Hereditas(Beijing) ›› 2021, Vol. 43 ›› Issue (12): 1121-1131.doi: 10.16288/j.yczz.21-205
• Review • Previous Articles Next Articles
Regulation of histone-to-protamine transition during spermiogenesis
Lu Yuan1,2( ), Tingting Ge1,2, Changmin Niu1,2, Wenhua Xu1,2, Ying Zheng1,2(
), Tingting Ge1,2, Changmin Niu1,2, Wenhua Xu1,2, Ying Zheng1,2( )
)
- 1. Department of Histology and Embryology, School of Medicine, Yangzhou University, Yangzhou 225009, China
2. Jiangsu Key Laboratory of Experimental & Translational Non-coding RNA Research, Yanghzou 225009, China;
-
Received:2021-06-10Revised:2021-10-27Online:2021-12-20Published:2021-11-10 -
Contact:Zheng Ying E-mail:2920054914@qq.com;yzzkl@163.com -
Supported by:Supported by the National Natural Science Foundation of China(82071696);Supported by the National Natural Science Foundation of China(82101674);the Key Scientific Project for Jiangsu Provincial Universities No(20KJA310002);the Postgraduate Scientific Research Innovation Program of Yangzhou University No(XKYCX20_35)
Cite this article
Lu Yuan, Tingting Ge, Changmin Niu, Wenhua Xu, Ying Zheng. Regulation of histone-to-protamine transition during spermiogenesis[J]. Hereditas(Beijing), 2021, 43(12): 1121-1131.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
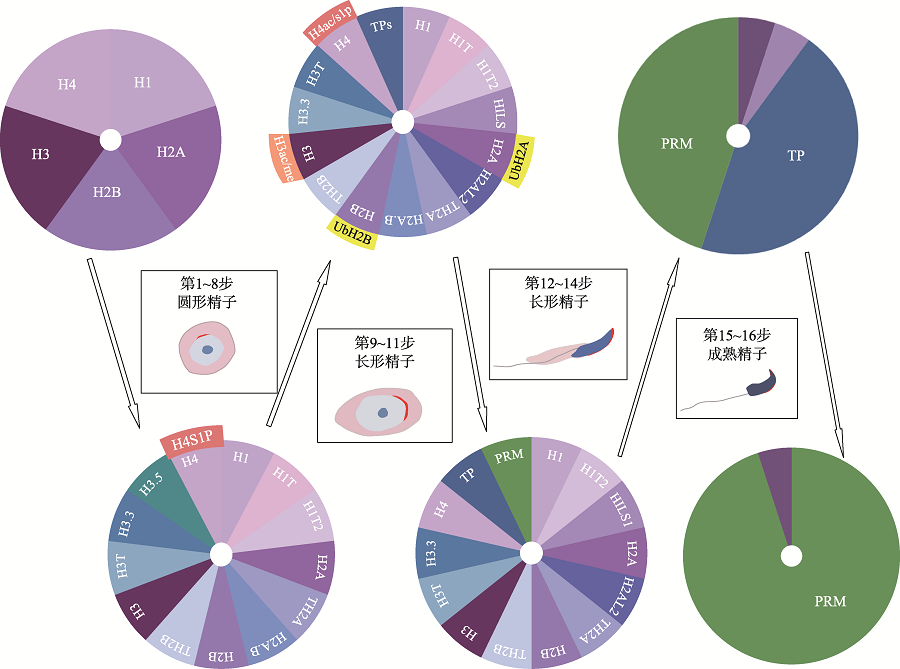
Table 1
Key genes related to the regulation mechanism of histone-to-protamine transition"
| 基因 | 基因敲除表型 | 功能 | 参考文献 |
|---|---|---|---|
| H1t | 雄性生育力正常 | 诱导局部染色质松弛 | [ |
| H1t2 | 雄性不育 | 参与PRM的掺入与染色质凝聚 | [ |
| Th2a/Th2b | 雄性不育 | 调节染色质开放或总组蛋白水平 | [ |
| H2al2 | 雄性不育 | 组装开放的核小体并允许TP掺入 | [ |
| H2a.b | 雄性生育力下降 | 调节H2AL2的解聚和TP1的掺入 | [ |
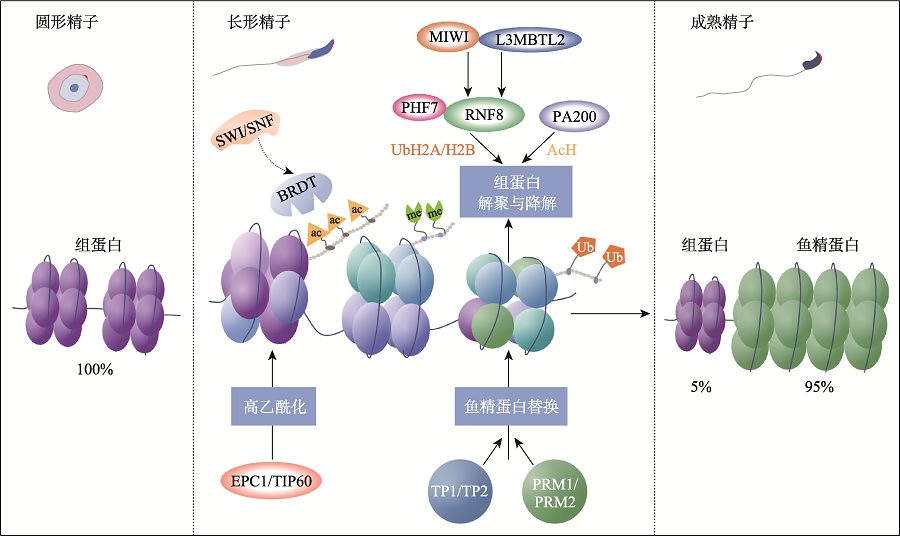
| H3f3a/H3f3b | 雄性不育 | 有助于形成开放的染色质构型 | [ |
| H3t | 雄性不育 | - | [ |
| Epc1 | 雄性不育 | 参与组蛋白的超乙酰化 | [ |
| Tip60 | 雄性不育 | 参与组蛋白的超乙酰化 | [ |
| Brdt | 雄性不育 | 介导乙酰化组蛋白的替换 | [ |
| Rnf8 | 雄性不育 | 单泛素化H2A和H2B,调节组蛋白的解聚 | [ |
| Miwi | 雄性不育 | 对RNF8的核转位必不可少,并促进组蛋白泛素化和解聚 | [ |
| L3mbtl2 | 雄性生育力下降 | 参与PRM1的沉积和染色质凝聚 | [ |
| Phf7 | 雄性不育 | 识别H3K4me2/me3并催化H2A泛素化以促进组蛋白的解聚 | [ |
| Pa200 | 雄性生育力下降 | 通过蛋白酶体介导乙酰化的组蛋白降解 | [ |
| Tnp1/Tnp2 | 雄性生育力下降 | PRM2的掺入和染色质凝聚所必需 | [ |
| Dazl | 雄性不育 | 调节TP mRNA的表达 | [ |
| Jmjd1a | 雄性不育 | 调节精子基因组包装和染色质凝聚 | [ |
| Prm1/Prm2 | 雄性不育 | 染色质凝聚所必需 | [ |
| Camk4 | 雄性不育 | 介导PRM2磷酸化 | [ |
| Ppp1cc2 | 雄性不育 | 使鱼精蛋白2第56位丝氨酸(Prm2Ser56)去磷酸化 | [ |
| Ip6k1 | 雄性不育 | 调控TP2和PRM2的表达 | [ |
| [1] | Hao SL, Ni FD, Yang WX. The dynamics and regulation of chromatin remodeling during spermiogenesis. Gene, 2019,706:201-210. |
| [2] | Lin Q, Sirotkin A, Skoultchi AI. Normal spermatogenesis in mice lacking the testis-specific linker histone H1t. Mol Cell Biol, 2000,20(6):2122-2128. |
| [3] | Mahadevan IA, Kumar S, Rao MRS. Linker histone variant H1t is closely associated with repressed repeat- element chromatin domains in pachytene spermatocytes. Epigenetics Chromatin, 2020,13(1):9. |
| [4] | Martianov I, Brancorsini S, Catena R, Gansmuller A, Kotaja N, Parvinen M, Sassone-Corsi P, Davidson I. Polar nuclear localization of H1T2, a histone H1 variant, required for spermatid elongation and DNA condensation during spermiogenesis. Proc Natl Acad Sci USA, 2005,102(8):2808-2813. |
| [5] | Tanaka H, Iguchi N, Isotani A, Kitamura K, Toyama Y, Matsuoka Y, Onishi M, Masai K, Maekawa M, Toshimori K, Okabe M, Nishimune Y. HANP1/H1T2, a novel histone H1-like protein involved in nuclear formation and sperm fertility. Mol Cell Biol, 2005,25(16):7107-7119. |
| [6] | Shalini V, Bhaduri U, Ravikkumar AC, Rengarajan A, Satyanarayana RMR. Genome-wide occupancy reveals the localization of H1T2 (H1fnt) to repeat regions and a subset of transcriptionally active chromatin domains in rat spermatids. Epigenetics Chromatin, 2021,14(1):3. |
| [7] | Mishra LN, Shalini V, Gupta N, Ghosh K, Suthar N, Bhaduri U, Rao MRS. Spermatid-specific linker histone HILS1 is a poor condenser of DNA and chromatin and preferentially associates with LINE-1 elements. Epigenetics Chromatin, 2018,11(1):43. |
| [8] | Shinagawa T, Huynh LM, Takagi T, Tsukamoto D, Tomaru C, Kwak HG, Dohmae N, Noguchi J, Ishii S. Disruption of Th2a and Th2b genes causes defects in spermatogenesis. Development, 2015,142(7):1287-1292. |
| [9] | Montellier E, Boussouar F, Rousseaux S, Zhang K, Buchou T, Fenaille F, Shiota H, Debernardi A, Héry P, Curtet S, Jamshidikia M, Barral S, Holota H, Bergon A, Lopez F, Guardiola P, Pernet K, Imbert J, Petosa C, Tan MJ, Zhao YM, Gérard M, Khochbin S. Chromatin-to- nucleoprotamine transition is controlled by the histone H2B variant TH2B. Genes Dev, 2013,27(15):1680-1692. |
| [10] | Barral S, Morozumi Y, Tanaka H, Montellier E, Govin J, de Dieuleveult M, Charbonnier G, Couté Y, Puthier D, Buchou T, Boussouar F, Urahama T, Fenaille F, Curtet S, Héry P, Fernandez-Nunez N, Shiota H, Gérard M, Rousseaux S, Kurumizaka H, Khochbin S. Histone variant H2A.L.2 guides transition protein-dependent protamine assembly in male germ cells. Mol Cell, 2017, 66(1): 89-101.e8. |
| [11] | Hoghoughi N, Barral S, Curtet S, Chuffart F, Charbonnier G, Puthier D, Buchou T, Rousseaux S, Khochbin S. RNA-guided genomic localization of H2A.L.2 histone variant. Cells, 2020,9(2):474. |
| [12] | Soboleva TA, Nekrasov M, Pahwa A, Williams R, Huttley GA, Tremethick DJ. A unique H2A histone variant occupies the transcriptional start site of active genes. Nat Struct Mol Biol, 2011,19(1):25-30. |
| [13] | Soboleva TA, Parker BJ, Nekrasov M, Hart-Smith G, Tay YJ, Tng WQ, Wilkins M, Ryan D, Tremethick DJ. A new link between transcriptional initiation and pre-mRNA splicing: the RNA binding histone variant H2A.B. PLoS Genet, 2017,13(2):e1006633. |
| [14] | Anuar ND, Kurscheid S, Field M, Zhang L, Rebar E, Gregory P, Buchou T, Bowles J, Koopman P, Tremethick DJ, Soboleva TA. Gene editing of the multi-copy H2A.B gene and its importance for fertility. Genome Biol, 2019,20(1):23. |
| [15] | Molaro A, Wood AJ, Janssens D, Kindelay SM, Eickbush MT, Wu S, Singh P, Muller CH, Henikoff S, Malik HS. Biparental contributions of the H2A.B histone variant control embryonic development in mice. PLoS Biol, 2020,18(12):e3001001. |
| [16] | Tang MCW, Jacobs SA, Mattiske DM, Soh YM, Graham AN, Tran A, Lim SL, Hudson DF, Kalitsis P, O'Bryan MK, Wong LH, Mann JR. Contribution of the two genes encoding histone variant H3.3 to viability and fertility in mice. PLoS Genet, 2015,11(2):e1004964. |
| [17] | Yuen BTK, Bush KM, Barrilleaux BL, Cotterman R, Knoepfler PS. Histone H3.3 regulates dynamic chromatin states during spermatogenesis. Development, 2014,141(18):3483-3494. |
| [18] | Tachiwana H, Kagawa W, Osakabe A, Kawaguchi K, Shiga T, Hayashi-Takanaka Y, Kimura H, Kurumizaka H. Structural basis of instability of the nucleosome containing a testis-specific histone variant, human H3T. Proc Natl Acad Sci USA, 2010,107(23):10454-10459. |
| [19] | Ueda J, Harada A, Urahama T, Machida S, Maehara K, Hada M, Makino Y, Nogami J, Horikoshi N, Osakabe A, Taguchi H, Tanaka H, Tachiwana H, Yao T, Yamada M, Iwamoto T, Isotani A, Ikawa M, Tachibana T, Okada Y, Kimura H, Ohkawa Y, Kurumizaka H, Yamagata K. Testis-specific histone variant H3t gene is essential for entry into spermatogenesis. Cell Rep, 2017,18(3):593-600. |
| [20] | Urahama T, Harada A, Maehara K, Horikoshi N, Sato K, Sato Y, Shiraishi K, Sugino N, Osakabe A, Tachiwana H, Kagawa W, Kimura H, Ohkawa Y, Kurumizaka H. Histone H3.5 forms an unstable nucleosome and accumulates around transcription start sites in human testis. Epigenetics Chromatin, 2016,9:2. |
| [21] | Shiraishi K, Shindo A, Harada A, Kurumizaka H, Kimura H, Ohkawa Y, Matsuyama H. Roles of histone H3.5 in human spermatogenesis and spermatogenic disorders. Andrology, 2018,6(1):158-165. |
| [22] | Luense LJ, Wang XS, Schon SB, Weller AH, Lin Shiao E, Bryant JM, Bartolomei MS, Coutifaris C, Garcia BA, Berger SL. Comprehensive analysis of histone post- translational modifications in mouse and human male germ cells. Epigenetics Chromatin, 2016,9:24. |
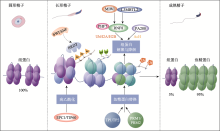
| [23] | Bao JQ, Bedford MT. Epigenetic regulation of the histone to protamine transition during spermiogenesis. Reproduction, 2016,151(5):R55-R70. |
| [24] | Ketchum CC, Larsen CD, McNeil A, Meyer-Ficca ML, Meyer RG. Early histone H4 acetylation during chromatin remodeling in equine spermatogenesis. Biol Reprod, 2018,98(1):115-129. |
| [25] | Dong YX, Isono KI, Ohbo K, Endo TA, Ohara O, Maekawa M, Toyama Y, Ito C, Toshimori K, Helin K, Ogonuki N, Inoue K, Ogura A, Yamagata K, Kitabayashi I, Koseki H. EPC1/TIP60-mediated histone acetylation facilitates spermiogenesis in mice. Mol Cell Biol, 2017,37(19):e00082-e00117. |
| [26] | Lu LY, Wu JX, Ye L, Gavrilina GB, Saunders TL, Yu XC. RNF8-dependent histone modifications regulate nucleosome removal during spermatogenesis. Dev Cell. 2010,18(3):371-384. |
| [27] | Guo YL, Song YF, Guo Z, Hu MJ, Liu B, Duan HY, Wang L, Yuan TX, Wang DG. Function of RAD6B and RNF8 in spermatogenesis. Cell Cycle, 2018,17(2):162-173. |
| [28] | Gou LT, Kang JY, Dai P, Wang X, Li F, Zhao S, Zhang M, Hua MM, Lu Y, Zhu Y, Li Z, Chen H, Wu LG, Li DS, Fu XD, Li JS, Shi HJ, Liu MF. Ubiquitination-deficient mutations in human Piwi cause male infertility by impairing histone-to-protamine exchange during spermiogenesis. Cell, 2017, 169(6): 1090-1104.e13. |
| [29] | Meng CL, Liao JY, Zhao DF, Huang HH, Qin JZ, Lee TL, Chen DG, Chan WY, Xia Y. L3MBTL2 regulates chromatin remodeling during spermatogenesis. Cell Death Differ, 2019,26(11):2194-2207. |
| [30] | Sin HS, Barski A, Zhang F, Kartashov AV, Nussenzweig A, Chen JJ, Andreassen PR, Namekawa SH. RNF8 regulates active epigenetic modifications and escape gene activation from inactive sex chromosomes in post-meiotic spermatids. Genes Dev, 2012,26(24):2737-2748. |
| [31] | Abe H, Meduri R, Li ZW, Andreassen PR, Namekawa SH. RNF8 is not required for histone-to-protamine exchange in spermiogenesis. Biol Reprod, 2021,ioab132. |
| [32] | Berkovits BD, Wolgemuth DJ. The role of the double bromodomain-containing BET genes during mammalian spermatogenesis. Curr Top Dev Biol, 2013,102:293-326. |
| [33] | Miller TCR, Simon B, Rybin V, Grötsch H, Curtet S, Khochbin S, Carlomagno T, Müller CW. A bromodomain- DNA interaction facilitates acetylation-dependent bivalent nucleosome recognition by the BET protein BRDT. Nat Commun, 2016,7:13855. |
| [34] | Gaucher J, Boussouar F, Montellier E, Curtet S, Buchou T, Bertrand S, Hery P, Jounier S, Depaux A, Vitte AL, Guardiola P, Pernet K, Debernardi A, Lopez F, Holota H, Imbert J, Wolgemuth DJ, Gérard M, Rousseaux S, Khochbin S. Bromodomain-dependent stage-specific male genome programming by Brdt. EMBO J, 2012,31(19):3809-3820. |
| [35] | Dhar S, Thota A, Rao MRS. Insights into role of bromodomain, testis-specific (Brdt) in acetylated histone H4-dependent chromatin remodeling in mammalian spermiogenesis. J Biol Chem, 2012,287(9):6387-6405. |
| [36] | Guo YL, Song YF, Guo Z, Hu MJ, Liu B, Duan HY, Wang L, Yuan TX, Wang DG. Function of RAD6B and RNF8 in spermatogenesis. Cell Cycle, 2018,17(2):162-173. |
| [37] | Wang XK, Kang JY, Wei LX, Yang XG, Sun HD, Yang SM, Lu L, Yan M, Bai MZ, Chen YY, Long JJ, Li N, Li DS, Huang J, Lei M, Shao Z, Yuan W, Zuo EW, Lu KH, Liu MF, Li JS. PHF7 is a novel histone H2A E3 ligase prior to histone-to-protamine exchange during spermiogenesis. Development, 2019, 146(13): dev175547. |
| [38] | Kim CR, Noda T, Kim H, Kim G, Park S, Na Y, Oura S, Shimada K, Bang I, Ahn JY, Kim YR, Oh SK, Choi HJ, Kim JS, Jung I, Lee H, Okada Y, Ikawa M, Baek SH. PHF7 modulates BRDT stability and histone-to-protamine exchange during spermiogenesis. Cell Rep, 2020,32(4):107950. |
| [39] | Berruti G. Destruction or reconstruction: a subtle liaison between the proteolytic and signaling role of protein ubiquitination in spermatogenesis. Adv Exp Med Biol, 2021,1288:215-240. |
| [40] | Liu ZQ, Miao DS, Xia QW, Hermo L, Wing SS. Regulated expression of the ubiquitin protein ligase, E3(histone)/ LASU1/Mule/ARF-BP1/HUWE1, during spermatogenesis. Dev Dyn, 2007,236(10):2889-2898. |
| [41] | Haraguchi CM, Mabuchi T, Hirata S, Shoda T, Tokumoto T, Hoshi K, Yokota S. Possible function of caudal nuclear pocket: degradation of nucleoproteins by ubiquitin- proteasome system in rat spermatids and human sperm. J Histochem Cytochem, 2007,55(6):585-595. |
| [42] | Khor B, Bredemeyer AL, Huang CY, Turnbull IR, Evans R, Maggi LB, White JM, Walker LM, Carnes K, Hess RA, Sleckman BP. Proteasome activator PA200 is required for normal spermatogenesis. Mol Cell Biol, 2006,26(8):2999-3007. |
| [43] | Qian MX, Pang Y, Liu CH, Haratake K, Du BY, Ji DY, Wang GF, Zhu QQ, Song W, Yu YD, Zhang XX, Huang HT, Miao SY, Chen LB, Zhang ZH, Liang YN, Liu S, Cha H, Yang D, Zhai YG, Komatsu T, Tsuruta F, Li HT, Cao C, Li W, Li GH, Cheng YF, Chiba T, Wang LF, Goldberg AL, Shen Y, Qiu XB. Acetylation-mediated proteasomal degradation of core histones during DNA repair and spermatogenesis. Cell, 2013,153(5):1012-1024. |
| [44] | Zhang ZH, Jiang TX, Chen LB, Zhou WH, Liu YX, Gao F, Qiu XB. Proteasome subunit α4s is essential for formation of spermatoproteasomes and histone degradation during meiotic DNA repair in spermatocytes. J Biol Chem, 2021,296:100130. |
| [45] | Rathke C, Baarends WM, Awe S, Renkawitz-Pohl R. Chromatin dynamics during spermiogenesis. Biochim Biophys Acta, 2014,1839(3):155-168. |
| [46] | Yu YE, Zhang Y, Unni E, Shirley CR, Deng JM, Russell LD, Weil MM, Behringer RR, Meistrich ML. Abnormal spermatogenesis and reduced fertility in transition nuclear protein 1-deficient mice. Proc Natl Acad Sci USA, 2000,97(9):4683-4688. |
| [47] | Zhao M, Shirley CR, Yu YE, Mohapatra B, Zhang Y, Unni E, Deng JM, Arango NA, Terry NH, Weil MM, Russell LD, Behringer RR, Meistrich ML. Targeted disruption of the transition protein 2 gene affects sperm chromatin structure and reduces fertility in mice. Mol Cell Biol, 2001,21(21):7243-7255. |
| [48] | Shirley CR, Hayashi S, Mounsey S, Yanagimachi R, Meistrich ML. Abnormalities and reduced reproductive potential of sperm from Tnp1- and Tnp2-null double mutant mice. Biol Reprod, 2004,71(4):1220-1229. |
| [49] | Tseden K, Topaloglu O, Meinhardt A, Dev A, Adham I, Müller C, Wolf S, Böhm D, Schlüter G, Engel W, Nayernia K. Premature translation of transition protein 2 mRNA causes sperm abnormalities and male infertility. Mol Reprod Dev, 2007,74(3):273-279. |
| [50] | Phillips BT, Williams JG, Atchley DT, Xu XJ, Li JL, Adams AL, Johnson KL, Hall TMT. Mass spectrometric identification of candidate RNA-binding proteins associated with transition nuclear protein mRNA in the mouse testis. Sci Rep, 2019,9(1):13618. |
| [51] | Okada Y, Scott G, Ray MK, Mishina Y, Zhang Y. Histone demethylase JHDM2A is critical for Tnp1 and Prm1 transcription and spermatogenesis. Nature, 2007,450(7166):119-123. |
| [52] | Liu ZL, Zhou SL, Liao L, Chen X, Meistrich M, Xu JM. Jmjd1a demethylase-regulated histone modification is essential for cAMP-response element modulator-regulated gene expression and spermatogenesis. J Biol Chem, 2010,285(4):2758-2770. |
| [53] | Eelaminejad Z, Favaedi R, Sodeifi N, Sadighi Gilani MA, Shahhoseini M. Deficient expression of JMJD1A histone demethylase in patients with round spermatid maturation arrest. Reprod Biomed Online, 2017,34(1):82-89. |
| [54] | Bao JQ, Bedford MT. Epigenetic regulation of the histone-to-protamine transition during spermiogenesis. Reproduction, 2016,151(5):R55-R70. |
| [55] | Cho C, Willis WD, Goulding EH, Jung-Ha H, Choi YC, Hecht NB, Eddy EM. Haploinsufficiency of protamine-1 or -2 causes infertility in mice. Nat Genet, 2001,28(1):82-86. |
| [56] | Brunner AM, Nanni P, Mansuy IM. Epigenetic marking of sperm by post-translational modification of histones and protamines. Epigenetics Chromatin, 2014,7(1):2. |
| [57] | Wu JY, Ribar TJ, Cummings DE, Burton KA, McKnight GS, Means AR. Spermiogenesis and exchange of basic nuclear proteins are impaired in male germ cells lacking Camk4. Nat Genet, 2000,25(4):448-452. |
| [58] | Itoh K, Kondoh G, Miyachi H, Sugai M, Kaneko Y, Kitano S, Watanabe H, Maeda R, Imura A, Liu Y, Ito C, Itohara S, Toshimori K, Fujita J. Dephosphorylation of protamine 2 at serine 56 is crucial for murine sperm maturation in vivo. Sci Signal, 2019, 12(574): eaao7232. |
| [59] | Malla AB, Bhandari R. IP6K1 is essential for chromatoid body formation and temporal regulation of Tnp2 and Prm2 expression in mouse spermatids. J Cell Sci, 2017,130(17):2854-2866. |
| [1] | Tingting Ge, Lu Yuan, Wenhua Xu, Ying Zheng. Role and mechanism of intraflagellar transport in mammalian spermiogenesis [J]. Hereditas(Beijing), 2021, 43(11): 1038-1049. |
| [2] | Lian Ren, Xiushan Wu, Yongqing Li. The mechanism underlying histone deacetylases regulating cardiac hypertrophy [J]. Hereditas(Beijing), 2020, 42(6): 536-547. |
| [3] | Longxiang Xie, Zhaoxiao Yu, Siyao Guo, Ping Li, Abualgasim Elgaili Abdalla, Jianping Xie. The roles of epigenetics and protein post-translational modifications in bacterial antibiotic resistance [J]. HEREDITAS(Beijing), 2015, 37(8): 793-800. |
| [4] | Zexian Liu, Yudong Cai, Xuejiang Guo, Ao Li, Tingting Li, Jianding Qiu, Jian Ren, Shaoping Shi, Jiangning Song, Minghui Wang, Lu Xie, Yu Xue, Ziding Zhang, Xingming Zhao. Post-translational modification (PTM) bioinformatics in China: progresses and perspectives [J]. HEREDITAS(Beijing), 2015, 37(7): 621-634. |
| [5] | Qiang Pu,Jia Luo,Linyuan Shen,Xuewei Li,Shunhua Zhang,Li Zhu. Application of modification-specific proteomics in the meat-quality study [J]. HEREDITAS(Beijing), 2015, 37(4): 327-335. |
| [6] | GUO Min-Xia, FU Yong-Fu. Advances on SUMO substrates in Arabidopsis [J]. HEREDITAS, 2013, 35(6): 727-734. |
| [7] | ZHANG Jun-Fang ZHU Hua-Bin ZHANG Liu-Guang HAO Hai-Sheng ZHAO Xue-Ming QIN Tong LU Yong-Qiang WANG Dong. Advance on research of gene expression during spermiogenesis at transcription level [J]. HEREDITAS, 2013, 35(5): 587-594. |
| [8] | HUANG Di, LI Jie, HE Li-Qun. Influence of Tripterygium wilfordii on [J]. HEREDITAS, 2009, 31(9): 941-946. |
| [9] | NIE Jing, IAN Chun-Yan, HANG Ling-Qiang. Progress in regulation of activity and stability of ubiquitin protein ligase MDM2 [J]. HEREDITAS, 2009, 31(10): 993-998. |
| [10] | GUO Yan-He, LIU Li, CAI Rong, QIAN Cheng. piRNA: A novel member of small RNA family [J]. HEREDITAS, 2008, 30(1): 28-34. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||