Hereditas(Beijing) ›› 2023, Vol. 45 ›› Issue (10): 922-932.doi: 10.16288/j.yczz.23-120
• Research Article • Previous Articles Next Articles
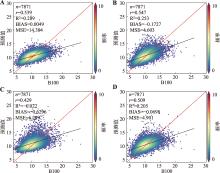
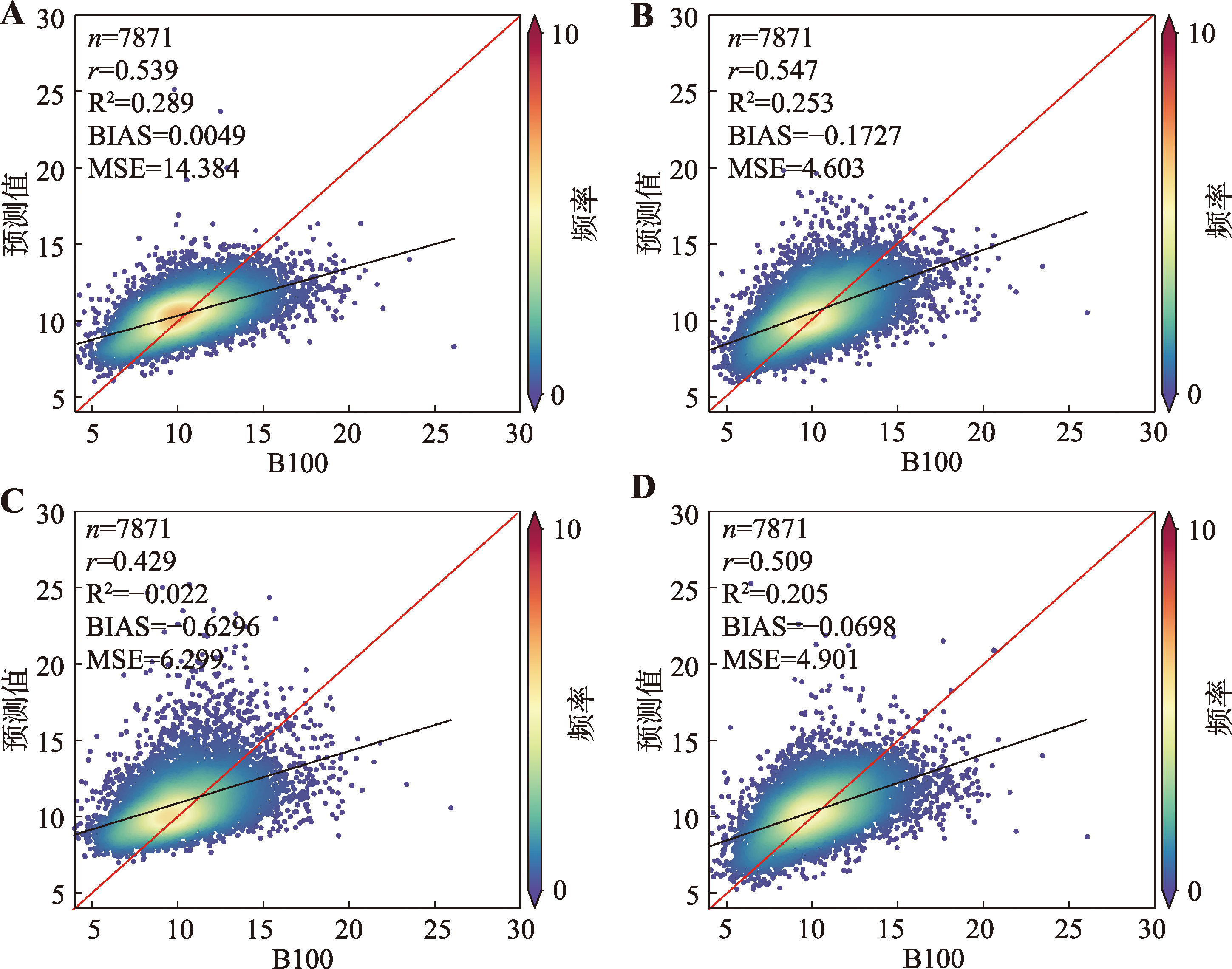
Genomic prediction of pig growth traits based on machine learning
Dong Chen1,2,3( ), Shujie Wang1,2,3, Zhenjian Zhao1,2,3, Xiang Ji1,2,3, Qi Shen1,2,3, Yang Yu1,2,3, Shengdi Cui1,2,3, Junge Wang1,2,3, Ziyang Chen1,2,3, Jinyong Wang4, Zongyi Guo4, Pingxian Wu4, Guoqing Tang1,2,3(
), Shujie Wang1,2,3, Zhenjian Zhao1,2,3, Xiang Ji1,2,3, Qi Shen1,2,3, Yang Yu1,2,3, Shengdi Cui1,2,3, Junge Wang1,2,3, Ziyang Chen1,2,3, Jinyong Wang4, Zongyi Guo4, Pingxian Wu4, Guoqing Tang1,2,3( )
)
- 1. Key Laboratory of Livestock and Poultry Multi-omics, Ministry of Agriculture and Rural Affairs, College of Animal Science and Technology, Sichuan Agricultural University, Chengdu 611130, China
2. Farm Animal Genetic Resources Exploration and Innovation Key Laboratory of Sichuan Province, Sichuan Agricultural University, Chengdu 611130, China
3. State Key Laboratory of Swine and Poultry Breeding Industry, College of Animal Science and Technology, Sichuan Agricultural University, Chengdu 611130, China
4. National Center of Technology Innovation for Pigs, Chongqing 402460, China
-
Received:2023-04-26Revised:2023-08-14Online:2023-10-20Published:2023-08-16 -
Contact:Guoqing Tang E-mail:1123278154@qq.com;tyq003@163.com -
Supported by:Strategic Priority Research Program of the National Center of Technology Innovation for Pigs(NCTIP-XD/B01);Sichuan Science and Technology Program(2020YFN0024);Sichuan Science and Technology Program(2021ZDZX0008);Sichuan Science and Technology Program(2021YFYZ0030);Sichuan Innovation Team of Pig(sccxtd-2022-08)
Cite this article
Dong Chen, Shujie Wang, Zhenjian Zhao, Xiang Ji, Qi Shen, Yang Yu, Shengdi Cui, Junge Wang, Ziyang Chen, Jinyong Wang, Zongyi Guo, Pingxian Wu, Guoqing Tang. Genomic prediction of pig growth traits based on machine learning[J]. Hereditas(Beijing), 2023, 45(10): 922-932.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
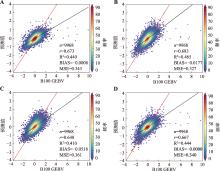
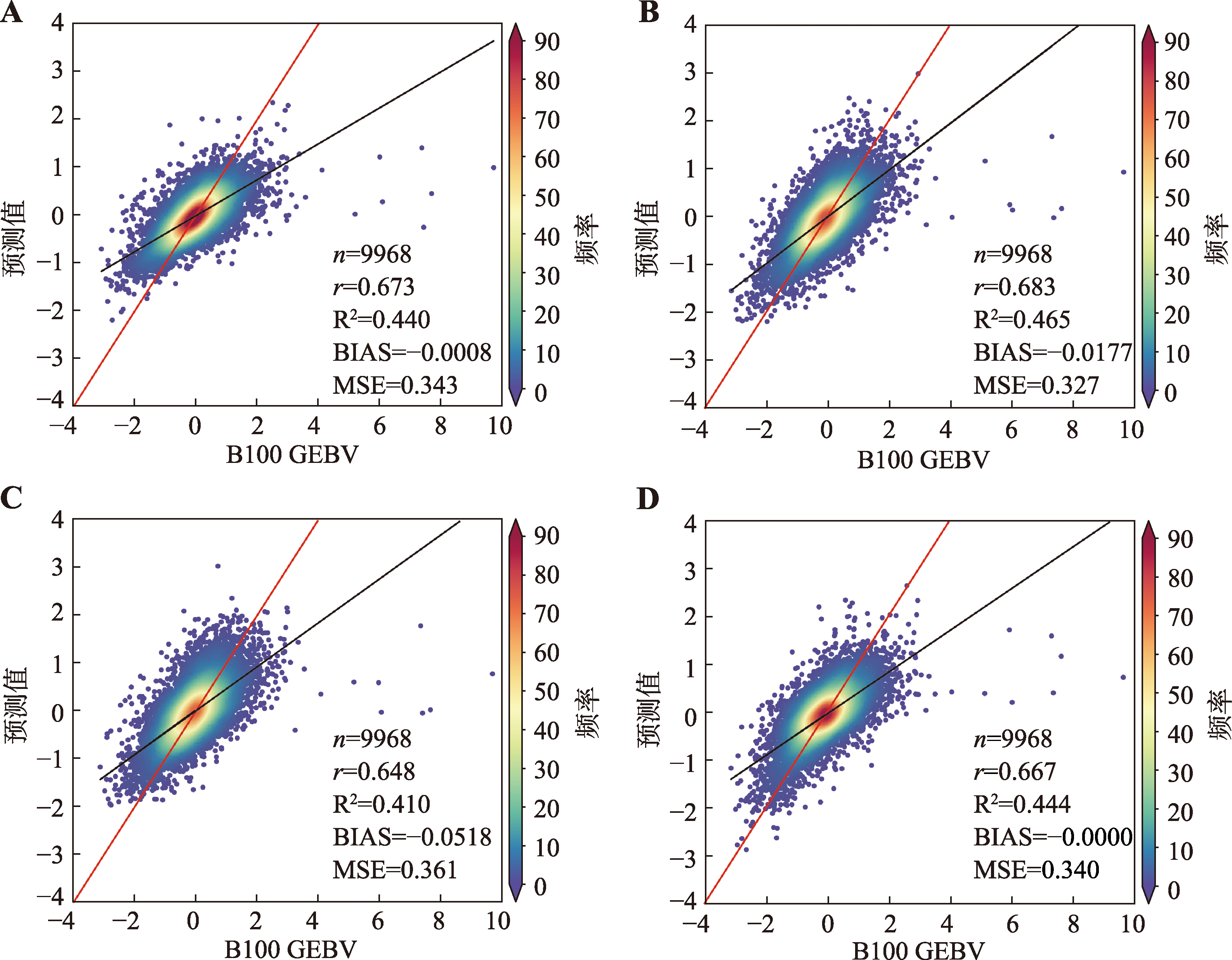
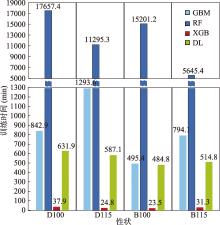
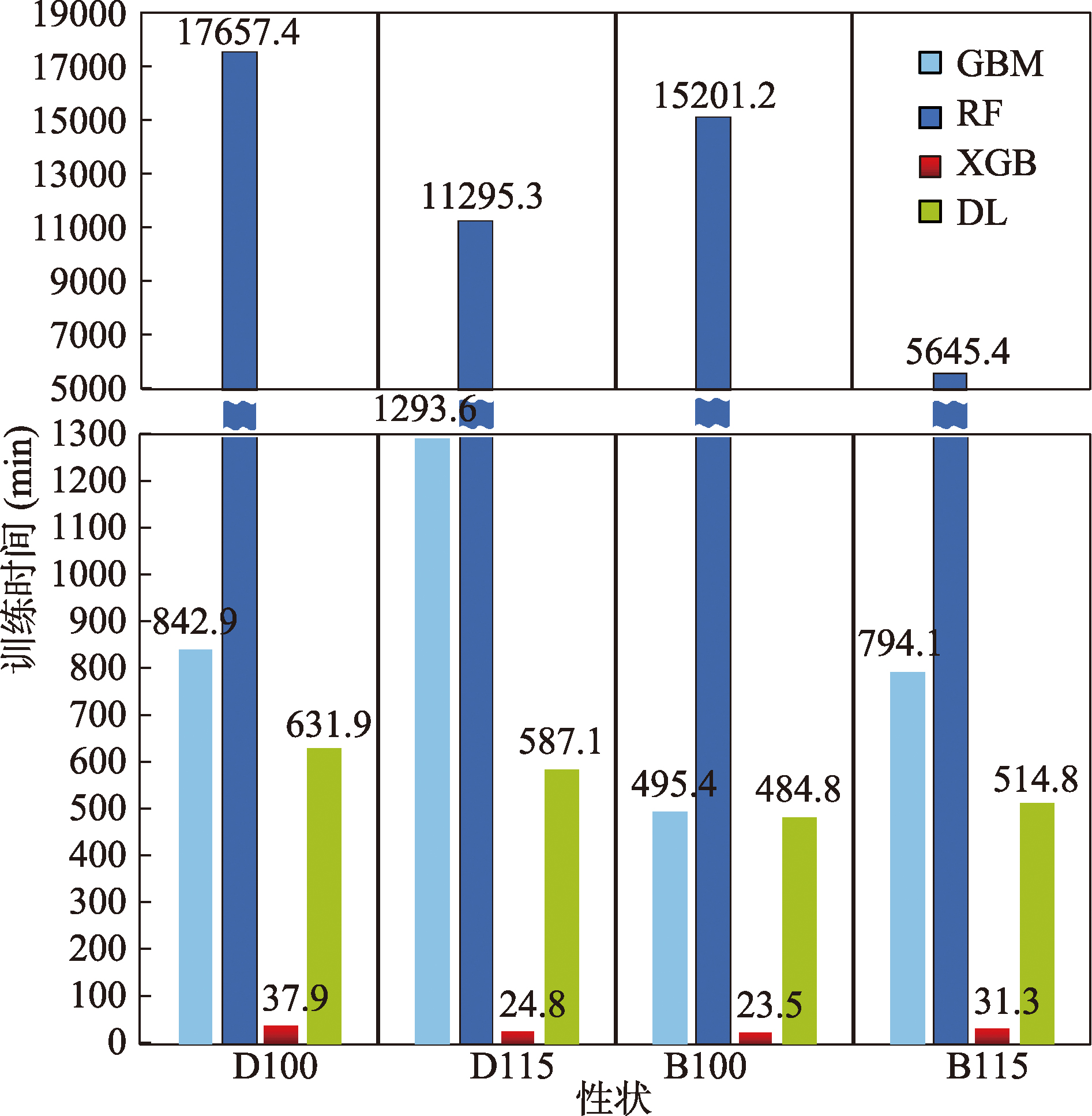
Table 4
Hyperparameter table for XGB model"
| 参数名 | GEBV估计参数 | 表型预测参数 |
|---|---|---|
| distribution | gaussian | gaussian |
| categorical_encoding | OneHotInternal | OneHotInternal |
| ntrees | 111 | 71 |
| max_depth | 5 | 10 |
| learn_rate | 0.3 | 5 |
| col_sample_rate | 0.8 | 0.8 |
| colsample_bylevel | 0.8 | 0.8 |
| tree_method | exact | exact |
| booster | gbtree | dart |
| reg_lambda | 0.001 | 100 |
| reg_alpha | 0.1 | 1 |
| dmatrix_type | sparse | sparse |
| backend | cpu | cpu |
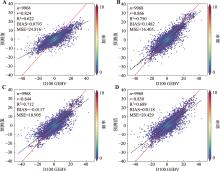
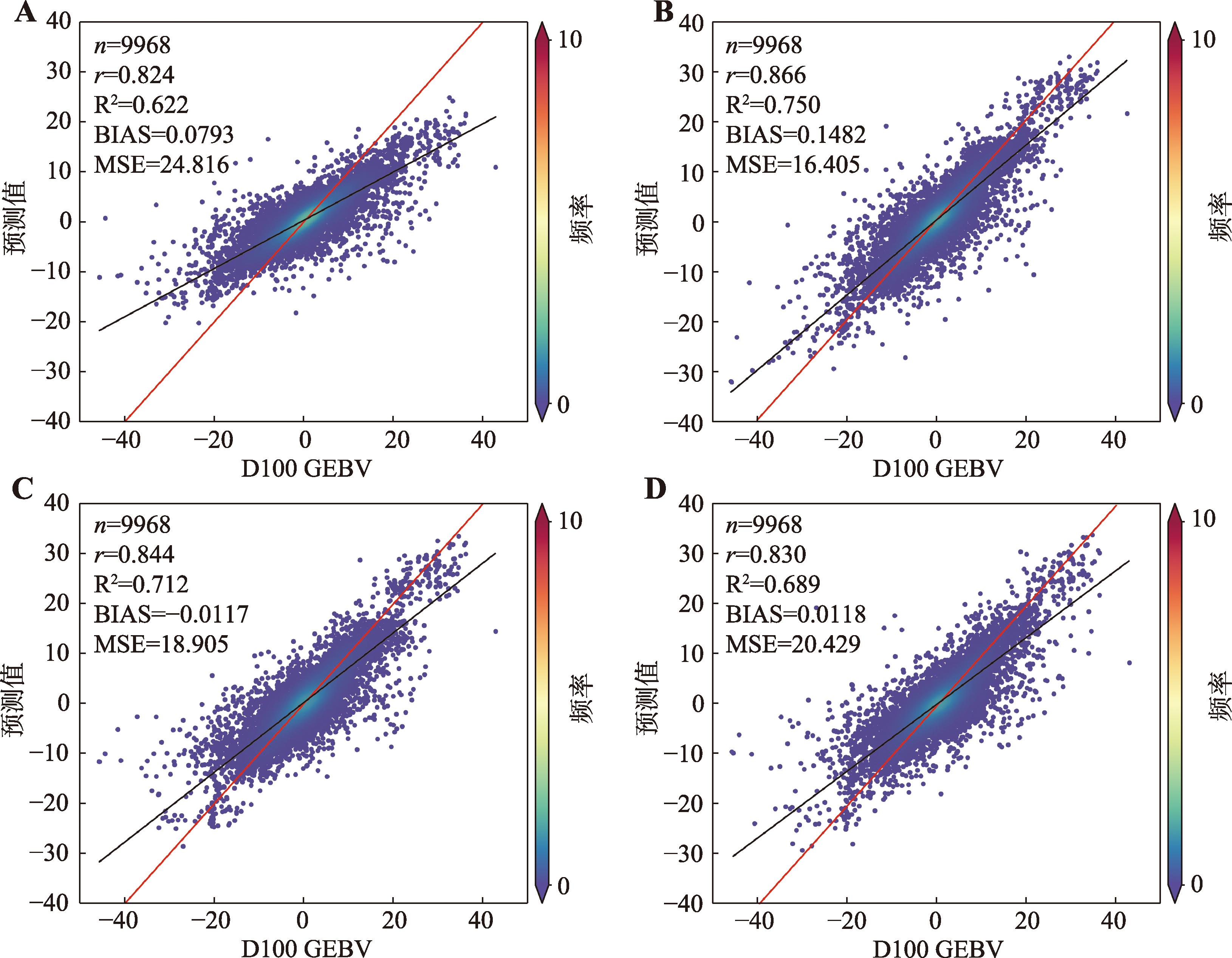
Table 5
Hyperparameter table for DL model"
| 参数 | GEBV估计参数 | 表型预测参数 |
|---|---|---|
| activation | RectifierWithDropout | RectifierWithDropout |
| hidden | [100,100,100] | [100, 100, 100] |
| epochs | 222 | 315 |
| epsilon | 1.00E-09 | 1.00E-08 |
| rate | 0.005 | 0.005 |
| input_dropout_ratio | 0.2 | 0.05 |
| hidden_dropout_ratios | [0.2, 0.2, 0.2] | [0.2, 0.2, 0.2] |
| distribution | gaussian | gaussian |
| tweedie_power | 1.5 | 1.5 |
| categorical_encoding | OneHotInternal | OneHotInternal |
| [1] | Wang KJ, Liu YF, Xu Q, Liu CK, Wang J, Ding C, Fang MY. A post-GWAS confirming GPAT3 gene associated with pig growth and a significant SNP influencing its promoter activity. Anim Genet, 2017, 48(4): 478-482. |
| [2] | Guo YM, Huang YX, Hou LJ, Ma JW, Chen CY, Ai HS, Huang LS, Ren J. Genome-wide detection of genetic markers associated with growth and fatness in four pig populations using four approaches. Genet Sel Evol, 2017, 49(1): 21. |
| [3] | Ding RR, Yang M, Wang XW, Quan JP, Zhuang ZW, Zhou SP, Li SY, Xu Z, Zheng EQ, Cai GY, Liu DW, Huang W, Yang J, Wu ZF. Genetic architecture of feeding behavior and feed efficiency in a Duroc pig population. Front Genet, 2018, 9: 220. |
| [4] | Vanraden PM. Symposium review:How to implement genomic selection. J Dairy Sci, 2020, 103(6): 5291-5301. |
| [5] | Meuwissen THE, Hayes BJ, Goddard ME. Prediction of total genetic value using genome-wide dense marker maps. Genetics, 2001, 157(4): 1819-1829. |
| [6] | Guo P, Zhang JB, Cao S. Study on Bayesian method and machine learning genome-wide selection of milk production traits in dairy cows. Heilongjiang Anim Sci Vet Med, 2023(5): 56-60+64. |
| 郭鹏, 张建斌, 曹晟. 奶牛产奶性状贝叶斯方法与机器学习全基因组选择研究. 黑龙江畜牧兽医, 2023(5): 56-60+64. | |
| [7] | Akanno EC, Schenkel FS, Sargolzaei M, Friendship RM, Robinson JAB. Opportunities for genome-wide selection for pig breeding in developing countries. J Anim Sci, 2013, 91(10): 4617-4627. |
| [8] | Samorè AB, Fontanesi L. Genomic selection in pigs: State of the art and perspectives. Ital J Anim Sci, 2016, 15: 211-232. |
| [9] | Marchini J, Howie B. Genotype imputation for genome- wide association studies. Nat Rev Genet, 2010, 11(7): 499-511. |
| [10] | Das S, Forer L, Schönherr S, Sidore C, Locke AE, Kwong A, Vrieze SI, Chew EY, Levy S, Mcgue M, Schlessinger D, Stambolian D, Loh PR, Iacono WG, Swaroop A, Scott LJ, Cucca F, Kronenberg F, Boehnke M, Abecasis GR, Fuchsberger C. Next-generation genotype imputation service and methods. Nat Genet, 2016, 48(10): 1284-1287. |
| [11] | Sollero BP, Howard JT, Spangler ML.The impact of reducing the frequency of animals genotyped at higher density on imputation and prediction accuracies using ssGBLUP1. J Anim Sci, 2019, 97(7): 2780-2792. |
| [12] | Fernandes Júnior GA, Carvalheiro R, de Oliveira HN, Sargolzaei M, Costilla R, Ventura RV, Fonseca LFS, Neves HHR, Hayes BJ, Imputation accuracy to whole-genome sequence in Nellore cattle. Genet Sel Evol, 2021, 53(1): 27. |
| [13] | Alves AAC, Espigolan R, Bresolin T, Costa RM, Fernandes Júnior GA, Ventura RV, Carvalheiro R, Albuquerque LG. Genome-enabled prediction of reproductive traits in Nellore cattle using parametric models and machine learning methods. Anim Genet, 2021, 52(1): 32-46. |
| [14] | Zhao W, Lai XS, Liu DY, Zhang ZY, Ma PP, Wang QS, Zhang Z, Pan YC. Applications of support vector machine in genomic prediction in pig and maize populations. Front Genet, 2020, 11: 598318. |
| [15] | Montesinos López OA, Montesinos López A, Crossa J.Random Forest for Genomic Prediction. In: Multivariate Statistical Machine Learning Methods for Genomic Prediction. Cham: Springer International Publishing, 2022, 633-681. |
| [16] | Liang M.Research on genome-wide selection based on machine learning algorithm[Dissertation]. Chinese Academy of Agricultural Sciences, 2021. |
| 梁忙.基于机器学习算法的全基因组选择研究[学位论文]. 中国农业科学院, 2021. | |
| [17] | Barreiro E, Munteanu CR, Cruz-Monteagudo M, Pazos A, González-Díaz H. Net-Net Auto Machine Learning (AutoML) Prediction of Complex Ecosystems. Scientific reports. 2018; 8(1): 12340. |
| [18] | Romero RAA, Deypalan MNY, Mehrotra S, Jungao JT, Sheils NE, Manduchi E, Moore JH. Benchmarking AutoML frameworks for disease prediction using medical claims. BioData Min, 2022, 15(1): 15. |
| [19] | Friedman JH. Greedy function approximation: A gradient boosting machine. Ann Statist, 2001, 29(5): 1189-1232. |
| [20] | Natekin A, Knoll A. Gradient boosting machines, a tutorial. Front Neurorobot, 2013, 7: 21. |
| [21] | Abdollahi-Arpanahi R, Gianola D, Peñagaricano F. Deep learning versus parametric and ensemble methods for genomic prediction of complex phenotypes. Genet Sel Evol, 2020, 52(1): 12. |
| [22] | Breiman L. Random Forests. Machine Learning, 2001, 45: 5-32. |
| [23] | González-recio O, Forni S. Genome-wide prediction of discrete traits using Bayesian regressions and machine learning. Genet Sel Evol, 2011, 43(1): 7. |
| [24] | Wang FY, Wang YC, Ji XK, Wang ZP. Effective macrosomia prediction using random forest algorithm. Int J Environ Res Public Health, 2022, 19(6): 3245. |
| [25] | Chen TQ, Guestrin C. XGBoost: a scalable tree boosting system. In:Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, 2016, 785-794. |
| [26] | Hou NZ, Li MZ, He L, Xie B, Wang L, Zhang RM, Yu Y, Sun XD, Pan ZS, Wang K. Predicting 30-days mortality for MIMIC-III patients with sepsis-3: a machine learning approach using XGboost. J Transl Med, 2020, 18(1): 462. |
| [27] | Tan XF, Li GS. Overview of deep learning development. In: The 15th National Security Geophysics Symposium. 2019. |
| 谭笑枫, 李广帅.深度学习发展综述. 见: 第十五届国家安全地球物理专题研讨会. 2019. | |
| [28] | Nayeri S, Sargolzaei M, Tulpan D. A review of traditional and machine learning methods applied to animal breeding. Anim Health Res Rev, 2019, 20(1): 31-46. |
| [29] | Zheye Peng, Zijun Tang, Minzhu Xie. Research progress in machine learning methods for gene-gene interaction detection. Hereditas(Beijing), 2018, 40(3): 218-226. |
| 彭哲也, 唐紫珺, 谢民主. 机器学习方法在基因交互作用探测中的研究进展. 遗传, 2018, 40(3): 218-226. | |
| [30] | Li B, Zhang NX, Wang YG, George AW, Reverter A, Li YT. Genomic prediction of breeding values using a subset of snps identified by three machine learning methods. Front Genet, 2018, 9: 237. |
| [1] | Chonglong Wang, Xiangdong Ding, Jianfeng Liu, Zongjun Yin, Qin Zhang. Bayesian methods for genomic breeding value estimation [J]. HEREDITAS, 2014, 36(2): 111-118. |
| [2] | WANG Chun-Ling MENG Chen-Ling CAO Shao-Xian ZHANG Jun MENG Chun-Hua WANG Hui-Li FANG Yong-Fei ZHU Dong-Dong MAO Da-Gan. Single nuclear polymorphisms in exon 3 of POMC gene and the asso-ciation with growth traits in Hu sheep and East Friesian × Hu crossbred sheep [J]. HEREDITAS, 2013, 35(9): 1095-1100. |
| [3] | WANG Peng-Yu, GUANQUE Zha-Xi, QI Quan-Qing, DE Mao, ZHANG Wen-Guang, LI Jin-Quan. Impact of maternal genetic effect on genetic parameter estimation of production traits for Qinghai fine-wool sheep [J]. HEREDITAS, 2012, 34(5): 584-590. |
| [4] | ZHU Yuan-Yuan, LIANG Hong-Wei, LI Zhong, LUO Xiang-Zhong, LI Lin, ZHANG Zhi-Wei, ZOU Gui-Wei. Polymorphism of MSTN gene and its association with growth traits in yellow catfish (Pelteobagruse fulvidraco) [J]. HEREDITAS, 2012, 34(1): 72-78. |
| [5] |
FAN Jia-Jia, BAI Jun-Jie, LI Xiao-Hui, HE Xiao-Ping, HE Xiao-Yan, |
| [6] | MAO Hai-Xia, CHEN Hong, CHEN Fu-Ying, ZHANG Chun-Lei, WANG Xin-Zhuang, WANG Ju-Qiang. Association of polymorphisms of DGAT2 gene with growth traits in Jiaxian red cattle [J]. HEREDITAS, 2008, 30(3): 329-332. |
| [7] | LI Shi-Peng, DU Zhi-Heng, NING Fang-Yong, SUN Hong-Xia, BAI Xiu-Juan. Correlation analysis between MC3R and MC4R gene polymorphism and growth traits in pigeon [J]. HEREDITAS, 2008, 30(10): 1333-1340. |
| [8] | . The modulation and control on growth and development traits of Nanyang Cattle in different phase [J]. HEREDITAS, 2006, 28(8): 927-932. |
| [9] | WANG Qing-Lai, WANG Ai-Guo, WU Zhen-Fang, ZHANG Hao, LUO Xu-Fang. Using VCE4.0 package to estimate genetic parameters on growth traits in Landrace [J]. HEREDITAS, 2004, 26(6): 811-814. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||