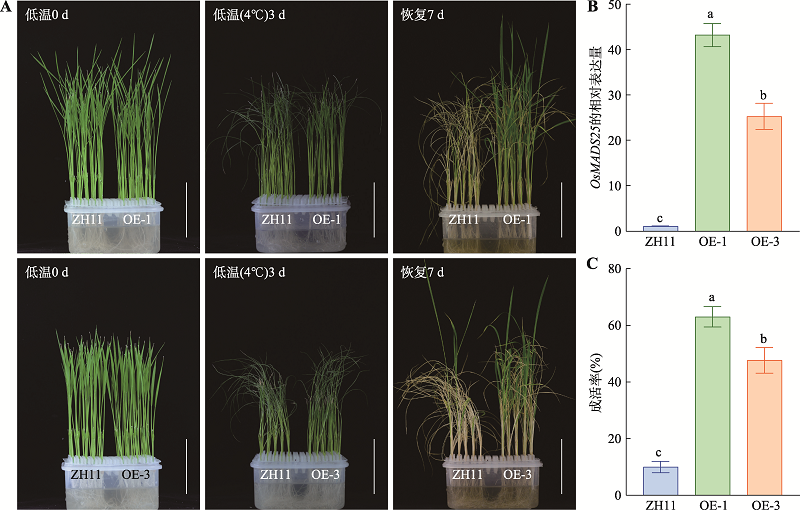
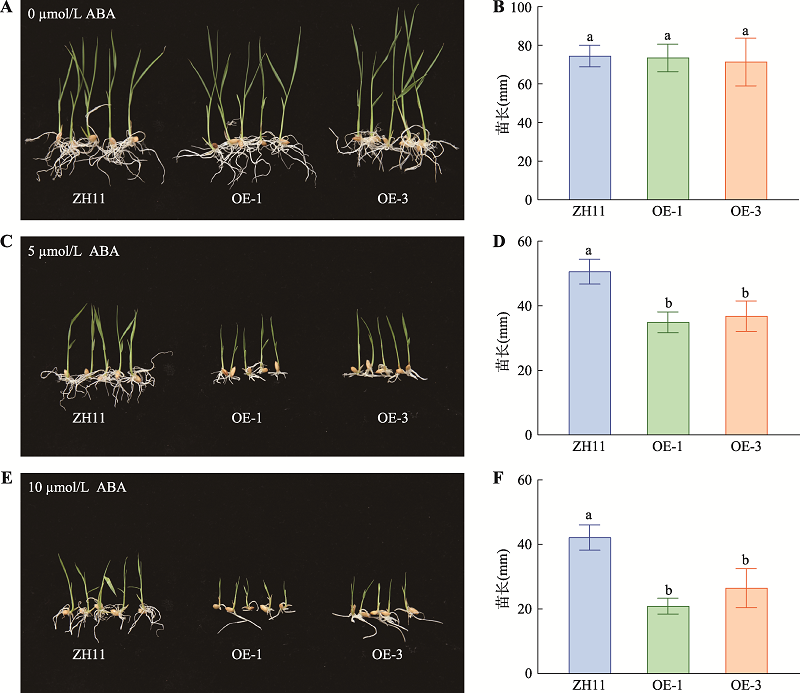
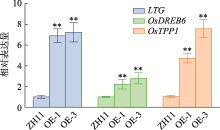
Hereditas(Beijing) ›› 2021, Vol. 43 ›› Issue (11): 1078-1087.doi: 10.16288/j.yczz.21-217
• Orginal Articles • Previous Articles Next Articles
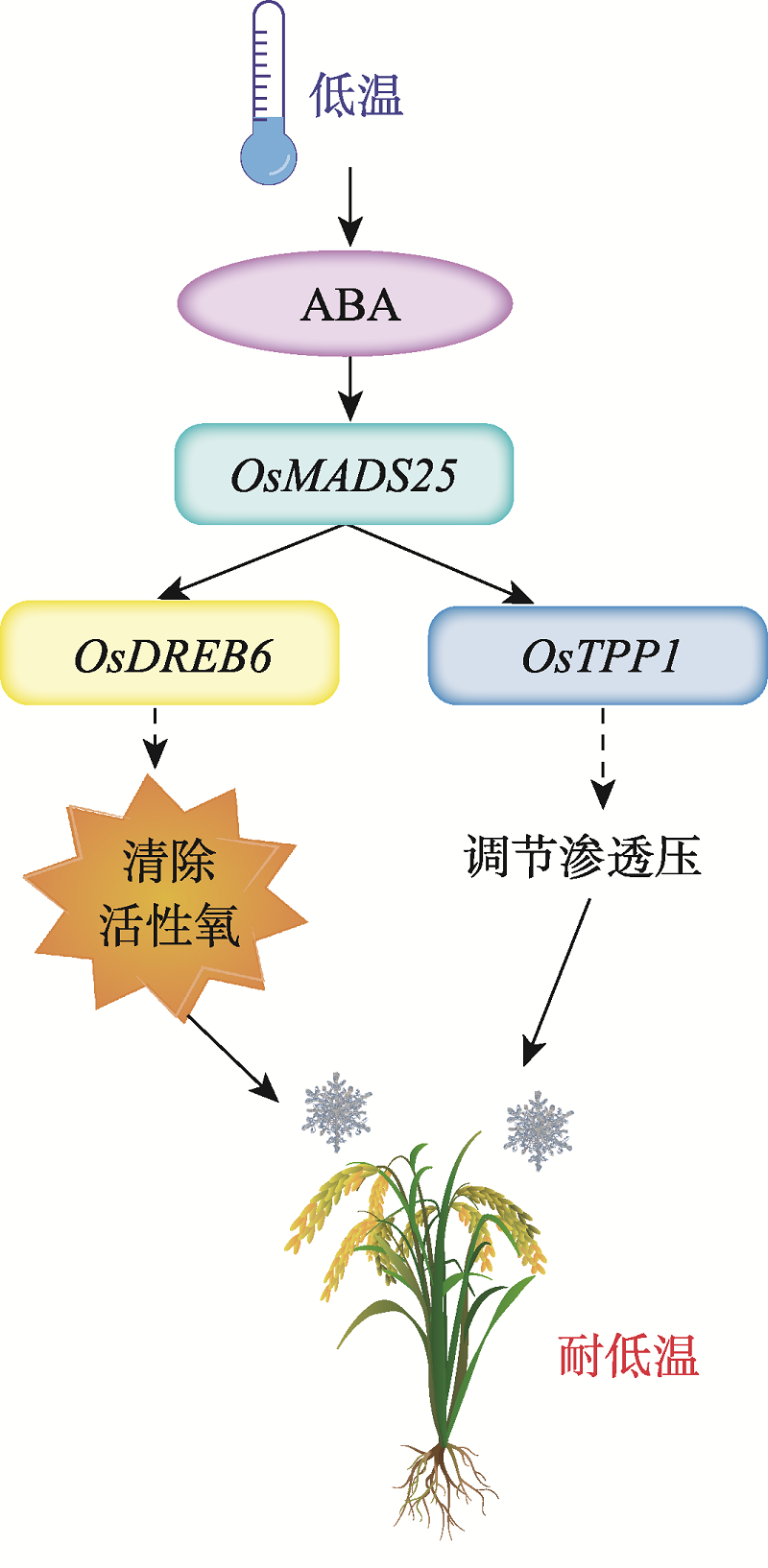
Transcription factor OsMADS25 improves rice tolerance to cold stress
Lingyue Yan1( ), Haojian Zhang1, Yuqing Zheng1, Yunqi Cong1, Citao Liu1, Fan Fan1, Cheng Zheng1, Guilong Yuan2, Gen Pan3, Dingyang Yuan2(
), Haojian Zhang1, Yuqing Zheng1, Yunqi Cong1, Citao Liu1, Fan Fan1, Cheng Zheng1, Guilong Yuan2, Gen Pan3, Dingyang Yuan2( ), Meijuan Duan1(
), Meijuan Duan1( )
)
- 1. Hunan Provincial Key Laboratory of Rice Stress Biology, College of Agriculture, Hunan Agricultural University, Changsha 410128, China
2. State Key Laboratory of Hybrid Rice, Hunan Hybrid Rice Research Center, Changsha 410125, China
3. Institute of Bast Fiber Crops, Chinese Academy of Agricultural Sciences, Changsha 410205, China
-
Received:2021-06-20Revised:2021-08-21Online:2021-11-20Published:2021-10-12 -
Contact:Yuan Dingyang,Duan Meijuan E-mail:yanlingyue0203@163.com;yuandingyang@hhrrc.ac.cn;duanmeijuan@163.com -
Supported by:Supported by the Hunan Science and Technology Major Project No(2018NK1010);the Hunan Science and Technology Talents Support Project No(2019TJ-Q08);the Research Initiation Fund of Hunan Agricultural University No(20154/5407419002);the Open Research Fund of the State Key Laboratory of Hybrid Rice, Hybrid Rice Research Center No(2020KF05);the Hunan Province Natural Science Fund No(2019JJ50714)
Cite this article
Lingyue Yan, Haojian Zhang, Yuqing Zheng, Yunqi Cong, Citao Liu, Fan Fan, Cheng Zheng, Guilong Yuan, Gen Pan, Dingyang Yuan, Meijuan Duan. Transcription factor OsMADS25 improves rice tolerance to cold stress[J]. Hereditas(Beijing), 2021, 43(11): 1078-1087.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
Table 1
Sequences of primers used for qRT-PCR"
| 引物名称 | 引物序列(5ʹ→3ʹ) |
|---|---|
| OsMADS25 | F: GAGGATCGACAACACGATGAA |
| R: GGTGCAGGAGAAGACAATGA | |
| LTG1 | F: CGTGCTGAATGGGCTGATAA |
| R: GTTGAGGTTGATGCCAAGGA | |
| OsEREB6 | F: TCCGGTTTGTTCCCAGTTTAG |
| R: GCCTGGATGTAGTGCATCTG | |
| OsTPP1 | F: CCATCTACATTGGAGACGACAG |
| R: CCTTGGGAACCTGTGAAACTA | |
| Ubiqitin | F: GCTCCGTGGCGGTATCAT |
| R: CGGCAGTTGACAGCCCTAG |
| [1] | Maclean JL, Dawe DC, Hardy B, Hettel GP. Rice almanac: source book for the most important economic activity on earth (3rd edition). Oxon, U.K.: CABI Pub., 2002. |
| [2] |
Gross BL, Zhao ZJ. Archaeological and genetic insights into the origins of domesticated rice. Proc Natl Acad Sci USA, 2014, 111(17):6190-6197.
doi: 10.1073/pnas.1308942110 |
| [3] |
Zhang ZY, Li JJ, Pan YH, Li JL, Zhou L, Shi HL, Zeng YW, Guo HF, Yang SM, Zheng WW, Yu JP, Sun XM, Li GL, Ding YL, Ma L, Shen SQ, Dai LY, Zhang HL, Yang SH, Guo Y, Li ZC. Natural variation in CTB4a enhances rice adaptation to cold habitats. Nat Commun, 2017, 8:14788.
doi: 10.1038/ncomms14788 |
| [4] |
Wang D, Liu JL, Li CG, Kang HX, Wang Y, Tan XQ, Liu MH, Deng YF, Wang ZL, Liu Y, Zhang DY, Xiao YH, Wang GL. Genome-wide association mapping of cold tolerance genes at the seedling stage in rice. Rice, 2016, 9(1):61.
doi: 10.1186/s12284-016-0133-2 pmid: 27848161 |
| [5] |
Schläppi MR, Jackson AK, Eizenga GC, Wang AJ, Chu CC, Shi Y, Shimoyama N, Boykin DL. Assessment of five chilling tolerance traits and GWAS mapping in rice using the USDA Mini-Core collection. Front Plant Sci, 2017, 8:957.
doi: 10.3389/fpls.2017.00957 pmid: 28642772 |
| [6] |
Zhao JL, Zhang SH, Yang TF, Zeng ZC, Huang ZH, Liu Q, Wang XF, Leach J, Leung H, Liu B. Global transcriptional profiling of a cold-tolerant rice variety under moderate cold stress reveals different cold stress response mechanisms. Physiol Plant, 2015, 154(3):381-394.
doi: 10.1111/ppl.2015.154.issue-3 |
| [7] |
Pan YH, Zhang HL, Zhang DL, Li JJ, Xiong HY, Yu JP, Li JL, Rashid MAR, Li GL, Ma XD, Cao GL, Han LZ, Li ZC. Genetic analysis of cold tolerance at the germination and booting stages in rice by association mapping. PLoS One, 2015, 10(3):e0120590.
doi: 10.1371/journal.pone.0120590 |
| [8] |
Zhu YJ, Chen K, Mi XF, Chen TX, Ali J, Ye GY, Xu JL, Li ZK. Identification and fine mapping of a stably expressed QTL for cold tolerance at the booting stage using an interconnected breeding population in rice. PLoS One, 2015, 10(12):e0145704.
doi: 10.1371/journal.pone.0145704 |
| [9] |
Fujino K, Sekiguchi H, Matsuda Y, Sugimoto K, Ono K, Yano M. Molecular identification of a major quantitative trait locus, qLTG3-1, controlling low-temperature germinability in rice. Proc Natl Acad Sci USA, 2008, 105(34):12623-12628.
doi: 10.1073/pnas.0805303105 |
| [10] | Liu CT, Schläppi MR, Mao BG, Wang W, Wang AJ, Chu CC. The bZIP73 transcription factor controls rice cold tolerance at the reproductive stage. Plant Biotechnol J, 2019, 17(9):1834-1849. |
| [11] |
Ma Y, Dai XY, Xu YY, Luo W, Zheng XM, Zeng DL, Pan YJ, Lin XL, Liu HH, Zhang DJ, Xiao J, Guo XY, Xu SJ, Niu YD, Jin JB, Zhang H, Xu X, Li LG, Wang W, Qian Q, Ge S, Chong K. COLD1 confers chilling tolerance in rice. Cell, 2015, 160(6):1209-1221.
doi: 10.1016/j.cell.2015.01.046 |
| [12] |
Zhao JL, Zhang SH, Dong JF, Yang TF, Mao XX, Liu Q, Wang XF, Liu B. A novel functional gene associated with cold tolerance at the seedling stage in rice. Plant Biotechnol J, 2017, 15(9):1141-1148.
doi: 10.1111/pbi.2017.15.issue-9 |
| [13] |
Kim SI, Andaya VC, Tai TH. Cold sensitivity in rice (Oryza sativa L.) is strongly correlated with a naturally occurring I99V mutation in the multifunctional glutathione transferase isoenzyme GSTZ2. Biochem J, 2011, 435(2):373-380.
doi: 10.1042/BJ20101610 |
| [14] |
Mao DH, Xin YY, Tan YJ, Hu XJ, Bai JJ, Liu ZY, Yu YL, Li LY, Peng C, Fan T, Zhu YX, Guo YL, Wang SH, Lu DP, Xing YZ, Yuan LP, Chen CY. Natural variation in the HAN1 gene confers chilling tolerance in rice and allowed adaptation to a temperate climate. Proc Natl Acad Sci USA, 2019, 116(9):3494-3501.
doi: 10.1073/pnas.1819769116 |
| [15] |
Lu GW, Wu FQ, Wu WX, Wang HJ, Zheng XM, Zhang YH, Chen XL, Zhou KN, Jin MN, Cheng ZJ, Li XY, Jiang L, Wang HY, Wan JM. Rice LTG1 is involved in adaptive growth and fitness under low ambient temperature. Plant J, 2014, 78(3):468-480.
doi: 10.1111/tpj.12487 |
| [16] |
Saito K, Hayano-Saito Y, Kuroki M, Sato Y. Map-based cloning of the rice cold tolerance gene Ctb1. Plant Sci, 2010, 179(1-2):97-102.
doi: 10.1016/j.plantsci.2010.04.004 |
| [17] |
Liu CT, Ou SJ, Mao BG, Tang JY, Wang W, Wang HR, Cao SY, Schläppi MR, Zhao BR, Xiao GY, Wang XP, Chu CC. Early selection of bZIP73 facilitated adaptation of japonica rice to cold climates. Nat Commun, 2018, 9(1):3302.
doi: 10.1038/s41467-018-05753-w |
| [18] |
Xiao N, Gao Y, Qian HJ, Gao Q, Wu YY, Zhang DP, Wang Z, Zhang XX, Yu L, Li YH, Pan CH, Liu GQ, Zhou CH, Jiang M, Huang NS, Dai ZY, Liang CZ, Chen Z, Chen JM, Li AH. Identification of genes related to cold tolerance and a functional allele that confers cold tolerance. Plant Physiol, 2018, 177(3):1108-1123.
doi: 10.1104/pp.18.00209 |
| [19] |
Yang SJ, Vanderbeld B, Wan JX, Huang YF. Narrowing down the targets: towards successful genetic engineering of drought-tolerant crops. Mol Plant, 2010, 3(3):469-490.
doi: 10.1093/mp/ssq016 |
| [20] |
Seo E, Lee H, Jeon J, Park H, Kim J, Noh YS, Lee I. Crosstalk between cold response and flowering inArabidopsis is mediated through the flowering-time gene SOC1 and its upstream negative regulator FLC. Plant Cell, 2009, 21(10):3185-3197.
doi: 10.1105/tpc.108.063883 |
| [21] |
Yu LH, Wu J, Zhang ZS, Miao ZQ, Zhao PX, Wang Z, Xiang CB. Arabidopsis MADS-box transcription factor AGL21 acts as environmental surveillance of seed germination by regulating ABI5 expression. Mol Plant, 2017, 10(6):834-845.
doi: 10.1016/j.molp.2017.04.004 |
| [22] |
Wei B, Cai T, Zhang RZ, Li AL, Huo NX, Li S, Gu YQ, Vogel J, Jia JZ, Qi YJ, Mao L. Novel microRNAs uncovered by deep sequencing of small RNA transcriptomes in bread wheat (Triticum aestivum L.) and Brachypodium distachyon(L.) Beauv. Funct Integr Genomics, 2009, 9(4):499-511.
doi: 10.1007/s10142-009-0128-9 |
| [23] | Khong GN, Pati PK, Richaud F, Parizot B, Bidzinski P, Mai CD, Bès M, Bourrié I, Meynard D, Beeckman T, Selvaraj MG, Manabu I, Genga AM, Brugidou C, Nang Do V, Guiderdoni E, Morel JB, Gantet P. OsMADS26 negatively regulates resistance to pathogens and drought tolerance in rice. Plant Physiol, 2015, 169(4):2935-2949. |
| [24] |
Chen C, Begcy K, Liu K, Folsom JJ, Wang Z, Zhang C, Walia H. Heat stress yields a unique MADS box transcription factor in determining seed size and thermal sensitivity. Plant Physiol, 2016, 171(1):606-622.
doi: 10.1104/pp.15.01992 pmid: 26936896 |
| [25] |
Zhang GP, Xu N, Chen HL, Wang GX, Huang JL. OsMADS25 regulates root system development via auxin signalling in rice. Plant J, 2018, 95(6):1004-1022.
doi: 10.1111/tpj.2018.95.issue-6 |
| [26] |
Xu N, Chu YL, Chen HL, Li XX, Wu Q, Jin L, Wang GX, Huang JL. Rice transcription factor OsMADS25 modulates root growth and confers salinity tolerance via the ABA-mediated regulatory pathway and ROS scavenging. PLoS Genet, 2018, 14(10):e1007662.
doi: 10.1371/journal.pgen.1007662 |
| [27] |
Yu CY, Liu YH, Zhang AD, Su S, Yan A, Huang LL, Ali I, Liu Y, Forde BG, Gan YB. MADS-box transcription factor OsMADS25 regulates root development through affection of nitrate accumulation in rice. PLoS One, 2015, 10(8):e0135196.
doi: 10.1371/journal.pone.0135196 |
| [28] | Liu CT, Wang W, Mao BG, Chu CC. Cold stress tolerance in rice: physiological changes, molecular mechanism, and future prospects. Hereditas(Beijing), 2018, 40(3):171-185. |
| 刘次桃, 王威, 毛毕刚, 储成才. 水稻耐低温逆境研究: 分子生理机制及育种展望. 遗传, 2018, 40(3):171-185. | |
| [29] |
Ke YG, Yang ZJ, Yu SW, Li TF, Wu JH, Gao H, Fu YP, Luo LJ. Characterization of OsDREB6 responsive to osmotic and cold stresses in rice. J Plant Biol, 2014, 57(3):150-161.
doi: 10.1007/s12374-013-0480-0 |
| [30] |
Habibur Rahman Pramanik M, Imai R. Functional identification of a trehalose 6-phosphate phosphatase gene that is involved in transient induction of trehalose biosynthesis during chilling stress in rice. Plant Mol Biol, 2005, 58(6):751-762.
doi: 10.1007/s11103-005-7404-4 pmid: 16240171 |
| [31] |
Ge LF, Chao DY, Shi M, Zhu MZ, Gao JP, Lin HX. Overexpression of the trehalose-6-phosphate phosphatase gene OsTPP1 confers stress tolerance in rice and results in the activation of stress responsive genes. Planta, 2008, 228(1):191-201.
doi: 10.1007/s00425-008-0729-x |
| [32] |
Zhang ZY, Li JH, Li F, Liu HH, Yang WS, Chong K, Xu YY. OsMAPK3 phosphorylates OsbHLH002/OsICE1 and inhibits its ubiquitination to activate OsTPP1 and enhances rice chilling tolerance. Dev Cell, 2017, 43(6): 731-743.e5.
doi: 10.1016/j.devcel.2017.11.016 |
| [1] | Gaohua Zhang, Shutao Yu, He Wang, Xuda Wang. Transcriptome profiling of high oleic peanut under low temperatureduring germination [J]. Hereditas(Beijing), 2019, 41(11): 1050-1059. |
| [2] | Citao Liu, Wei Wang, Bigang Mao, Chengcai Chu. Cold stress tolerance in rice: physiological changes, molecular mechanism, and future prospects [J]. Hereditas(Beijing), 2018, 40(3): 171-185. |
| [3] | Feifei Yu,Qi Xie. Ubiquitination modification precisely modulates the ABA signaling pathway in plants [J]. Hereditas(Beijing), 2017, 39(8): 692-706. |
| [4] | SONG Bing, WANG Pi-Wu, FU Yong-Ping, FAN Xu-Hong, XIA Hai-Feng, GAO-Wei, HONG Yang, WANG He, ZHANG Zhuo, MA Jian. Cloning and functional analysis of SCTF-1 encoding a C2H2-type zinc finger protein from soybean [J]. HEREDITAS(Beijing), 2012, 34(6): 749-756. |
| [5] | YAN Qi, BANG Jin-Xia, CUI Liang, XIE Da-Xiang, WANG Zhi-Wei, LI Kui, CHEN Xiao-Han. Molecular cloning of Litopenaeus vannamei TCP-1-eta gene and analysis on its relationship with cold tolerance [J]. HEREDITAS, 2011, 33(2): 168-174. |
| [6] | GAO Guo-Jiang, CHANG Yu-Mei, HAN Qi-Xia, CHE Bing-Jie, LI Meng-Yun, XUE Liang-Xi, LIANG Li-Qun. Screening of microsatellite markers associated with cold tolerance of large yellow croaker (Pseudosciaena crocea R.) [J]. HEREDITAS, 2010, 32(3): 248-253. |
| [7] | GUI Min, ZENG Ya-Wen, DU Juan, PU Xiao-Ying, SHEN Shi-Quan, YANG Shu-Ming, ZHANG Hao. Evaluation of morphological traits on Near-isogenic lines of cold tolerance and molecular validation at booting stage in Japonica Rice [J]. HEREDITAS, 2006, 28(8): 972-976. |
| [8] | ZHONG Ke-Ya, YE Miao-Shui, HU Xin-Wen, GUO Jian-Chun. Role of the Transcription Factors CBF in Plant Cold Tolerance [J]. HEREDITAS, 2006, 28(2): 249-254. |
| [9] | HUANG Xue-Wen, ZHAO Qi, CHEN Dao-Zhen, ZHANG Li-Shan. Mutations in the D-Loop Region of Mitochondrial DNA and the ROS Level in the Tissue of Hepatocellular Carcinoma [J]. HEREDITAS, 2005, 27(1): 14-20. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||