Hereditas(Beijing) ›› 2021, Vol. 43 ›› Issue (12): 1149-1158.doi: 10.16288/j.yczz.21-221
• Research Article • Previous Articles Next Articles
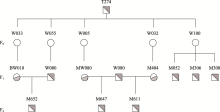
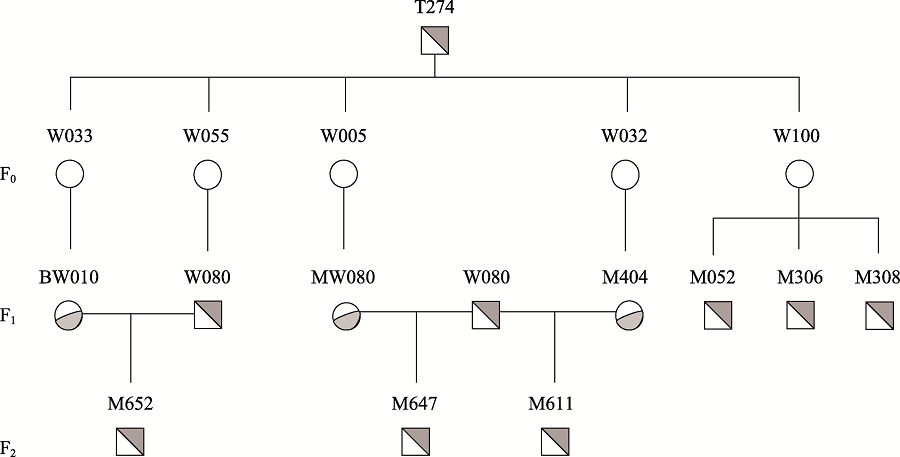
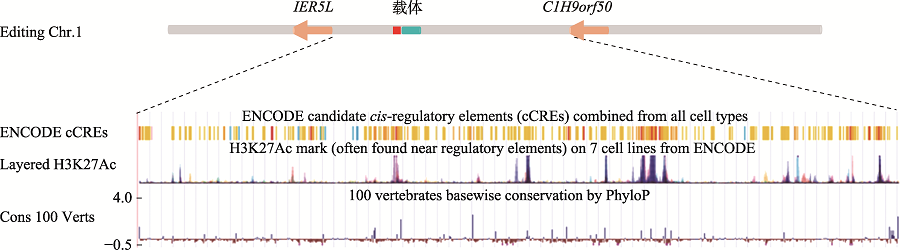
Identification of genomic insertion of dominant-negative GHR mutation transgenes in Wuzhishan pig using whole genome sequencing method
Qiang Wei1,2( ), Yan Ao3, Manman Yang1,2, Tao Chen1,2, Hu Han1,2, Xingju Zhang1,2, Ran Wang1,2, Qiuju Xia1, Fangfang Jiang1,2, Yong Li1,2(
), Yan Ao3, Manman Yang1,2, Tao Chen1,2, Hu Han1,2, Xingju Zhang1,2, Ran Wang1,2, Qiuju Xia1, Fangfang Jiang1,2, Yong Li1,2( )
)
- 1. BGI Institute of Applied Agriculture, BGI-Shenzhen, Shenzhen 518120, China
2. Shenzhen Engineering Laboratory for Genomics-Assisted Animal Breeding, BGI-Shenzhen, Shenzhen 518083, China
3. Key Laboratory of Agricultural Animal Genetics, Breeding and Reproduction of Ministry of Education, Huazhong Agricultural University, Wuhan 430070, China
-
Received:2021-06-24Revised:2021-08-30Online:2021-12-20Published:2021-10-18 -
Contact:Li Yong E-mail:weiqiang@genomics.cn;liyong3@genomics.cn -
Supported by:Supported by the Science and Technology Innovation Strategy Projects of Guangdong Province No(2018B020203002);Guangdong Province Basic and Applied Basic Research Fund No(2019B1515210028)
Cite this article
Qiang Wei, Yan Ao, Manman Yang, Tao Chen, Hu Han, Xingju Zhang, Ran Wang, Qiuju Xia, Fangfang Jiang, Yong Li. Identification of genomic insertion of dominant-negative GHR mutation transgenes in Wuzhishan pig using whole genome sequencing method[J]. Hereditas(Beijing), 2021, 43(12): 1149-1158.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
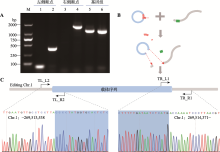
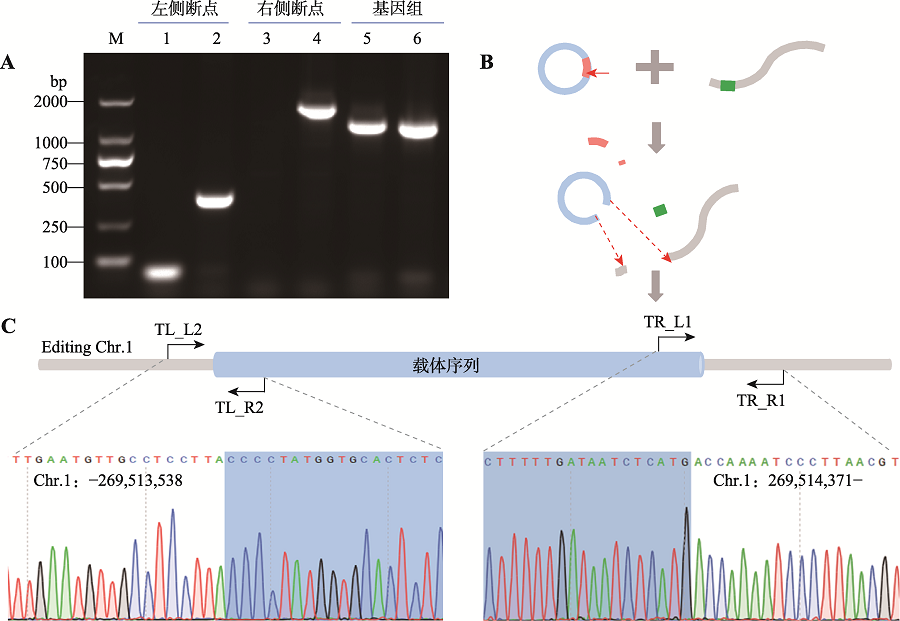
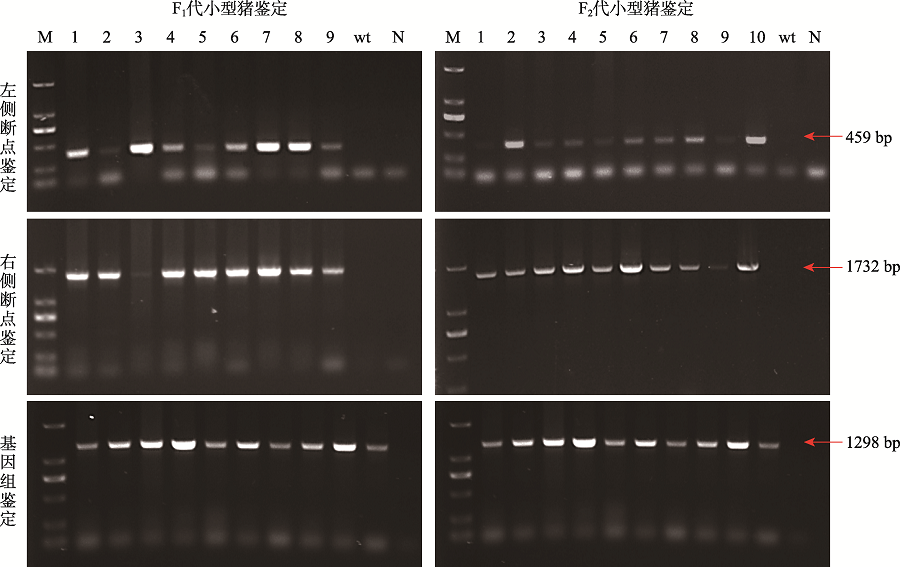
Table 1
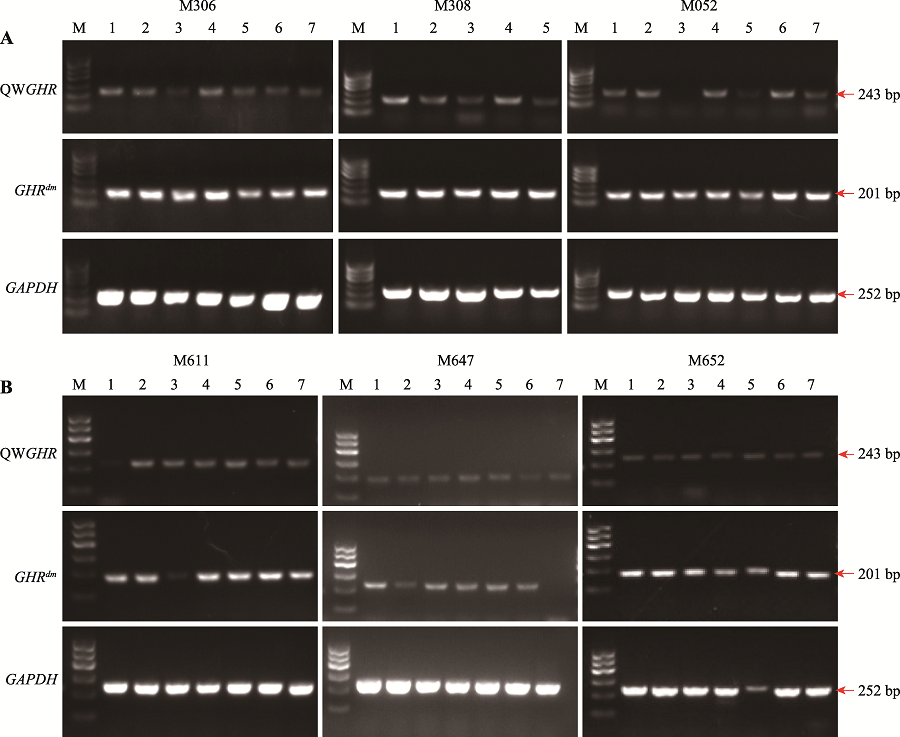
Specific primers used for insert site identification"
| 检测位置 | 引物名称 | 序列(5’→3’) | 产物大小(bp) |
|---|---|---|---|
| 左侧断点 | TL274L1 | GGCTTCAGTTACGGAGGATG | 459 |
| TL274R2 | AACGCCTAACCCTAAGCAGAT | ||
| 右侧断点 | TR274L1 | AGCGTTGGCTACCCGTGATA | 1732 |
| TR274R1 | AGGTCCGTGACAAGACAGCA | ||
| 基因组序列 | TL274L1 | ATGGAAGACCTTGACATTGACC | 1298 |
| TR274R1 | AGGTCCGTGACAAGACAGCA |
| [1] | Maksimenko OG, Deykin AV, Khodarovich YM, Georgiev PG. Use of transgenic animals in biotechnology: prospects and problems. Acta Naturae, 2013,5(1):33-46. |
| [2] | Kong QR, Wu ML, Zhang L, Wang F, Yin Z, Mu YS, Liu ZH. Transgene insertion affects transcription and epigenetic modification of flanking host sequence in transgenic pigs. Cell Mol Biol (Noisy-le-grand), 2011,57 Suppl: OL1505-1512. |
| [3] | Clark J, Whitelaw B. A future for transgenic livestock. Nat Rev Genet, 2003,4(10):825-833. |
| [4] | Niemann H, Petersen B. The production of multi- transgenic pigs: update and perspectives for xenotransplantation. Transgenic Res, 2016,25(3):361-374. |
| [5] | Ruan JX, Xu J, Chen-Tsai RY, Li K. Genome editing in livestock: are we ready for a revolution in animal breeding industry? Transgenic Res, 2017,26(6):715-726. |
| [6] | Friedrich G, Soriano P. Promoter traps in embryonic stem cells: a genetic screen to identify and mutate developmental genes in mice. Genes Dev, 1991,5(9):1513-1523. |
| [7] | Li XP, Yang Y, Bu L, Guo XG, Tang CC, Song J, Fan NN, Zhao BT, Ouyang ZM, Liu ZM, Zhao Y, Yi XL, Quan LQ, Liu SC, Yang ZG, Ouyang HS, Chen YE, Wang Z, Lai LX. Rosa26-targeted swine models for stable gene over- expression and Cre-mediated lineage tracing. Cell Res, 2014,24(4):501-504. |
| [8] | Ruan JX, Li HG, Xu K, Wu TW, Wei JL, Zhou R, Liu ZG, Mu YL, Yang SL, Ouyang HS, Chen-Tsai RY, Li K. Highly efficient CRISPR/Cas9-mediated transgene knockin at the H11 locus in pigs. Sci Rep, 2015,5:14253. |
| [9] | Ma LY, Wang YZ, Wang HT, Hu YQ, Chen JY, Tan T, Hu M, Liu XJ, Zhang R, Xing YM, Zhao YQ, Hu XX, Li N. Screen and verification for transgene integration sites in pigs. Sci Rep, 2018,8(1):7433. |
| [10] | Yang DS, Wang CE, Zhao BT, Li W, Ouyang Z, Liu ZM, Yang HQ, Fan P, O'Neill A, Gu WW, Yi H, Li SH, Lai LX, Li XJ. Expression of Huntington's disease protein results in apoptotic neurons in the brains of cloned transgenic pigs. Hum Mol Genet, 2010,19(20):3983-3994. |
| [11] | Lu YF, Tian C, Deng JX. Research progress in identification technology of genetically modified animals. Prog Biotech, 2000,20(3):60-61. |
| 卢一凡, 田靫, 邓继先. 转基因动物鉴定技术的研究进展. 生物工程进展, 2000,20(3):60-61. | |
| [12] | Fujimoto S, Matsunaga S, Murata M. Mapping of T-DNA and Ac/Ds by TAIL-PCR to analyze chromosomal rearrangements. Methods Mol Biol, 2016,1469:207-216. |
| [13] | Groenen MAM, Archibald AL, Uenishi H, Tuggle CK, Takeuchi Y, Rothschild MF, Rogel-Gaillard C, Park C, Milan D, Megens HJ, Li ST, Larkin DM, Kim H, Frantz LAF, Caccamo M, Ahn H, Aken BL, Anselmo A, Anthon C, Auvil L, Badaoui B, Beattie CW, Bendixen C, Berman D, Blecha F, Blomberg J, Bolund L, Bosse M, Botti S, Bujie Z, Bystrom M, Capitanu B, Carvalho-Silva D, Chardon P, Chen C, Cheng R, Choi SH, Chow W, Clark RC, Clee C, Crooijmans RPMA, Dawson HD, Dehais P, De Sapio F, Dibbits B, Drou N, Du ZQ, Eversole K, Fadista J, Fairley S, Faraut T, Faulkner GJ, Fowler KE, Fredholm M, Fritz E, Gilbert JGR, Giuffra E, Gorodkin J, Griffin DK, Harrow JL, Hayward A, Howe K, Hu ZL, Humphray SJ, Hunt T, Hornshøj H, Jeon JT, Jern P, Jones M, Jurka J, Kanamori H, Kapetanovic R, Kim J, Kim JH, Kim KW, Kim TH, Larson G, Lee K, Lee KT, Leggett R, Lewin HA, Li YR, Liu WS, Loveland JE, Lu Y, Lunney JK, Ma J, Madsen O, Mann K, Matthews L, McLaren S, Morozumi T, Murtaugh MP, Narayan J, Nguyen DT, Ni PX, Oh SJ, Onteru S, Panitz F, Park EW, Park HS, Pascal G, Paudel Y, Perez-Enciso M, Ramirez-Gonzalez R, Reecy JM, Rodriguez-Zas S, Rohrer GA, Rund L, Sang YM, Schachtschneider K, Schraiber JG, Schwartz J, Scobie L, Scott C, Searle S, Servin B, Southey BR, Sperber G, Stadler P, Sweedler JV, Tafer H, Thomsen B, Wali R, Wang J, Wang J, White S, Xu X, Yerle M, Zhang GJ, Zhang JG, Zhang J, Zhao SH, Rogers J, Churcher C, Schook LB. Analyses of pig genomes provide insight into porcine demography and evolution. Nature, 2012,491(7424):393-398. |
| [14] | Meurens F, Summerfield A, Nauwynck H, Saif L, Gerdts V. The pig: a model for human infectious diseases. Trends Microbiol, 2012,20(1):50-57. |
| [15] | Park D, Park SH, Ban YW. Kim YS, Park KC, Kim NS, Kim JK, Choi IK. A bioinformatics approach for identifying transgene insertion sites using whole genome sequencing data. BMC Biotechnol, 2017,17(1):67. |
| [16] | Park D, Kim D, Jang G, Lim J, Shin YJ, Kim J, Seo MS, Park SH, Kim JK, Kwon TH, Choi IY. Efficiency to discovery transgenic loci in GM rice using next generation sequencing whole genome re-sequencing. Genomics Inform, 2015,13(3):81-85. |
| [17] | Niu L, He HL, Zhang YY, Yang J, Zhao QQ, Xing GJ, Zhong XF, Yang XD. Efficient identification of genomic insertions and flanking regions through whole-genome sequencing in three transgenic soybean events. Transgenic Res, 2021,30(1):1-9. |
| [18] | Guo BF, Guo Y, Hong HL, Qiu LJ. Identification of genomic insertion and flanking sequence of G2-EPSPS and GAT transgenes in soybean using whole genome sequencing method. Front Plant Sci, 2016,7:1009. |
| [19] | Ji Y, Abrams N, Zhu W, Salinas E, Yu ZY, Palmer DC, Jailwala P, Franco Z, Roychoudhuri R, Stahlberg E, Gattinoni L, Restifo NP. Identification of the genomic insertion site of Pmel-1 TCR α and β transgenes by next- generation sequencing. PLoS One, 2014,9(5):e96650. |
| [20] | Jacobsen JC, Erdin S, Chiang C, Hanscom C, Handley RR, Barker DD, Stortchevoi A, Blumenthal I, Reid SJ, Snell RG, MacDonald ME, Morton AJ, Ernst C, Gusella JF, Talkowski ME. Potential molecular consequences of transgene integration: the R6/2 mouse example. Sci Rep, 2017,7:41120. |
| [21] | Li FD, Li Y, Liu H, Zhang XJ, Liu CX, Tian K, Bolund L, Dou HW, Yang WX, Yang HM, Staunstrup NH, Du YT. Transgenic Wuzhishan minipigs designed to express a dominant-negative porcine growth hormone receptor display small stature and a perturbed insulin/IGF-1 pathway. Transgenic Res, 2015,24(6):1029-1042. |
| [22] | Liu W, Lu G. Progress in technology of transgenic animal models. Hereditas(Beijing), 2001,23(3):289-291. |
| 刘薇, 卢光. 转基因动物技术的研究进展. 遗传, 2001,23(3):289-291. | |
| [23] | Chandler KJ, Chandler RL, Broeckelmann EM, Hou Y, Southard-Smith EM, Mortlock DP. Relevance of BAC transgene copy number in mice: transgene copy number variation across multiple transgenic lines and correlations with transgene integrity and expression. Mamm Genome, 2007,18(10):693-708. |
| [24] | Le Saux A, Houdebine LM, Jolivet G. Chromosome integration of BAC (bacterial artificial chromosome): evidence of multiple rearrangements. Transgenic Res, 2010,19(5):923-931. |
| [25] | Hu W, Zhu ZY. Research progress of BAC and genetic modification. Prog Biotech, 2001,21(4):3-7. |
| 胡炜, 朱作言. BAC及其转基因研究进展. 生物工程进展, 2001,21(4):3-7. | |
| [26] | Kong QR, Wu ML, Wang ZK, Zhang XM, Li L, Liu XY, Mu YS, Liu ZH. Effect of trichostatin A and 5-Aza-2'- deoxycytidine on transgene reactivation and epigenetic modification in transgenic pig fibroblast cells. Mol Cell Biochem, 2011,355(1-2):157-165. |
| [27] | Yin Z, Kong QR, Zhao ZP, Wu ML, Mu YS, Hu K, Liu ZH. Position effect variegation and epigenetic modification of a transgene in a pig model. Genet Mol Res, 2012,11(1):355-369. |
| [28] | Kong QR, Liu ZH. Inheritance and expression stability of transgene in transgenic animals. Hereditas(Beijing), 2011,33(5):504-511. |
| 孔庆然, 刘忠华. 外源基因在转基因动物中遗传和表达的稳定性. 遗传, 2011,33(5):504-511. | |
| [29] | Laboulaye MA, Duan X, Qiao M, Whitney IE, Sanes JR. Mapping transgene insertion sites reveals complex interactions between mouse transgenes and neighboring endogenous genes. Front Mol Neurosci, 2018,11:385. |
| [30] | Niwa H, Yamamura K, Miyazaki J. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene, 1991,108(2):193-199. |
| [1] | Hongyuan Zheng, Lin Yan, Chao Yang, Yarong Wu, Jingliang Qin, Tongyu Hao, Dajin Yang, Yunchang Guo, Xiaoyan Pei, Tongyan Zhao, Yujun Cui. Population genomics study of Vibrio alginolyticus [J]. Hereditas(Beijing), 2021, 43(4): 350-361. |
| [2] | Wenwen Deng, Caiwu Li, Siyue Zhao, Rengui Li, Yongguo He, Daifu Wu, Shengzhi Yang, Yan Huang, Hemin Zhang, Likou Zou. Whole genome sequencing reveals the distribution of resistance and virulence genes of pathogenic Escherichia coli CCHTP from giant panda [J]. Hereditas(Beijing), 2019, 41(12): 1138-1147. |
| [3] | Wenwen Deng,Mei Long,Shengzhi Yang,Likou Zou. Evolution and the flanking sequences of β-lactamase resistance gene blaOKP [J]. Hereditas(Beijing), 2018, 40(7): 585-592. |
| [4] | Chao Yang, Ruifu Yang, Yujun Cui. Bacterial genome-wide association study: methodologies and applications [J]. Hereditas(Beijing), 2018, 40(1): 57-65. |
| [5] | Caifang Ren, Hongyan Sun, Lizhong Wang, Guomin Zhang, Yixuan Fan, Guangyao Yan, Dan Wang, Feng Wang. Reprogramming mechanism and genetic stability of induced pluripotent stem cells (iPSCs) [J]. HEREDITAS(Beijing), 2014, 36(9): 879-887. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||