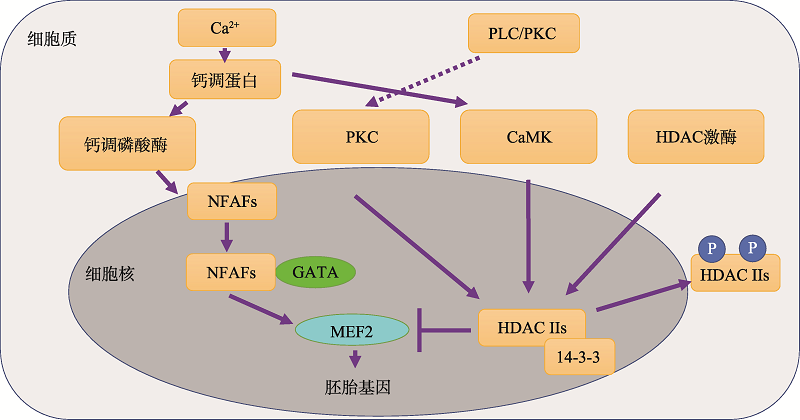
Hereditas(Beijing) ›› 2020, Vol. 42 ›› Issue (6): 536-547.doi: 10.16288/j.yczz.19-346
• Review • Previous Articles Next Articles
The mechanism underlying histone deacetylases regulating cardiac hypertrophy
Lian Ren, Xiushan Wu, Yongqing Li( )
)
- State Key Lab of Development Biology of Freshwater Fish, Key Laboratory of the Ministry of Education, Heart Development Research Center, College of Life Sciences, Hunan Normal University, Changsha 410081, China
-
Received:2020-01-18Revised:2020-04-13Online:2020-06-20Published:2020-04-27 -
Contact:Li Yongqing E-mail:liyongqing2002cn@aliyun.com -
Supported by:Supported by the National Natural Science Foundation of China No(81470377);Hunan Province Biological Development Engineering and New Product R & D Collaborative Innovation Center No(2013-448-6)
Cite this article
Lian Ren, Xiushan Wu, Yongqing Li. The mechanism underlying histone deacetylases regulating cardiac hypertrophy[J]. Hereditas(Beijing), 2020, 42(6): 536-547.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
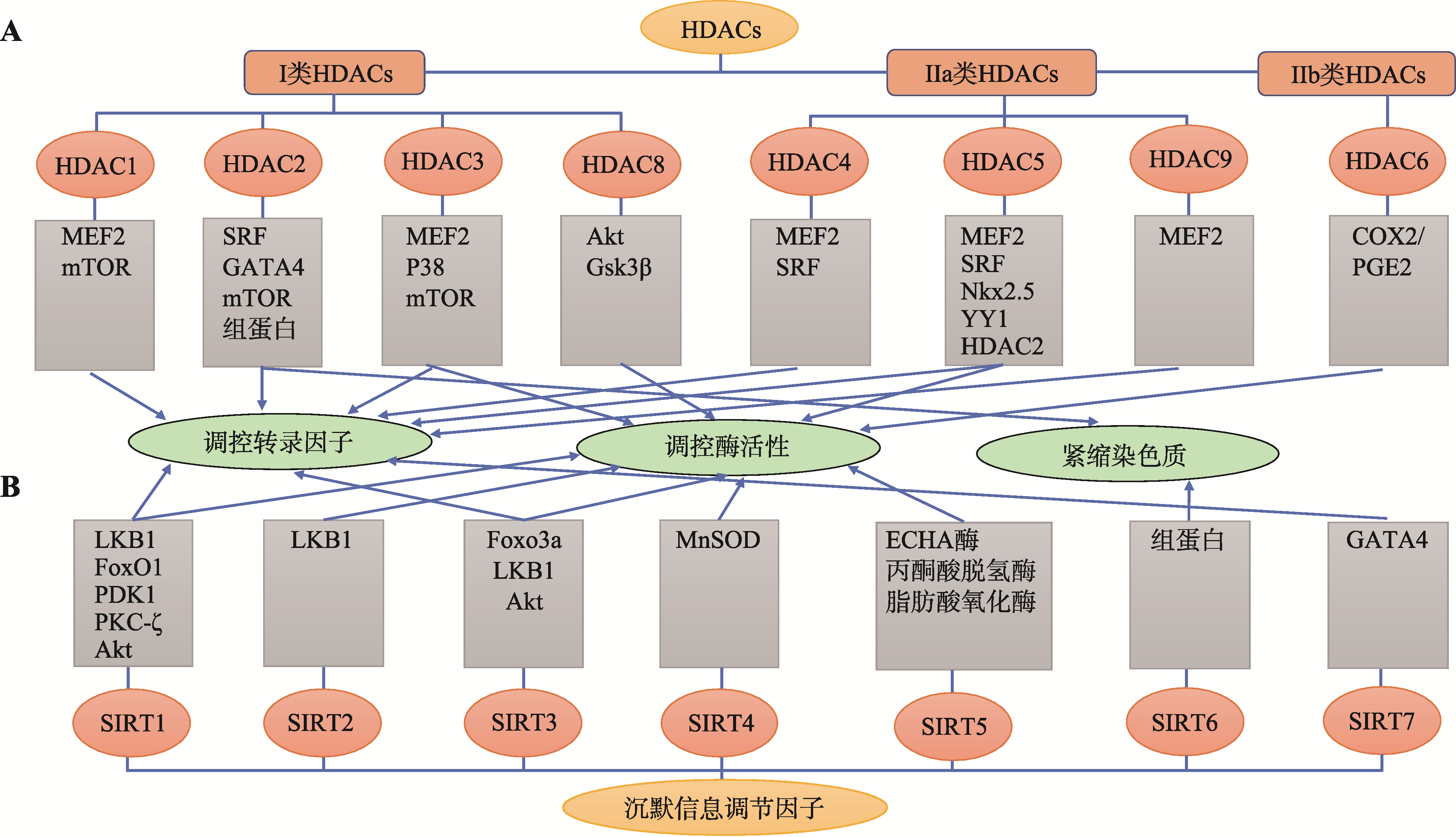
Table 1
Structural characteristics of HDACs subfamily"
| HDACs亚族 | 分子结构 | 研究模型 | 参考文献 |
|---|---|---|---|
| I类 | HDAC1 | HDAC1敲除小鼠模型 | [ |
| HDAC2 | HDAC2敲除/转基因小鼠模型 | [ | |
| HDAC3 | HDAC3转基因小鼠模型 | [ | |
| HDAC8 | 高血压大鼠模型 | [ | |
| IIa类 | HDAC4 | Dvl1转基因CaMKII敲除小鼠模型 | [ |
| HDAC5 | HDAC5敲除/转基因小鼠模型 | [ | |
| HDAC7 | - | ||
| HDAC9 | HDAC9敲除小鼠模型 | [ | |
| IIb类 | HDAC6 | HDAC6敲除小鼠模型 | [ |
| HDAC10 | - | ||
| IV类 | HDAC11 | - | |
| III类 | SIRT1 | SIRT1敲除小鼠模型 | [ |
| SIRT2 | SIRT2敲除/转基因小鼠模型 | [ | |
| SIRT3 | SIRT3敲除/转基因小鼠模型 | [ | |
| SIRT4 | SIRT4敲除/转基因小鼠模型 | [ | |
| SIRT5 | SIRT5敲除小鼠模型 | [ | |
| SIRT6 | SIRT6转基因小鼠模型 | [ | |
| SIRT7 | SIRT7心脏特异敲除小鼠模型 | [ |
| [1] | Hunter DJ, Reddy KS . Noncommunicable diseases. N Engl J Med, 2013,369(14):1336-1343. |
| [2] | Kouzarides T . Acetylation: a regulatory modification to rival phosphorylation? EMBO J, 2000,19(6):1176-1179. |
| [3] | Taunton J, Hassig CA, Schreiber SL . A mammalian histone deacetylase related to the yeast transcriptional regulator Rpd3p. Science, 1996,272(5260):408-411. |
| [4] | Jones P, Altamura S, De Francesco R, Gallinari P, Lahm A, Neddermann P, Rowley M, Serafini S, Steinkühler C . Probing the elusive catalytic activity of vertebrate class IIa histone deacetylases. Bioorg Med Chem Lett, 2008,18(6):1814-1819. |
| [5] | Duan BY, Ye D, Zhu SC, Jia WW, Lu CQ, Wang GY, Guo XD, Yu YY, Wu CY, Kang JH . HDAC10 promotes angiogenesis in endothelial cells through the PTPN22/ ERK axis. Oncotarget, 2017,8(37):61338-61349. |
| [6] | Houtkooper RH, Pirinen E, Auwerx J . Sirtuins as regulators of metabolism and healthspan. Nat Rev Mol Cell Biol, 2012,13(4):225-238. |
| [7] | Cao J, Sun L, Aramsangtienchai P, Spiegelman NA, Zhang XY, Seto E, Lin HN . DAC11 regulates type I interferon signaling through defatty-acylation of SHMT2. Proc Natl Acad Sci USA, 2019,116(12):5487-5492. |
| [8] | Morales CR, Li DL, Pedrozo Z, May HI, Jiang N, Kyrychenko V, Cho GW, Kim SY, Wang ZV, Rotter D, Rothermel BA, Schneider JW, Lavandero S, Gillette TG, Hill JA. Inhibition of class I histone deacetylases blunts cardiac hypertrophy through TSC2-dependent mTOR repression. Sci Signal, 2016, 9(422): ra34. |
| [9] | Lagger G, O'Carroll D, Rembold M, Khier H, Tischler J, Weitzer G, Schuettengruber B, Hauser C, Brunmeir R, Jenuwein T, Seiser C . Essential function of histone deacetylase 1 in proliferation control and CDK inhibitor repression. EMBO J, 2002,21(11):2672-2681. |
| [10] | Yang XJ, Seto E . Collaborative spirit of histone deacetylases in regulating chromatin structure and gene expression. Curr Opin Genet Dev, 2003,13(2):143-153. |
| [11] | Kesherwani V, Nandi SS, Sharawat SK, Shahshahan HR, Mishra PK . Hydrogen sulfide mitigates homocysteine- mediated pathological remodeling by inducing miR-133a in cardiomyocytes. Mol Cell Biochem, 2015,404(1-2):241-250. |
| [12] | Chandrasekaran S, Peterson RE, Mani SK, Addy B, Buchholz AL, Xu L, Thiyagarajan T, Kasiganesan H, Kern CB, Menick DR . Histone deacetylases facilitate sodium/ calcium exchanger up-regulation in adult cardiomyocytes. FASEB J, 2009,23(11):3851-3864. |
| [13] | Montgomery RL, Davis CA, Potthoff MJ, Haberland M, Fielitz J, Qi XX, Hill JA, Richardson JA, Olson EN . Histone deacetylases 1 and 2 redundantly regulate cardiac morphogenesis, growth, and contractility. Genes Dev, 2007,21(14):1790-1802. |
| [14] | Yuan YG, Peng WZ, Liu YX, Xu ZS . Palmatine attenuates isoproterenol-induced pathological hypertrophy via selectively inhibiting HDAC2 in rats. Int J Immunopathol Pharmacol, 2017,30(4):406-412. |
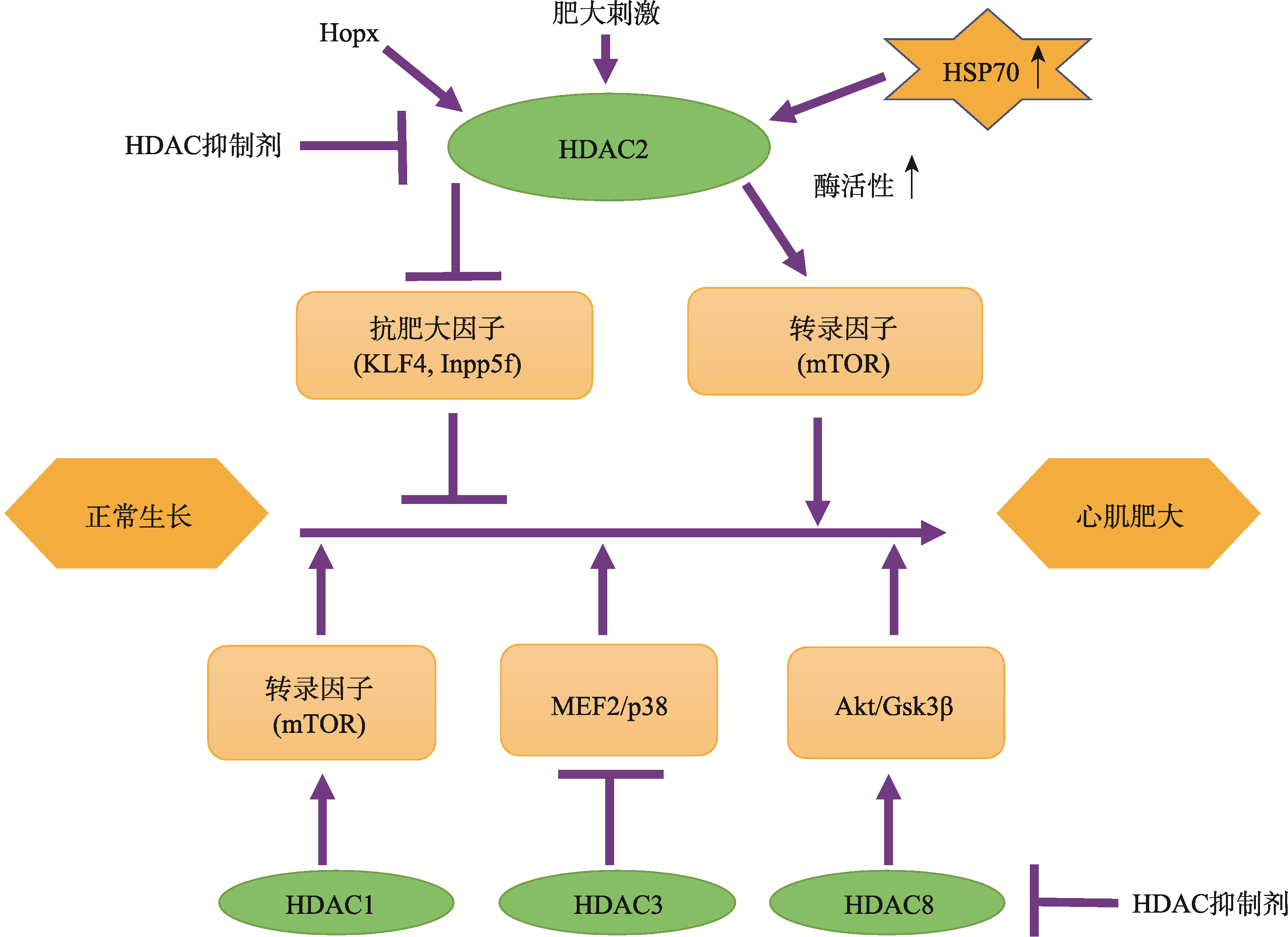
| [15] | Trivedi CM, Luo Y, Yin Z, Zhang MZ, Zhu WT, Wang T, Floss T, Goettlicher M, Noppinger PR, Wurst W, Ferrari VA, Abrams CS, Gruber PJ, Epstein JA . Hdac2 regulates the cardiac hypertrophic response by modulating Gsk3β activity. Nat Med, 2007,13(3):324-331. |
| [16] | Eom GH, Kook H . Role of histone deacetylase 2 and its posttranslational modifications in cardiac hypertrophy. BMB Rep, 2015,48(3):131-138. |
| [17] | Raghunathan S, Goyal RK, Patel BM . Selective inhibition of HDAC2 by magnesium valproate attenuates cardiac hypertrophy. Can J Physiol Pharmacol, 2017,95(3):260-267. |
| [18] | Kook H, Lepore JJ, Gitler AD, Lu MM, Wing-Man Yung W, Mackay J, Zhou R, Ferrari V, Gruber P, Epstein JA . Cardiac hypertrophy and histone deacetylase-dependent transcriptional repression mediated by the atypical homeodomain protein Hop. J Clin Invest, 2003,112(6):863-871. |
| [19] | Trivedi CM, Zhu W, Wang Q, Jia C, Kee HJ, Li L, Hannenhalli S, Epstein JA . Hopx and Hdac2 interact to modulate Gata4 acetylation and embryonic cardiac myocyte proliferation. Dev Cell, 2010,19(3):450-459. |
| [20] | Li RF, Cao SS, Fang WJ, Song Y, Luo XT, Wang HY, Wang JG . Roles of HDAC2 and HDAC8 in cardiac remodeling in renovascular hypertensive rats and the effects of valproic acid sodium. Pharmacology, 2017,99(1-2):27-39. |
| [21] | Grégoire S, Xiao L, Nie J, Zhang X, Xu M, Li J, Wong J, Seto E, Yang XJ . Histone deacetylase 3 interacts with and deacetylates myocyte enhancer factor 2. Mol Cell Biol, 2007,27(4):1280-95. |
| [22] | Pillai VB, Sundaresan NR, Samant SA, Wolfgeher D, Trivedi CM, Gupta MP . Acetylation of a conserved lysine residue in the ATP binding pocket of p38 augments its kinase activity during hypertrophy of cardiomyocytes. Mol Cell Biol, 2011,31(11):2349-2363. |
| [23] | Trivedi CM, Lu MM, Wang QH, Epstein JA . Transgenic over-expression of Hdac3 in the heart produces increased postnatal cardiac myocyte proliferation but does not induce hypertrophy. J Biol Chem, 2008,283(39):26484-26489. |
| [24] | Kee HJ, Bae EH, Park S, Lee KE, Suh SH, Kim SW, Jeong MH. HDAC inhibition suppresses cardiac hypertrophy and fibrosis in DOCA-salt hypertensive rats via regulation of HDAC6/HDAC8 enzyme activity. Kidney Blood Press Res, 2013,37(4-5):229-239. |
| [25] | Yan MW, Chen C, Gong W, Yin ZW, Zhou L, Chaugai S, Wang DW. miR-21-3p regulates cardiac hypertrophic response by targeting histone deacetylase-8. Cardiovasc Res, 2015,105(3):340-352. |
| [26] | Backs J, Backs T, Bezprozvannaya S, McKinsey TA, Olson EN. Histone deacetylase 5 acquires calcium/calmodulin- dependent kinase II responsiveness by oligomerization with histone deacetylase 4. Mol Cell Biol, 2008,28(10):3437-3445. |
| [27] | Backs J, Worst BC, Lehmann LH, Patrick DM, Jebessa Z, Kreusser MM, Sun Q, Chen L, Heft C, Katus HA, Olson EN . Selective repression of MEF2 activity by PKA- dependent proteolysis of HDAC4. J Cell Biol, 2011,195(3):403-415. |
| [28] | Sucharov CC, Dockstader K, McKinsey TA. YY1 protects cardiac myocytes from pathologic hypertrophy by interacting with HDAC5. Mol Biol Cell, 2008,19(10):4141-4153. |
| [29] | Eom GH, Nam YS, Oh JG, Choe N, Min HK, Yoo EK, Kang G, Nguyen VH, Min JJ, Kim JK, Lee IK, Bassel- Duby R, Olson EN, Park WJ, Kook H . Regulation of acetylation of histone deacetylase 2 by p300/CBP-Associated Factor/Histone deacetylase 5 in the development of cardiac hypertrophy. Circ Res, 2014,114(7):1133-1143. |
| [30] | Hu T, Schreiter FC, Bagchi RA, Tatman PD, Hannink M , McKinsey TA. HDAC5 catalytic activity suppresses cardiomyocyte oxidative stress and NRF2 target gene expression. J Biol Chem, 2019,294(21):8640-8652. |
| [31] | Zhang H, Shao Z, Alibin CP, Acosta C, Anderson HD . Liganded peroxisome proliferator-activated receptors (ppars) preserve nuclear histone deacetylase 5 levels in endothelin-treated sprague-dawley rat cardiac myocytes. PLoS One, 2014,9(12):e115258. |
| [32] | Haworth RS, Stathopoulou K, Candasamy AJ, Avkiran M . Neurohormonal regulation of cardiac histone deacetylase 5 nuclear localization by phosphorylation-dependent and phosphorylation-independent mechanisms. Circ Res, 2012,110(12):1585-1595. |
| [33] | Zhang LL, Deng MK, Lu AH, Chen YT, Chen Y, Wu C, Tan Z, Boini KM, Yang TX, Zhu Q, Wang L . Sodium butyrate attenuates angiotensin II-induced cardiac hypertrophy by inhibiting COX2/PGE2 pathway via a HDAC5/HDAC6-dependent mechanism. J Cell Mol Med, 2019,23(12):8139-8150. |
| [34] | Zhang CL, McKinsey TA, Chang S, Antos CL, Hill JA, Olson EN, . Class II histone deacetylases act as signal- responsive repressors of cardiac hypertrophy. Cell, 2002,110(4):479-488. |
| [35] | Zhang CL, McKinsey TA, Olson EN. The transcriptional corepressor MITR is a signal-responsive inhibitor of myogenesis. Proc Natl Acad Sci USA, 2001,98(13):7354-7359. |
| [36] | Kee HJ, Bae EH, Park S, Lee KE, Suh SH, Kim SW, Jeong MH. HDAC inhibition suppresses cardiac hypertrophy and fibrosis in DOCA-salt hypertensive rats via regulation of HDAC6/HDAC8 enzyme activity. Kidney Blood Press Res, 2013,37(4-5):229-239. |
| [37] | Demos-Davies KM, Ferguson BS, Cavasin MA, Mahaffey JH, Williams SM, Spiltoir JI, Schuetze KB, Horn TR, Chen B, Ferrara C, Scellini B, Piroddi N, Tesi C, Poggesi C, Jeong MY, McKinsey TA. HDAC6 contributes to pathological responses of heart and skeletal muscle to chronic angiotensin-II signaling. Am J Physiol Heart Circ Physiol, 2014,307(2):H252-258. |
| [38] | Ma S, Feng J, Zhang R, Chen JW, Han D, Li X, Yang B, Li XJ, Fan MM, Li CY, Tian ZH, Wang YB, Cao F . SIRT1 activation by resveratrol alleviates cardiac dysfunction via mitochondrial regulation in diabetic cardiomyopathy mice. Oxid Med Cell Longev, 2017,2017:4602715. |
| [39] | Sundaresan NR, Pillai VB, Wolfgeher D, Samant S, Vasudevan P, Parekh V, Raghuraman H, Cunningham JM, Gupta M, Gupta MP. The deacetylase sirt1 promotes membrane localization and activation of akt and pdk1 during tumorigenesis and cardiac hypertrophy. Sci Signal, 2011, 4(182): ra46. |
| [40] | Li JY, Huang JY, Lu J, Guo Z, Li ZM, Gao H, Wang PX, Luo WW, Cai SD, Hu YH, Guo KT, Wang LP, Li ZZ, Wang MH, Zhang XL, Liu PQ . Sirtuin 1 represses PKC-ζ activity through regulating interplay of acetylation and phosphorylation in cardiac hypertrophy. Br J Pharmacol, 2019,176(3):416-435. |
| [41] | Li ST, Zhu ZX, Xue M, Yi XC, Liang JJ, Niu C, Chen G, Shen YJ, Zhang HP, Zheng JY, Zhao CC, Liang YZ, Cong WT, Wang Y, Jin LT . Fibroblast growth factor 21 protects the heart from angiotensin II-induced cardiac hypertrophy and dysfunction via SIRT1. Biochim Biophys Acta Mol Basis Dis, 2019,1865(6):1241-1252. |
| [42] | Tang XQ, Chen XF, Wang NY, Wang XM, Liang ST, Zheng W, Lu YB, Zhao X, Hao DL, Zhang ZQ, Zou MH, Liu DP, Chen HZ . SIRT2 acts as a cardioprotective deacetylase in pathological cardiac hypertrophy. Circulation, 2017,136(21):2051-2067. |
| [43] | Pillai VB, Bindu S, Sharp W, Fang YH, Kim G, Gupta M, Samant S, Gupta MP . Sirt3 protects mitochondrial DNA damage and blocks the development of doxorubicin- induced cardiomyopathy in mice. Am J Physiol Heart Circ Physiol, 2016,310(8):H962-972. |
| [44] | Sundaresan NR, Gupta M, Kim G, Rajamohan SB, Isbatan A, Gupta MP . Sirt3 blocks the cardiac hypertrophic response by augmenting foxo3a-dependent antioxidant defense mechanisms in mice. J Clin Invest, 2009,119(9):2758-2771. |
| [45] | Pillai VB, Sundaresan NR, Kim G, Gupta M, Rajamohan SB, Pillai JB, Samant S, Ravindra PV, Isbatan A, Gupta MP . Exogenous nad blocks cardiac hypertrophic response via activation of the sirt3-lkb1-amp-activated kinase pathway. J Biol Chem, 2010,285(5):3133-3144. |
| [46] | Luo YX, Tang XQ, An XZ, Xie XM, Chen XF, Zhao X, Hao DL, Chen HZ, Liu DP . SIRT4 accelerates Ang II-induced pathological cardiac hypertrophy by inhibiting manganese superoxide dismutase activity. Eur Heart J, 2017,38(18):1389-1398. |
| [47] | Sadhukhan S, Liu XJ, Ryu D, Nelson OD, Stupinski JA, Li Z, Chen W, Zhang S, Weiss RS, Locasale JW, Auwerx J, Lin HN . Metabolomics-assisted proteomics identifies succinylation and SIRT5 as important regulators of cardiac function. Proc Natl Acad Sci USA, 2016,113(16):4320-4325. |
| [48] | Hershberger KA, Abraham DM, Martin AS, Mao L, Liu J, Gu H, Locasale JW, Hirschey MD . Sirtuin 5 is required for mouse survival in response to cardiac pressure overload. J Biol Chem, 2017,292(48):19767-19781. |
| [49] | Sundaresan NR, Vasudevan P, Zhong L, Kim G, Samant S, Parekh V, Pillai VB, Ravindra PV, Gupta M, Jeevanandam V, Cunningham JM, Deng CX, Lombard DB, Mostoslavsky R, Gupta MP . The sirtuin sirt6 blocks igf-akt signaling and development of cardiac hypertrophy by targeting c-jun. Nat Med, 2012,18(11):1643-1650. |
| [50] | Yamamura S, Izumiya Y, Araki S, Nakamura T, Kimura Y, Hanatani S, Yamada T, Ishida T, Yamamoto M, Onoue Y, Arima Y, Yamamoto E, Sunagawa Y, Yoshizawa T, Nakagata N, Bober E, Braun T, Sakamoto K, Kaikita K, Morimoto T, Yamagata K, Tsujita K. Cardiomyocyte Sirt(Sirtuin) 7 ameliorates stress-induced cardiac hypertrophy by interacting with and deacetylating GATA4. Hypertension, 2020,75(1):98-108. |
| [51] | Schueler M, Zhang Q, Schlesinger J, Tönjes M, Sperling SR . Dynamics of Srf, p300 and histone modifications during cardiac maturation in mouse. Mol Biosyst, 2012,8(2):495-503. |
| [52] | Schlesinger J, Schueler M, Grunert M, Fischer JJ, Zhang Q, Krueger T, Lange M, Tönjes M, Dunkel I, Sperling SR . The cardiac transcription network modulated by Gata4, Mef2a, Nkx2.5, Srf, histone modifications, and microRNAs. PLoS Genet, 2011,7(2):e1001313. |
| [53] | Mathias RA, Guise AJ, Cristea IM . Post-translational modifications regulate class IIa histone deacetylase (HDAC) function in health and disease. Mol Cell Proteomics, 2015,14(3):456-470. |
| [54] | Vega RB, Harrison BC, Meadows E, Roberts CR, Papst PJ, Olson EN , McKinsey TA. Protein kinases C and D mediate agonist-dependent cardiac hypertrophy through nuclear export of histone deacetylase 5. Mol Cell Biol, 2004,24(19):8374-8385. |
| [55] | McKinsey TA, Zhang CL, Lu J, Olson EN . Signal-dependent nuclear export of a histone deacetylase regulates muscle differentiation. Nature, 2000,408(6808):106-111. |
| [56] | Sparrow DB, Miska EA, Langley E, Reynaud-Deonauth S, Kotecha S, Towers N, Spohr G, Kouzarides T, Mohun TJ . MEF-2 function is modified by a novel co-repressor, MITR. EMBO J, 1999,18(18):5085-5098. |
| [57] | Morin S1, Charron F, Robitaille L, Nemer M . GATA- dependent recruitment of MEF2 proteins to target promoters. EMBO J, 2000,19(9):2046-2055. |
| [58] | Youn HD, Chatila TA, Liu JO . Integration of calcineurin and MEF2 signals by the coactivator p300 during T-cell apoptosis. EMBO J, 2000,19(16):4323-4331. |
| [59] | Zhang M, Hagenmueller M, Riffel JH, Kreusser MM, Bernhold E, Fan J, Katus HA, Backs J, Hardt SE . Calcium/Calmodulin-dependent protein kinase II couples Wnt signaling with histone deacetylase 4 and mediates dishevelled-induced cardiomyopathy. Hypertension, 2015,65(2):335-344. |
| [60] | Calalb MB, McKinsey TA, Newkirk S, Huynh K, Sucharov CC, Bristow MR. Increased phosphorylation- dependent nuclear export of class II histone deacetylases in failing human heart. Clin Transl Sci, 2009,2(5):325-332. |
| [61] | Cao DS, Wang ZG, Zhang CL, Oh J, Xing W, Li S, Richardson JA, Wang DZ, Olson EN . Modulation of smooth muscle gene expression by association of histone acetyltransferases and deacetylases with myocardin. Mol Cell Biol, 2005,25(1):364-376. |
| [62] | McDonald OG, Wamhoff BR, Hoofnagle MH, Owens GK . Control of SRF binding to CArG box chromatin regulates smooth muscle gene expression in vivo. J Clin Invest, 2006,116(1):36-48. |
| [63] | Fischle W, Dequiedt F, Hendzel MJ, Guenther MG, Lazar MA, Voelter W, Verdin E . Enzymatic activity associated with class II HDACs is dependent on a multiprotein complex containing HDAC3 and SMRT/N-CoR. Mol Cell, 2002,9(1):45-57. |
| [64] | Chang CW, Lee L, Yu D, Dao K, Bossuyt J, Bers DM . Acute β-adrenergic activation triggers nuclear import of histone deacetylase 5 and delays G(q)-induced transcriptional activation. J Biol Chem, 2013,288(1):192-204. |
| [65] | Mangmool S, Shukla AK, Rockman HA . β-arrestin- dependent activation of Ca 2+/Calmodulin kinase Ⅱ after β-adrenergic receptor stimulation . J Cell Biol, 2010,189(3):573-587. |
| [66] | Gurha P, Abreu-Goodger C, Wang T, Ramirez MO, AL, van Drumond S, Chen Dongen Y, Bartonicek N, Enright AJ, Lee B, Kelm RJ Jr, Reddy AK, Taffet GE, Bradley A, Wehrens XH, Entman ML, Rodriguez A,. Targeted deletion of microRNA-22 promotes stress-induced cardiac dilation and contractile dysfunction. Circulation, 2012,125(22):2751-2761. |
| [67] | Huang ZP, Chen JH, Seok HY, Zhang Z, Kataoka M, Hu XY, Wang DZ . MicroRNA-22 regulates cardiac hypertrophy and remodeling in response to stress. Circ Res, 2013,112(9):1234-1243. |
| [68] | Diniz GP, Lino CA, Moreno CR, Senger N, Barreto-Chaves MLM . MicroRNA-1 overexpression blunts cardiomyocyte hypertrophy elicited by thyroid hormone. J Cell Physiol, 2017,232(12):3360-3368. |
| [69] | Xiao Y, Zhang X, Fan S, Cui G, Shen Z . MicroRNA-497 inhibits cardiac hypertrophy by targeting Sirt4. PLoS One, 2016,11(12):e0168078. |
| [70] | Kee HJ, Sohn IS, Nam KI, Park JE, Qian YR, Yin Z, Ahn Y, Jeong MH, Bang YJ, Kim N, Kim JK, Kim KK, Epstein JA, Kook H . Inhibition of histone deacetylation blocks cardiac hypertrophy induced by angiotensin II infusion and aortic banding. Circulation, 2006,113(1):51-59. |
| [71] | Cao DJ, Wang ZV, Battiprolu PK, Jiang N, Morales CR, Kong Y, Rothermel BA, Gillette TG, Hill JA . Histone deacetylase (HDAC) inhibitors attenuate cardiac hypertrophy by suppressing autophagy. Proc Natl Acad Sci USA, 2011,108(10):4123-4128. |
| [72] | Antos CL, McKinsey TA, Dreitz M, Hollingsworth LM, Zhang CL, Schreiber K, Rindt H, Gorczynski RJ, Olson EN. Dose-dependent blockade to cardiomyocyte hypertrophy by histone deacetylase inhibitors. J Biol Chem, 2003,278(31):28930-28937. |
| [73] | Majumdar G, Adris P, Bhargava N, Chen H, Raghow R . Pan-histone deacetylase inhibitors regulate signaling pathways involved in proliferative and pro-inflammatory mechanisms in H9c2 cells. BMC Genomics, 2012,13:709. |
| [74] | Majumdar G, Rooney RJ, Johnson IM, Raghow R . Panhistone deacetylase inhibitors inhibit proinflammatory signaling pathways to ameliorate interleukin-18-induced cardiac hypertrophy. Physiol Genomics, 2011,43(24):1319-1333. |
| [75] | Kook H, Lepore JJ, Gitler AD, Lu MM, Wing-Man Yung W, Mackay J, Zhou R, Ferrari V, Gruber P, Epstein JA. Cardiac hypertrophy and histone deacetylase-dependent transcriptional repression mediated by the atypical homeodomain protein Hop. J Clin Invest, 2003,112(6):863-871. |
| [76] | Kong YQ, Tannous P, Lu GR, Berenji K, Rothermel BA, Olson EN, Hill JA . Suppression of class I and II histone deacetylases blunts pressure-overload cardiac hypertrophy. Circulation, 2006,113(22):2579-2588. |
| [77] | Iyer A, Fenning A, Lim J, Le GT, Reid RC, Halili MA, Fairlie DP, Brown L . Antifibrotic activity of an inhibitor of histone deacetylases in DOCA-salt hypertensive rats. Br J Pharmacol, 2010,159(7):1408-1417. |
| [78] | Kang SH, Seok YM, Song MJ, Lee HA, Kurz T, Kim I . Histone deacetylase inhibition attenuates cardiac hypertrophy and fibrosis through acetylation of mineralocorticoid receptor in spontaneously hypertensive rats. Mol Pharmacol, 2015,87(5):782-791. |
| [79] | Cho YK, Eom GH, Kee HJ, Kim HS, Choi WY, Nam KI, Ma JS, Kook H . Sodium valproate, a histone deacetylase inhibitor, but not captopril, prevents right ventricular hypertrophy in rats. Circ J, 2010,74(4):760-770. |
| [80] | Cardinale JP, Sriramula S, Pariaut R, Guggilam A, Mariappan N, Elks CM, Francis J . HDAC inhibition attenuates inflammatory, hypertrophic, and hypertensive responses in spontaneously hypertensive rats. Hypertension, 2010,56(3):437-444. |
| [81] | Gallo P, Latronico MV, Gallo P, Grimaldi S, Borgia F, Todaro M, Jones P, Gallinari P, De Francesco R, Ciliberto G, Steinkühler C, Esposito G, Condorelli G . Inhibition of class I histone deacetylase with an apicidin derivative prevents cardiac hypertrophy and failure. Cardiovasc Res, 2008,80(3):416-424. |
| [82] | Ooi JY, Tuano NK, Rafehi H, Gao XM, Ziemann M, Du XJ, El-Osta A . HDAC inhibition attenuates cardiac hypertrophy by acetylation and deacetylation of target genes. Epigenetics, 2015,10(5):418-430. |
| [83] | Cao JH, Liao WT, Wo C, Xu GR, Xu HX, Li PL, Tao Y, Wang P, Lin JR, Deng LR . Transcriptome screening and verification of genes related to metabolism affected by histone deacetylase inhibitors. Hereditas(Beijing), 2015,37(9):918-925. |
| 曹继红, 廖尉廷, 沃琤, 徐国荣, 徐焕新, 李平龙, 陶冶, 王鹏, 林加日, 邓连瑞 . 组蛋白去乙酰化酶抑制剂影响的代谢相关基因的组学筛查及验证. 遗传, 2015,37(9):918-925. |
| [1] | Yang Yang, Mingxing Chu, Qiuyue Liu. The mechanism of circadian clock and its influence on animal circannual rhythm [J]. Hereditas(Beijing), 2023, 45(5): 409-424. |
| [2] | Chengan Lv, Ruoran Wang, Zhuo-Xian Meng. Molecular mechanism of islet β-cell functional alternations during type 2 diabetes [J]. Hereditas(Beijing), 2022, 44(10): 840-852. |
| [3] | Shanshan Gao, Jinliang Li, Jiani Yang, Tong Zhou, Rui Liu, Xiaoping Wang, Li Yu. Progresses on adaptive evolution of gliding and flying ability in mammals [J]. Hereditas(Beijing), 2022, 44(1): 46-58. |
| [4] | Lu Yuan, Tingting Ge, Changmin Niu, Wenhua Xu, Ying Zheng. Regulation of histone-to-protamine transition during spermiogenesis [J]. Hereditas(Beijing), 2021, 43(12): 1121-1131. |
| [5] | Xianci Xue,Li Yu. Advances on polyphenism in insects [J]. Hereditas(Beijing), 2017, 39(9): 798-809. |
| [6] | Longxiang Xie, Zhaoxiao Yu, Siyao Guo, Ping Li, Abualgasim Elgaili Abdalla, Jianping Xie. The roles of epigenetics and protein post-translational modifications in bacterial antibiotic resistance [J]. HEREDITAS(Beijing), 2015, 37(8): 793-800. |
| [7] | Zexian Liu, Yudong Cai, Xuejiang Guo, Ao Li, Tingting Li, Jianding Qiu, Jian Ren, Shaoping Shi, Jiangning Song, Minghui Wang, Lu Xie, Yu Xue, Ziding Zhang, Xingming Zhao. Post-translational modification (PTM) bioinformatics in China: progresses and perspectives [J]. HEREDITAS(Beijing), 2015, 37(7): 621-634. |
| [8] | Qiang Pu,Jia Luo,Linyuan Shen,Xuewei Li,Shunhua Zhang,Li Zhu. Application of modification-specific proteomics in the meat-quality study [J]. HEREDITAS(Beijing), 2015, 37(4): 327-335. |
| [9] | Xiaohua Yang, Huafeng Zhang, Jianghua Lai. Alcohol dependence mediated by monoamine neurotransmitters in the central nervous system [J]. HEREDITAS, 2014, 36(1): 11-20. |
| [10] | XU Chen-Lu, SUN Xiao-Mei, ZHANG Shou-Gong. Mechanism on differential gene expression and heterosis formation [J]. HEREDITAS, 2013, 35(6): 714-726. |
| [11] | GUO Min-Xia, FU Yong-Fu. Advances on SUMO substrates in Arabidopsis [J]. HEREDITAS, 2013, 35(6): 727-734. |
| [12] | LI Ze-Qin LI Jing-Xiao ZHANG Gen-Fa. Expression regulation of plant ascorbate peroxidase and its tolerance to abiotic stresses [J]. HEREDITAS, 2013, 35(1): 45-54. |
| [13] | WANG Xiang-Meng, WANG Dan-Qiao, HONG Xiao-Yan. Function study advances of Parkinson¢s disease related genes [J]. HEREDITAS, 2010, 32(8): 779-784. |
| [14] | NIE Jing, IAN Chun-Yan, HANG Ling-Qiang. Progress in regulation of activity and stability of ubiquitin protein ligase MDM2 [J]. HEREDITAS, 2009, 31(10): 993-998. |
| [15] | ZHOU Yuan-Fei, XUE Qing-Zhong. Gene Cloning and Structure Characterization of CMS/Rf system in Plants [J]. HEREDITAS, 2005, 27(6): 1007-1012. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||