Hereditas(Beijing) ›› 2022, Vol. 44 ›› Issue (1): 80-91.doi: 10.16288/j.yczz.21-305
• Orginal Articles • Previous Articles Next Articles
Genome-wide identification and expression analysis of the PEBP genes in Arabidopsis pumila
Yongguang Li1,2( ), Yuhuan Jin1, Li Guo1, Hao Ai2, Ruining Li2, Xianzhong Huang2,3(
), Yuhuan Jin1, Li Guo1, Hao Ai2, Ruining Li2, Xianzhong Huang2,3( )
)
- 1. College of Life Sciences, Shihezi University, Shihezi 832003, China
2. College of Agriculture, Anhui Science and Technology University, Fengyang 233100, China
3. Plant Genomics Laboratory, College of Life Sciences, Shihezi University, Shihezi 832003, China
-
Received:2021-08-24Revised:2021-11-15Online:2022-01-20Published:2022-01-06 -
Contact:Huang Xianzhong E-mail:zongheng1476408@163.com;huangxz@ahstu.edu.cn -
Supported by:Supported by the National Natural Science Foundation of China No(U1303302);the International Science and Technology Cooperation Project of Shihezi University No(GJHZ201806);the Talent Introduction Start-up Fund Project of Anhui Science and Technology University(NXYJ202001)
Cite this article
Yongguang Li, Yuhuan Jin, Li Guo, Hao Ai, Ruining Li, Xianzhong Huang. Genome-wide identification and expression analysis of the PEBP genes in Arabidopsis pumila[J]. Hereditas(Beijing), 2022, 44(1): 80-91.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
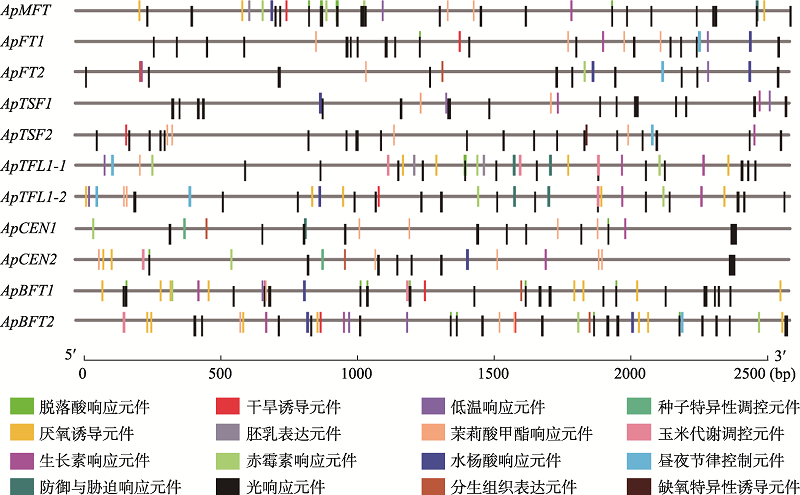
Supplementary Table 1
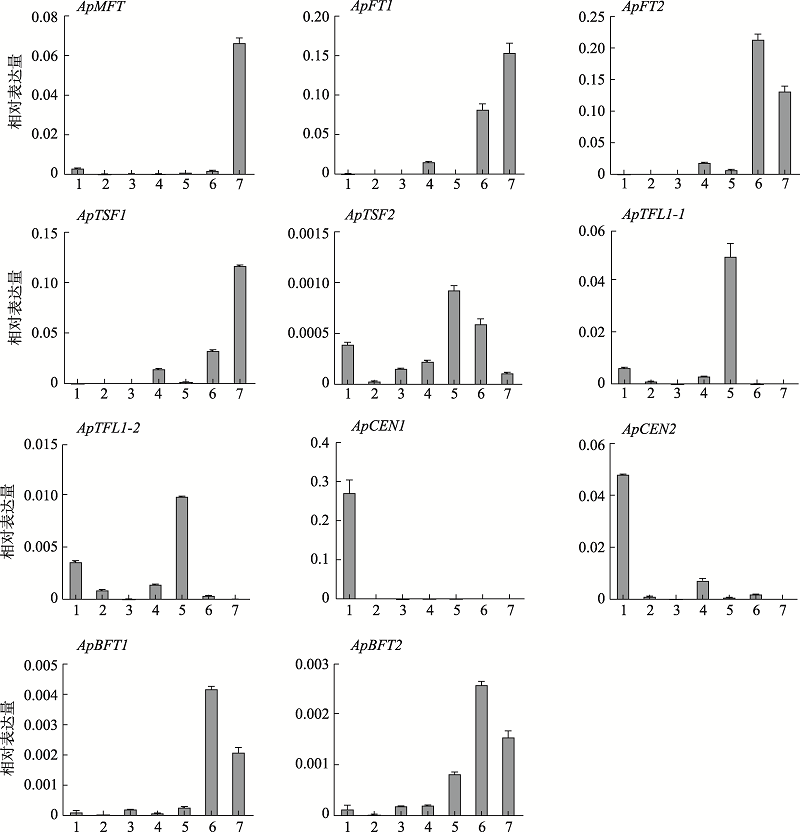
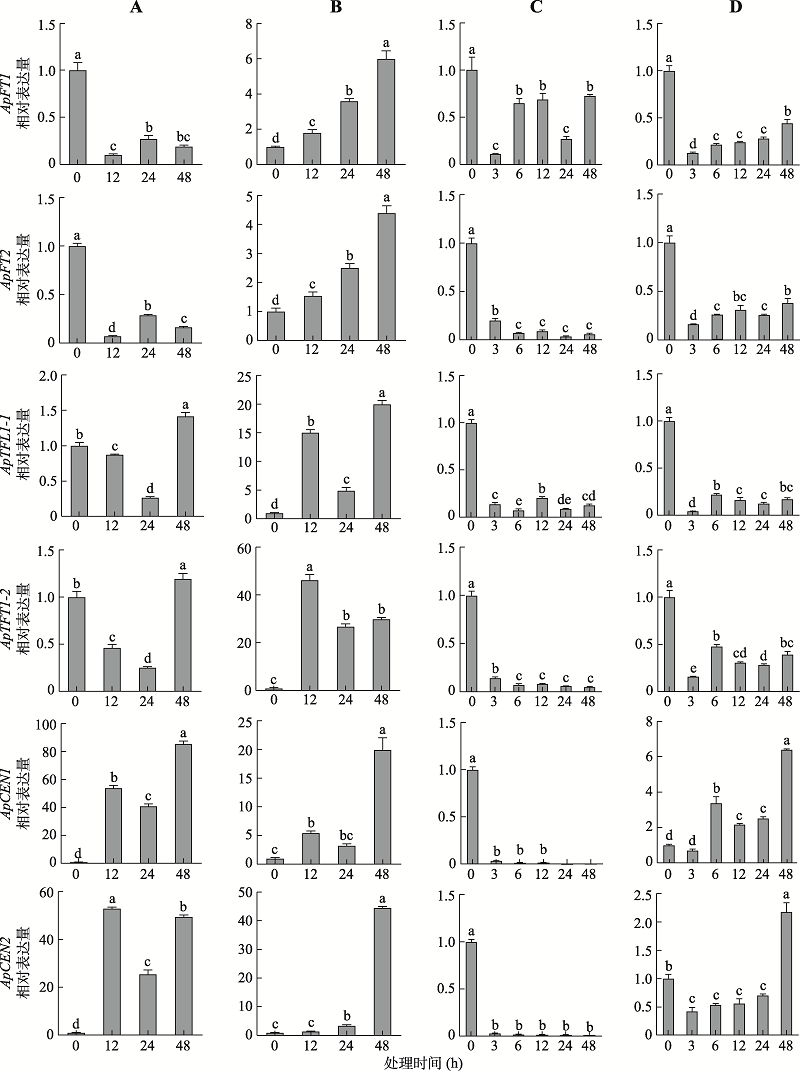
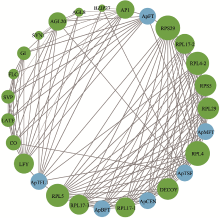
Primer sequences used in quantitative real-time PCR validation"
| 序列 | 引物名称 | 上游引物序列(5′→3′) | 下游引物序列(5′→3′) | 引物用途 |
|---|---|---|---|---|
| 1 | qRT-ApMFT | TGATCCTTTGGTGGTCGGAA | CAGACATATTGGCGGTTGGG | 实时定量PCR |
| 2 | qRT-ApFT1 | TAGTAAGCCGAGTTGTTGGAGA | AGAAGGCCTTAGATCCAAACCA | |
| 3 | qRT-ApFT2 | GCAGAGTTGTTGGAGACGTTC | AGGCCTTAGATCCAAGCCATT | |
| 4 | qRT-ApTSF1 | TAGGGTTACTTATGGCCAACGA | GTAGAAGTTCCTGAGGTCGTCT | |
| 5 | qRT-ApTSF2 | CTGGTTTGTGTTGGTGACTGAT | CTCGTAGCACACCATCTCATTG | |
| 6 | qRT-ApTFL1-1 | GGTGGTGATCTCAGATCCTTCT | GCTCACCACCTCTTTTCCAAAT | |
| 7 | qRT-ApTFL1-2 | TTCCTCTGTCTCCTCCAAACC | TCGTTACGATCCAGTGCAGAT | |
| 8 | qRT-ApBFT1 | CCTCTCTAAACCTCGCGTTG | CTAGGTGCATCAGGGTCCAT | |
| 9 | qRT-ApBFT2 | TCCAAGCGTGACAATGAGAG | CAACGCGAGGTTTAGAGAGG | |
| 10 | qRT-ApCEN1 | CTCGGCCAAACATAGGGATA | CGGTTCTGCTTGAACAACAA | |
| 11 | qRT-ApCEN2 | GCCTCGACCAAACATAGGAA | CGCAAATTCTCGAGTGTTGA | |
| 12 | qRT-ApGAPDH | CTCCCATGTTTGTTGTTGGTGTCA | CTTCCACCTCTCCAGTCCTTCATT | |
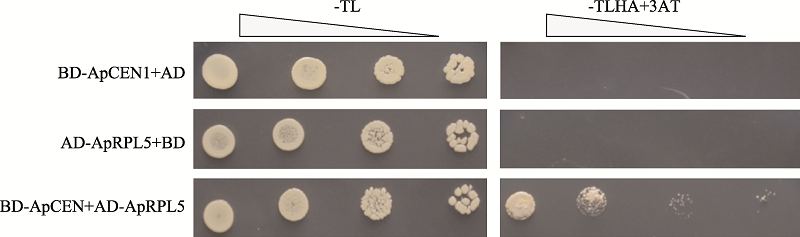
| 13 | BD-ApCEN1 | CGGAATTCATGGCTAGGATTTCCTCAGAC | CGGGATCCTCAACGGCGTCTAGCTGCGGT | 酵母双杂 |
| 14 | AD-ApRPL5 | CGGAATTCATGGCGTCTCCTTCGCTTCT | CGGGATCCTCATCTCTTTGTCTTTCCTTTTCC |
Supplementary Table 2
The information of the PEBP gene family in Arabidopsis pumila"
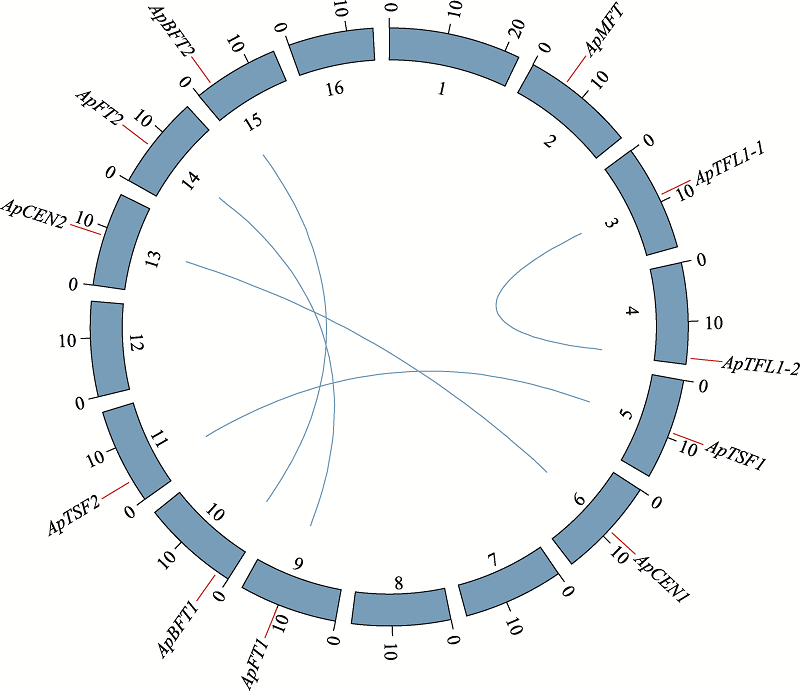
| 基因名称 | 基因ID | 染色体号 | 染色体上 位置(bp) | 氨基酸长度(aa) | 分子量(kDa) | 等电点 | 亚细胞定位 |
|---|---|---|---|---|---|---|---|
| ApBFT1 | evm.model.9__7_CONTIGS_16776222.1453 | Ap11 | 2193322-2194294 | 177 | 20.1 | 8.76 | 细胞核与细胞质 |
| ApBFT2 | evm.model.12__9_CONTIGS_14700693.579 | Ap16 | 2804281-2805282 | 177 | 20.1 | 8.97 | 细胞核与细胞质 |
| ApCEN1 | evm.model.7__7_CONTIGS_17117682.2650 | Ap7 | 8559887-8560979 | 175 | 19.83 | 7.02 | 细胞核与细胞质 |
| ApCEN2 | evm.model.10__7_CONTIGS_16195798.1995 | Ap14 | 8584783-8585893 | 175 | 19.83 | 7.02 | 细胞核与细胞质 |
| ApFT1 | evm.model.11__11_CONTIGS_16209776.2341 | Ap10 | 10023935-10026225 | 175 | 19.72 | 7.76 | 细胞核与细胞质 |
| ApFT2 | evm.model.3__5_CONTIGS_18323972.2179 | Ap15 | 6874127-6876615 | 175 | 19.71 | 7.75 | 细胞核与细胞质 |
| ApMFT | evm.model.1__5_CONTIGS_22361435.1653 | Ap2 | 6368463-6370804 | 173 | 19.12 | 7.93 | 细胞核与细胞质 |
| ApTFL1-1 | evm.model.4__12_CONTIGS_18100624.1250 | Ap4 | 8915426-8916419 | 177 | 20.16 | 9.59 | 细胞核与细胞质 |
| ApTFL1-2 | evm.model.6__5_CONTIGS_17268225.3213 | Ap5 | 16188501-16189686 | 178 | 20.25 | 9.59 | 细胞核与细胞质 |
| ApTSF1 | evm.model.0__8_CONTIGS_29314649.4152 | Ap6 | 9022426-9024468 | 175 | 19.67 | 7.75 | 细胞核与细胞质 |
| ApTSF2 | evm.model.0__8_CONTIGS_29314649.839 | Ap12 | 3822734-3824876 | 177 | 19.94 | 8.52 | 细胞核与细胞质 |
Supplementary Table 3
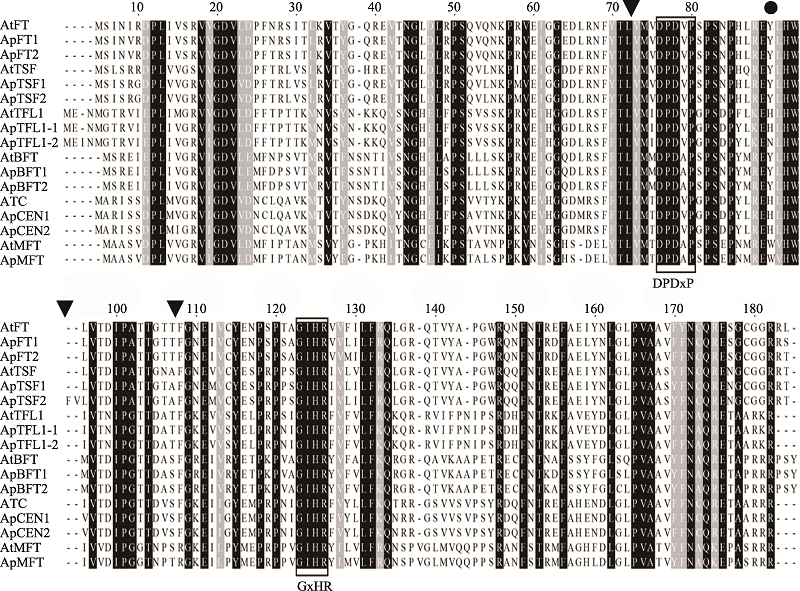
Conserved amino acid sequences of the PEBP proteins in Arabidopsis pumila"
| motif | 长度(aa) | 氨基酸保守序列 |
|---|---|---|
| 1 | 39 | JVTDIPGTTDASFGKEIVCYETPRPPIGIHRFVFVLFRQ |
| 2 | 36 | APGWRDQFNTREFAEEYBLGLPVAAVFFNCQRETAA |
| 3 | 27 | RSFFTLVMVDPDVPSPSBPYLREYLHW |
| 4 | 26 | KZVTNGHELRPSQVLNKPRVEIGGGD |
| 5 | 26 | DPLIVGRVIGDVLDPFTPSVTMRVTY |
| 6 | 10 | IEIRDGLRVI |
| [1] |
Banfield MJ, Barker JJ, Perry AC, Brady RL. Function from structure? The crystal structure of human phosphatidylethanolamine-binding protein suggests a role in membrane signal transduction. Structure, 1998, 6(10):1245-1254.
pmid: 9782050 |
| [2] |
Jin S, Nasim Z, Susila H, Ahn JH. Evolution and functional diversification of FLOWERING LOCUS T/TERMINAL FLOWER 1 family genes in plants. Semin Cell Dev Biol, 2021, 109:20-30.
doi: 10.1016/j.semcdb.2020.05.007 |
| [3] |
Karlgren A, Gyllenstrand N, Källman T, Sundström JF, Moore D, Lascoux M, Lagercrantz U. Evolution of the PEBP gene family in plants: functional diversification in seed plant evolution. Plant Physiol, 2011, 156(4):1967-1977.
doi: 10.1104/pp.111.176206 pmid: 21642442 |
| [4] |
Xi WY, Liu C, Hou XL, Yu H. MOTHER OF FT AND TFL1 regulates seed germination through a negative feedback loop modulating ABA signaling in Arabidopsis. Plant Cell, 2010, 22(6):1733-1748.
doi: 10.1105/tpc.109.073072 |
| [5] |
Higuchi Y. Florigen and anti-florigen: flowering regulation in horticultural crops. Breed Sci, 2018, 68(1):109-118.
doi: 10.1270/jsbbs.17084 |
| [6] |
Eshed Y, Lippman ZB. Revolutions in agriculture chart a course for targeted breeding of old and new crops. Science, 2019, 366(6466): eaax0025.
doi: 10.1126/science.aax0025 |
| [7] |
Kinoshita T, Ono N, Hayashi Y, Morimoto S, Nakamura S, Soda M, Kato Y, Ohnishi M, Nakano T, Inoue SI, Shimazaki KI. FLOWERING LOCUS T regulates stomatal opening. Curr Biol, 2011, 21(14):1232-1238.
doi: 10.1016/j.cub.2011.06.025 |
| [8] |
Krieger U, Lippman ZB, Zamir D. The flowering gene SINGLE FLOWER TRUSS drives heterosis for yield in tomato. Nat Genet, 2010, 42(5):459-463.
doi: 10.1038/ng.550 pmid: 20348958 |
| [9] |
Navarro C, Abelenda JA, Cruz-Oró E, Cuéllar CA, Tamaki S, Silva J, Shimamoto K, Prat S. Control of flowering and storage organ formation in potato by FLOWERING LOCUS T. Nature, 2011, 478(7367):119-122.
doi: 10.1038/nature10431 |
| [10] |
Lee R, Baldwin S, Kenel F, McCallum J, Macknight R,. FLOWERING LOCUS T genes control onion bulb formation and flowering. Nat Commun, 2013, 4:2884.
doi: 10.1038/ncomms3884 |
| [11] |
Abelenda JA, Bergonzi S, Oortwijn M, Sonnewald S, Du MR, Visser RGF, Sonnewald U, Bachem CWB. Source- sink regulation is mediated by interaction of an FT homolog with a SWEET protein in potato. Curr Biol, 2019, 29(7): 1178-1186.e6.
doi: S0960-9822(19)30157-5 pmid: 30905604 |
| [12] |
Bradley D, Ratcliffe O, Vincent C, Carpenter R, Coen E. Inflorescence commitment and architecture in Arabidopsis. Science, 1997, 275(5296):80-83.
pmid: 8974397 |
| [13] |
Zhang B, Li CX, Li Y, Yu H. Mobile TERMINAL FLOWER1 determines seed size in Arabidopsis. Nat Plants, 2020, 6(9):1146-1157.
doi: 10.1038/s41477-020-0749-5 pmid: 32839516 |
| [14] |
Corbesier L, Vincent C, Jang S, Fornara F, Fan QZ, Searle I, Giakountis A, Farrona S, Gissot L, Turnbull C, Coupland G. FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science, 2007, 316(5827):1030-1033.
pmid: 17446353 |
| [15] |
Tamaki S, Matsuo S, Wong HL, Yokoi S, Shimamoto K. Hd3a protein is a mobile flowering signal in rice. Science, 2007, 316(5827):1033-1036.
doi: 10.1126/science.1141753 |
| [16] |
Abe M, Kobayashi Y, Yamamoto S, Daimon Y, Yamaguchi A, Ikeda Y, Ichinoki H, Notaguchi M, Goto K, Araki T. FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science, 2005, 309(5737):1052-1056.
doi: 10.1126/science.1115983 |
| [17] |
Wigge PA, Kim MC, Jaeger KE, Busch W, Schmid M, Lohmann JU, Weigel D. Integration of spatial and temporal information during floral induction in Arabidopsis. Science, 2005, 309(5737):1056-1059.
doi: 10.1126/science.1114358 |
| [18] |
Hanano S, Goto K. Arabidopsis TERMINAL FLOWER1 is involved in the regulation of flowering time and inflorescence development through transcriptional repression. Plant Cell, 2011, 23(9):3172-3184.
doi: 10.1105/tpc.111.088641 |
| [19] |
Collani S, Neumann M, Yant L, Schmid M. FT modulates genome-wide DNA-binding of the bZIP transcription factor FD. Plant Physiol, 2019, 180(1):367-380.
doi: 10.1104/pp.18.01505 pmid: 30770462 |
| [20] |
Romera-Branchat M, Severing E, Pocard C, Ohr H, Vincent C, Née G, Martinez-Gallegos R, Jang S, Andrés F, Madrigal P, Coupland G. Functional divergence of the Arabidopsis florigen-interacting bZIP transcription factors FD and FDP. Cell Rep, 2020, 31(9):107717.
doi: S2211-1247(20)30694-X pmid: 32492426 |
| [21] |
Zhu Y, Klasfeld S, Jeong CW, Jin R, Goto K, Yamaguchi N, Wagner D. TERMINAL FLOWER 1-FD complex target genes and competition with FLOWERING LOCUS T. Nat Commun, 2020, 11(1):5118.
doi: 10.1038/s41467-020-18782-1 |
| [22] | Mao ZM. Early spring ephemeral plant area characteristics. Arid Zone Res, 1992, 9(1):11-12. |
| 毛祖美. 早春短命植物区系特点. 干旱区研究, 1992, 9(1):11-12. | |
| [23] | Yuan SF, Tang HP. Research advances in the eco-physiological characteristics of ephemerals adaptation to habitats. Acta Prataculturae Sinica, 2010, 19(1):240-247. |
| 袁素芬, 唐海萍. 短命植物生理生态特性对生境的适应性研究进展. 草业学报, 2010, 19(1):240-247. | |
| [24] | Mao ZM, An ZX, Zhou GL, Yang CY, Han YL, Li XY, Zhang YF. Flora of Xinjiang (vol. 2, part 2). Urumqi: Xinjiang Science and Technology Health Press, 1995, 145-146. |
| 毛祖美, 安争夕, 周桂玲, 杨昌友, 韩英兰, 李学禹, 张彦福. 《新疆植物志》(第二卷第二分册). 乌鲁木齐: 新疆科技卫生出版社, 1995, 145-146. | |
| [25] |
Huang XZ, Yang LF, Jin YH, Lin J, Liu F. Generation, annotation, and analysis of a large-scale expressed sequence tag library fromArabidopsis pumila to explore salt-responsive genes. Front Plant Sci, 2017, 8:955.
doi: 10.3389/fpls.2017.00955 |
| [26] |
Jin YH, Liu F, Huang W, Sun Q, Huang XZ. Identification of reliable reference genes for qRT-PCR in the ephemeral plant Arabidopsis pumila based on full-length transcriptome data. Sci Rep, 2019, 9(1):8408.
doi: 10.1038/s41598-019-44849-1 |
| [27] | Li XC, Kang KC, Huang XZ, Fan YB, Song MM, Huang YJ, Ding JJ. Genome-wide identification, phylogenetic analysis and expression profiling of the MKK gene family in Arabidopsis pumila. Hereditas(Beijing), 2020, 42(4):403-421. |
| 李晓翠, 康凯程, 黄先忠, 范永斌, 宋苗苗, 黄韵杰, 丁佳佳. 小拟南芥MKK基因家族全基因组鉴定及进化和表达分析. 遗传, 2020, 42(4):403-421. | |
| [28] | Thompson JD, Gibson TJ, Higgins DG. Multiple sequence alignment using ClustalW and Clustal X. Curr Protoc Bioinformatics, 2002, Chapter 2: Unit 2.3. |
| [29] |
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol, 2013, 30(12):2725-2729.
doi: 10.1093/molbev/mst197 |
| [30] |
Wang YP, Tang HB, DeBarry JD, Tan X, Li JP, Wang XY, Lee TH, Jin HZ, Marler B, Guo H, Kissinger JC, Paterson AH. MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res, 2012, 40(7):e49.
doi: 10.1093/nar/gkr1293 |
| [31] |
Chen CJ, Chen H, Zhang Y, Thomas HR, Frank MH, He YH, Xia R. TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol Plant, 2020, 13(8):1194-1202.
doi: 10.1016/j.molp.2020.06.009 |
| [32] |
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2 (-Delta Delta C(T)) method. Methods, 2001, 25(4):402-408.
pmid: 11846609 |
| [33] |
Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res, 2003, 13(11):2498-2504.
pmid: 14597658 |
| [34] |
Michaels SD, Amasino RM. FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell, 1999, 11(5):949-956.
pmid: 10330478 |
| [35] |
Hartmann U, Höhmann S, Nettesheim K, Wisman E, Saedler H, Huijser P. Molecular cloning of SVP: a negative regulator of the floral transition in Arabidopsis. Plant J, 2000, 21(4):351-360.
pmid: 10758486 |
| [36] |
Weigel D, Alvarez J, Smyth DR, Yanofsky MF, Meyerowitz EM. LEAFY controls floral meristem identity in Arabidopsis. Cell, 1992, 69(5):843-859.
pmid: 1350515 |
| [37] |
Fowler S, Lee K, Onouchi H, Samach A, Richardson K, Morris B, Coupland G, Putterill J. GIGANTEA: a circadian clock-controlled gene that regulates photoperiodic flowering in Arabidopsis and encodes a protein with several possible membrane-spanning domains. EMBO J, 1999, 18(17):4679-4688.
pmid: 10469647 |
| [38] | Peng FY, Hu ZQ, Yang RC. Genome-wide comparative analysis of flowering-related genes in arabidopsis, wheat, and barley. Int J Plant Genomics, 2015, 2015:874361. |
| [39] |
Nakagawa M, Shimamoto K, Kyozuka J. Overexpression of RCN1 and RCN2, rice TERMINAL FLOWER 1/CENTRORADIALIS homologs, confers delay of phase transition and altered panicle morphology in rice. Plant J, 2002, 29(6):743-750.
pmid: 12148532 |
| [40] |
Liu YY, Yang KZ, Wei XX, Wang XQ. Revisiting the phosphatidylethanolamine-binding protein (PEBP) gene family reveals crypticFLOWERING LOCUS T gene homologs in gymnosperms and sheds new light on functional evolution. New Phytol, 2016, 212(3):730-744.
doi: 10.1111/nph.14066 pmid: 27375201 |
| [41] |
Zhang TC, Qiao Q, Novikova PY, Wang Q, Yue JP, Guan YL, Ming SP, Liu TM, De J, Liu YX, Al-Shehbaz IA, Sun H, Van Montagu M, Huang JL, Van de Peer Y, Qiong L,. Genome of Crucihimalaya himalaica, a close relative of Arabidopsis, shows ecological adaptation to high altitude. Proc Natl Acad Sci USA, 2019, 116(14):7137-7146.
doi: 10.1073/pnas.1817580116 |
| [42] |
Mager WH. Control of ribosomal protein gene expression. Biochim Biophys Acta, 1988, 949(1):1-15.
pmid: 3275463 |
| [43] |
Van Minnebruggen A, Neyt P, De Groeve S, Coussens G, Ponce MR, Micol JL, Van Lijsebettens M. The ang3 mutation identified the ribosomal protein gene RPL5B with a role in cell expansion during organ growth. Physiol Plant, 2010, 138(1):91-101.
doi: 10.1111/j.1399-3054.2009.01301.x pmid: 19878482 |
| [44] |
Danilevskaya ON, Meng X, Hou ZL, Ananiev EV, Simmons CR. A genomic and expression compendium of the expanded PEBP gene family from maize. Plant Physiol, 2008, 146(1):250-264.
doi: 10.1104/pp.107.109538 pmid: 17993543 |
| [45] |
Kobayashi Y, Kaya H, Goto K, Iwabuchi M, Araki T. A pair of related genes with antagonistic roles in mediating flowering signals. Science, 1999, 286(5446):1960-1962.
pmid: 10583960 |
| [46] |
Bradley D, Carpenter R, Copsey L, Vincent C, Rothstein S, Coen E. Control of inflorescence architecture in Antirrhinum. Nature, 1996, 379(6568):791-797.
doi: 10.1038/379791a0 |
| [47] |
Conti L, Bradley D. TERMINAL FLOWER1 is a mobile signal controlling Arabidopsis architecture. Plant Cell, 2007, 19(3):767-778.
doi: 10.1105/tpc.106.049767 |
| [48] |
Chung KS, Yoo SY, Yoo SJ, Lee JS, Ahn JH. BROTHER OF FT AND TFL1 (BFT), a member of the FT/TFL1 family, shows distinct pattern of expression during the vegetative growth of Arabidopsis. Plant Signal Behav, 2010, 5(9):1102-1104.
doi: 10.4161/psb.5.9.12415 |
| [1] | Qianbin Zhu, Zhicheng Gan, Xiaocui Li, Yingjie Zhang, Heming Zhao, Xianzhong Huang. Genome-wide identification, phylogenetic and expression of MAPKKK gene family in Arabidopsis pumila [J]. Hereditas(Beijing), 2022, 44(11): 1044-1055. |
| [2] | Xiaocui Li, Kaicheng Kang, Xianzhong Huang, Yongbin Fan, Miaomiao Song, Yunjie Huang, Jiajia Ding. Genome-wide identification, phylogenetic analysis and expression profiling of the MKK gene family in Arabidopsis pumila [J]. Hereditas(Beijing), 2020, 42(4): 403-421. |
| [3] | DANG Yong-Hui, YAN Chun-Xia, CHEN Teng. Epigenetic mechanisms of the influence of maternal care on off spring development [J]. HEREDITAS, 2011, 33(9): 919-924. |
| [4] | SHU Li-Ping, XU Zhuang, JU Cui-Xia, LI Qiu-Chi. Plant stress-inducible promoters and their function [J]. HEREDITAS, 2010, 32(3): 229-234. |
| [5] | SUN Chang-Hui, DENG Xiao-Jian, FANG Jun, CHU Cheng-Cai. An overview of flowering transition in higher plants [J]. HEREDITAS, 2007, 29(10): 1182-1182―1190. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||