes using long read sequence information . BMC Bioinformatics, 2014,15:211.
doi: 10.1186/1471-2105-15-211 pmid: 24950923 |
| [89] |
Quinlan AR, Hall IM . BEDTools: a flexible suite of utilities for comparing genomic features . Bioinformatics, 2010,26(6):841-842.
doi: 10.1093/bioinformatics/btq033 pmid: 20110278 |
| [90] |
Luo R, Liu B, Xie Y, Li Z, Huang W, Yuan J, He G, Chen Y, Pan Q, Liu Y, Tang J, Wu G, Zhang H, Shi Y, Liu Y, Yu C, Wang B, Lu Y, Han C, Cheung DW, Yiu SM, Peng S, Xiaoqian Z, Liu G, Liao X, Li Y, Yang H, Wang J, Lam TW, Wang J . SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler . Gigascience, 2012,1(1):18.
doi: 10.1186/2047-217X-1-18 pmid: 23587118 |
| [91] |
Zerbino DR, Birney E . Velvet: algorithms for de novo short read assembly using de Bruijn graphs . Genome Res, 2008,18(5):821-829.
doi: 10.1101/gr.074492.107 pmid: 18349386 |
| [92] |
Zhang TW, Luo YF, Chen YP, Li XN, Yu J . BIGrat: a repeat resolver for pyrosequencing-based re-sequencing with Newbler . BMC Res Notes, 2012,5:567.
doi: 10.1186/1756-0500-5-567 pmid: 23069129 |
| [93] |
Xie Y, Wu G, Tang J, Luo R, Patterson J, Liu S, Huang W, He G, Gu S, Li S, Zhou X, Lam TW, Li Y, Xu X, Wong GK, Wang J . SOAPdenovo-Trans: de novo transcriptome assembly with short RNA-Seq reads . Bioinformatics, 2014,30(12):1660-1666.
doi: 10.1093/bioinformatics/btu077 |
| [94] |
Haas BJ, Papanicolaou A, Yassour M, Grabherr M, Blood PD, Bowden J, Couger MB, Eccles D, Li B, Lieber M, Macmanes MD, Ott M, Orvis J, Pochet N, Strozzi F, Weeks N, Westerman R, William T, Dewey CN, Henschel R, Leduc RD, Friedman N, Regev A . De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis . Nat Protoc, 2013,8(8):1494-1512.
doi: 10.1038/nprot.2013.084 pmid: 23845962 |
| [95] |
Maccallum I, Przybylski D, Gnerre S, Burton J, Shlyakhter I, Gnirke A, Malek J, Mckernan K, Ranade S, Shea TP, Williams L, Young S, Nusbaum C, Jaffe DB . ALLPATHS 2: small genomes assembled accurately and with high continuity from short paired reads . Genome Biol, 2009,10(10):R103.
doi: 10.1186/gb-2009-10-10-r103 pmid: 19796385 |
| [96] |
Kajitani R, Yoshimura D, Okuno M, Minakuchi Y, Kagoshima H, Fujiyama A, Kubokawa K, Kohara Y, Toyoda A, Itoh T . Platanus-allee is a de novo haplotype assembler enabling a comprehensive access to divergent heterozygous regions . Nat Commun, 2019,10(1):1702.
doi: 10.1038/s41467-019-09575-2 pmid: 30979905 |
| [97] |
Lee HO, Choi JW, Baek JH, Oh JH, Lee SC, Kim CK . Assembly of the mitochondrial genome in the campanulaceae family using Illumina low-coverage sequencing . Genes, 2018,9(8):383.
doi: 10.3390/genes9080383 pmid: 30061537 |
| [98] |
Li D, Liu CM, Luo R, Sadakane K, Lam TW . MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph . Bioinformatics, 2015,31(10):1674-1676.
doi: 10.1093/bioinformatics/btv033 pmid: 25609793 |
| [99] | Plese B, Rossi ME, Kenny NJ, Taboada S, Koutsouveli V, Riesgo A . Trimitomics: an efficient pipeline for mitochondrial assembly from transcriptomic reads in non-model species . bioRxiv, 2018,19(5):1230-1239. |
| [100] |
Bolger AM, Lohse M, Usadel B . Trimmomatic: a flexible trimmer for Illumina sequence data . Bioinformatics, 2014,30(15):2114-2120.
doi: 10.1093/bioinformatics/btu170 |
| [101] |
Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, Chen Z, Mauceli E, Hacohen N, Gnirke A, Rhind N, Di Palma F, Birren BW, Nusbaum C, Lindblad-Toh K, Friedman N, Regev A . Full-length transcriptome assembly from RNA-Seq data without a reference genome . Nat Biotechnol, 2011,29(7):644-652.
doi: 10.1038/nbt.1883 pmid: 21572440 |
| [102] |
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ . Basic local alignment search tool . J Mol Biol, 1990,215(3):403-410.
doi: 10.1016/S0022-2836(05)80360-2 pmid: 2231712 |
| [103] |
Li M, Schroeder R, Ko A, Stoneking M . Fidelity of capture-enrichment for mtDNA genome sequencing: influence of NUMTs . Nucleic Acids Res, 2012,40(18):e137.
doi: 10.1093/nar/gks499 pmid: 22649055 |
| [104] | Li Y, Li X, Chen Y . Research summary of mitochondria pseudogene. J Mianyang Norm Univ, 2012,31(05):68-75. |
| 李艳, 黎霞, 陈艳 . 线粒体假基因研究综述. 绵阳师范学院学报, 2012,31(05):68-75. | |
| [105] |
Velozo Timbó R, Coiti Togawa R, Costa MMC, Andow DA, Paula DP . Mitogenome sequence accuracy using different elucidation methods . PLoS One, 2017,12(6):e0179971.
doi: 10.1371/journal.pone.0179971 pmid: 28662089 |
| [106] |
Peters JL, Bolender KA, Pearce JM . Behavioural vs. molecular sources of conflict between nuclear and mitochondrial DNA: the role of male-biased dispersal in a Holarctic sea duck . Mol Ecol, 2012,21(14):3562-3575.
doi: 10.1111/j.1365-294X.2012.05612.x |
| [107] |
Ekblom R, Smeds L, Ellegren H . Patterns of sequencing coverage bias revealed by ultra-deep sequencing of vertebrate mitochondria . BMC Genomics, 2014,15:467.
doi: 10.1186/1471-2164-15-467 pmid: 24923674 |
| [1] | Qianzhi Shao, Yi Jiang, Jinyu Wu. Whole-genome sequencing and its application in the research and diagnoses of genetic diseases [J]. Hereditas(Beijing), 2014, 36(11): 1087-1098. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||