Hereditas(Beijing) ›› 2023, Vol. 45 ›› Issue (11): 963-975.doi: 10.16288/j.yczz.23-215
• Review • Previous Articles Next Articles
Progresses on genetic susceptibility of COVID-19
Yuanfeng Li1,2( ), Tianzhun Wu3(
), Tianzhun Wu3( ), Shunqi Chen2, Yuting Wang2, Tao Zeng4,5, Ruofan Li4,5, Gangqiao Zhou1,6(
), Shunqi Chen2, Yuting Wang2, Tao Zeng4,5, Ruofan Li4,5, Gangqiao Zhou1,6( )
)
- 1. State Key Laboratory of Proteomics, National Center for Protein Sciences at Beijing, Beijing Institute of Radiation Medicine, Beijing 100850, China
2. College of Chemistry and Environmental Science, Hebei University, Baoding 071002, China
3. Guangxi Medical University Cancer Hospital, Nanning 530021, China
4. Medical School of Chinese PLA, Beijing 100853, China
5. Faculty of Hepato-Pancreato-Biliary Surgery, First Medical Center, Chinese PLA General Hospital, Beijing 100853, China
6. Center for Global Health, School of Public Health, Nanjing Medical University, Nanjing 211166, China
-
Received:2023-08-07Revised:2023-09-21Online:2023-11-20Published:2023-10-12 -
Contact:Gangqiao Zhou E-mail:liyf_snp@163.com;wutianzhun@stu.gxmu.edu.cn;zhougq114@126.com -
Supported by:Emergency Key Program of Guangzhou Laboratory(EKPG21-19);Open Cooperation Project of Nanjing Medical University Global Health Center(JX103SYL202200313)
Cite this article
Yuanfeng Li, Tianzhun Wu, Shunqi Chen, Yuting Wang, Tao Zeng, Ruofan Li, Gangqiao Zhou. Progresses on genetic susceptibility of COVID-19[J]. Hereditas(Beijing), 2023, 45(11): 963-975.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
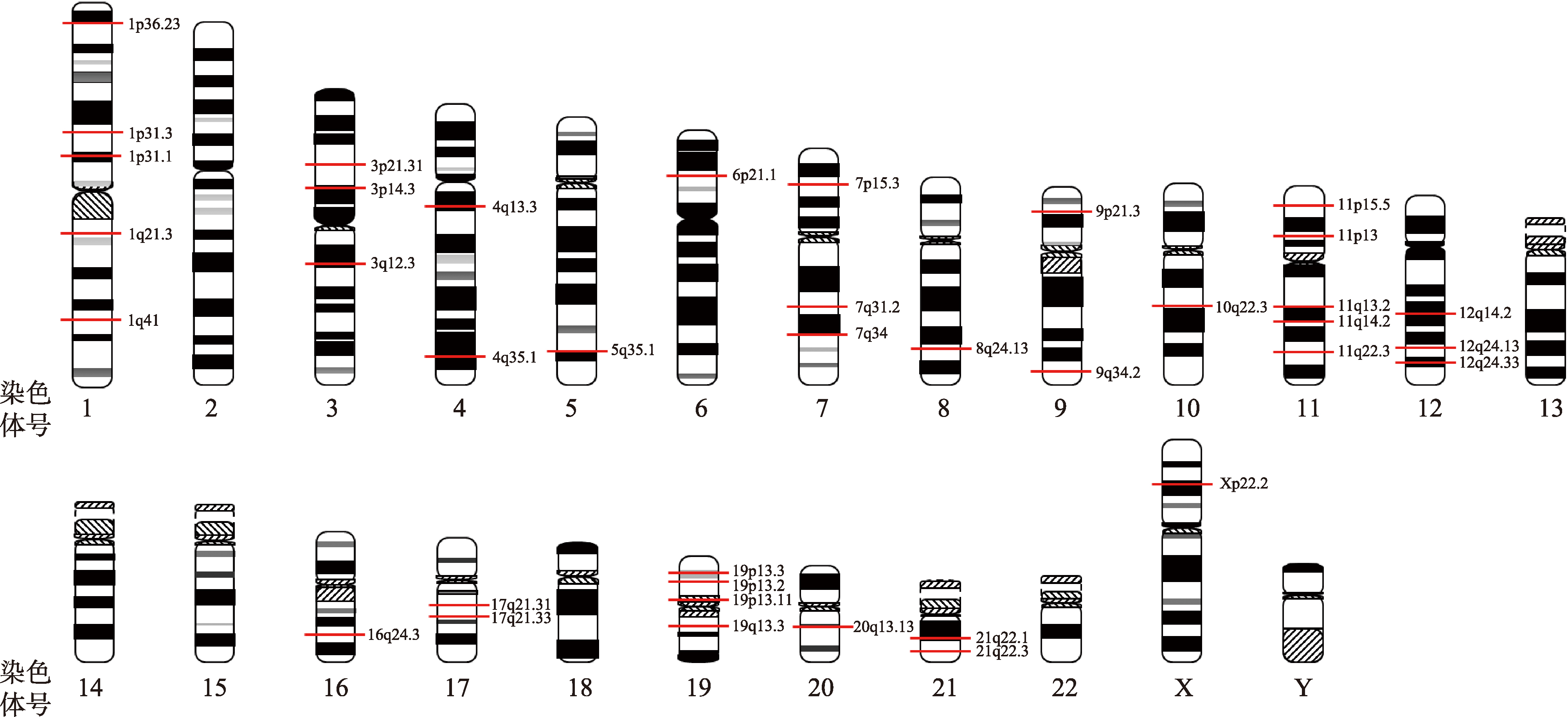
Table 1
Genome-wide association studies of COVID-19"
| 人群来源 | 病例/对照的数量 | 多态性位点或基因 | 主要科学发现 | 参考文献 |
|---|---|---|---|---|
| 意大利和 西班牙 | 意大利:835名危重症患者和1255名对照;西班牙:775名危重症患者和950名对照 | rs11385942、rs657152 | 发现3p21.31和9q34.2是危重型COVID-19的易感基因区域 | [ |
| 英国 | 2244名危重症患者与匹配人群对照组 | rs10735079、rs2109069、rs74956615、rs2236757 | 新发现4个危重型COVID-19的易感基因区域 | [ |
| 全球 | 7491名危重症患者和48,400名对照个体 | rs114301457、rs7528026、rs41264915、rs1123573、rs2271616、rs73064425等 | 新发现16个位点与危重型COVID-19显著相关 | [ |
| 全球 | 24,202名危重症患者与匹配人群对照组 | rs2478868、rs12046291、rs71658797、rs114301457、rs7528026、rs41264915等 | 新发现49个与COVID-19重症化显著关联的基因区域 | [ |
| 全球 | 15,434名新冠阳性患者与匹配人群对照组 | rs9411378、rs8176719、rs13078854 | 验证了ABO基因和3p21.31区域与COVID-19显著相关 | [ |
| 全球 | 49,562名病例和200万对照个体 | rs10490770、rs1886814、rs72711165、rs2109069、rs74956615、rs10774671等 | 发现13个位点与SARS-CoV-2的感染或COVID-19的严重程度相关 | [ |
| 全球 | 125,584名新冠病例和超过250万对照个体 | rs721917、rs117169628、rs35705950等 | 鉴定了7个与SARS-CoV-2感染相关的位点和16个与COVID-19疾病严重程度相关的位点 | [ |
| 全球 | 52,630名新冠患者和70,4016名对照个体 | rs7310667、rs13050728、rs505922、rs35081325等 | 定义了8种COVID-19相关风险表型 | [ |
| 全球 | 47,298名阳性感染者且味觉或嗅觉丧失的新冠病例和22,543名阳性感染者且味觉或嗅觉正常的新冠对照组 | rs7688383 | 首次发现SARS-CoV-2感染所致嗅觉或味觉丧失的遗传易感因素 | [ |
| 中国 | 332名新冠患者和匹配的对照个体 | rs6020298 | 首次在中国人群中开展的COVID-19遗传关联研究 | [ |
| 中国 | 885名新冠重症或危重症患者和546名轻症或无症状患者 | rs1712779、rs10831496 | 在中国人群中鉴定了2个与COVID-19重症化显著相关的位点 | [ |
| 中国 | 1401名新冠患者与948名对照个体 | rs1853837、rs8176719、rs74490654 | 新鉴定3个与COVID-19相关的易感基因区域(FOXP4-AS1、ABO和 MEF2B) | [ |
| 中国 | 632名新冠患者与3021名对照个体 | rs2069837 | 鉴定了中国人群COVID-19易感位点rs2069837,该位点通过IL-6影响COVID-19的重症化风险 | [ |
| 日本 | 2393名新冠患者与3289名对照个体 | rs60200309 | 首篇日本人群COVID-19的GWAS。鉴定了5q35区域的DOCK2基因与COVID-19患者的重症化显著相关 | [ |
| 全球 | 659名危重症患者和534 名无症状或轻症患者 | TLR3、IRF7和IRF9等 | 鉴定了与COVID-19重症化相关的13个罕见变异位点 | [ |
| 全球 | 1202名男性患者与331名对照个体 | TLR7 | 发现X连锁隐性遗传的TLR7缺陷是重症COVID-19的遗传风险因素之一 | [ |
| 全球 | 20,952名新冠患者和565,205名对照个体 | rs769102632、rs1256764500、rs754119466、rs761377603、rs2287960 | 发现了8个与COVID-19显著相关的罕见变异 | [ |
| 中国 | 451名新冠患者 | 无显著关联位点 | 揭示了可能在重症COVID-19患者中富集的信号通路、HC-pLoF变异和候选基因 | [ |
| 全球 | 52,630名新冠患者和704,016名对照个体 | rs190509934 | 新发现ACE2基因的罕见变异与COVID-19易感性相关 | [ |
| [1] |
Huang CL, Wang YM, Li XW, Ren LL, Zhao JP, Hu Y, Zhang L, Fan GH, Xu JY, Gu XY, Cheng ZS, Yu T, Xia JA, Wei Y, Wu WJ, Xie XL, Yin W, Li H, Liu M, Xiao Y, Gao H, Guo L, Xie JG, Wang GF, Jiang RM, Gao ZC, Jin Q, Wang JW, Cao B.Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet, 2020, 395(10223): 497-506.
doi: S0140-6736(20)30183-5 pmid: 31986264 |
| [2] | Wu ZY, McGoogan JM.Characteristics of and important lessons from the coronavirus disease 2019(COVID-19) outbreak in China:summary of a report of 72,314 cases from the Chinese center for disease control and prevention. JAMA, 2020, 323(13): 1239-1242. |
| [3] |
Zhang XN, Tan Y, Ling Y, Lu G, Liu F, Yi ZG, Jia XF, Wu M, Shi BS, Xu SB, Chen J, Wang W, Chen B, Jiang L, Yu ST, Lu J, Wang JZ, Xu MZ, Yuan ZH, Zhang Q, Zhang XX, Zhao GP, Wang SY, Chen SJ, Lu HZ.Viral and host factors related to the clinical outcome of COVID-19. Nature, 2020, 583(7816): 437-440.
doi: 10.1038/s41586-020-2355-0 |
| [4] |
Buniello A, MacArthur JAL, Cerezo M, Harris LW, Hayhurst J, Malangone C, McMahon A, Morales J, Mountjoy E, Sollis E, Suveges D, Vrousgou O, Whetzel PL, Amode R, Guillen JA, Riat HS, Trevanion SJ, Hall P, Junkins H, Flicek P, Burdett T, Hindorff LA, Cunningham F, Parkinson H. The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res, 2019, 47(D1): D1005-D1012.
doi: 10.1093/nar/gky1120 |
| [5] |
Velavan TP, Pallerla SR, Rüter J, Augustin Y, Kremsner PG, Krishna S, Meyer CG. Host genetic factors determining COVID-19 susceptibility and severity. EBioMedicine, 2021, 72: 103629.
doi: 10.1016/j.ebiom.2021.103629 |
| [6] |
Kousathanas A, Pairo-Castineira E, Rawlik K, Stuckey A, Odhams CA, Walker S, Russell CD, Malinauskas T, Wu Y, Millar J, Shen X, Elliott KS, Griffiths F, Oosthuyzen W, Morrice K, Keating S, Wang B, Rhodes D, Klaric L, Zechner M, Parkinson N, Siddiq A, Goddard P, Donovan S, Maslove D, Nichol A, Semple MG, Zainy T, Maleady-Crowe F, Todd L, Salehi S, Knight J, Elgar G, Chan G, Arumugam P, Patch C, Rendon A, Bentley D, Kingsley C, Kosmicki JA, Horowitz JE, Baras A, Abecasis GR, Ferreira MAR, Justice A, Mirshahi T, Oetjens M, Rader DJ, Ritchie MD, Verma A, Fowler TA, Shankar-Hari M, Summers C, Hinds C, Horby P, Ling L, McAuley D, Montgomery H, Openshaw PJM, Elliott P, Walsh T, Tenesa A, GenOMICC investigators, 23andMe investigators, COVID-19 Human Genetics Initiativ, Fawkes A, Murphy L, Rowan K, Ponting CP, Vitart V, Wilson JF, Yang J, Bretherick AD, Scott RH, Hendry SC, Moutsianas L, Law A, Caulfield MJ, Baillie JK,. Whole- genome sequencing reveals host factors underlying critical COVID-19. Nature, 2022, 607(7917): 97-103.
doi: 10.1038/s41586-022-04576-6 |
| [7] |
Severe Covid-19 GWAS Group, Ellinghaus D, Degenhardt F, Bujanda L, Buti M, Albillos A, Invernizzi P, Fernández J, Prati D, Baselli G, Asselta R, Grimsrud MM, Milani C, Aziz F, Kässens J, May S, Wendorff M, Wienbrandt L, Uellendahl-Werth F, Zheng TH, Yi XL, de Pablo R, Chercoles AG, Palom A, Garcia-Fernandez AE, Rodriguez-Frias F, Zanella A, Bandera A, Protti A, Aghemo A, Lleo A, Biondi A, Caballero-Garralda A, Gori A, Tanck A, Carreras Nolla A, Latiano A, Fracanzani AL, Peschuck A, Julià A, Pesenti A, Voza A, Jiménez D, Mateos B, Nafria Jimenez B, Quereda C, Paccapelo C, Gassner C, Angelini C, Cea C, Solier A, Pestaña D, Muñiz-Diaz E, Sandoval E, Paraboschi EM, Navas E, García Sánchez F, Ceriotti F, Martinelli-Boneschi F, Peyvandi F, Blasi F, Téllez L, Blanco-Grau A, Hemmrich-Stanisak G, Grasselli G, Costantino G, Cardamone G, Foti G, Aneli S, Kurihara H, ElAbd H, My I, Gálvan-Femenia I, Martín J, Erdmann J, Ferrusquía- Acosta J, Garcia-Etxebarria K, Izquierdo-Sanchez L, Bettini LR, Sumoy L, Terranova L, Moreira L, Santoro L, Scudeller L, Mesonero F, Roade L, Rühlemann MC, Schaefer M, Carrabba M, Riveiro-Barciela M, Figuera Basso ME, Valsecchi MG, Hernandez-Tejero M, Acosta-Herrera M, D'Angiò M, Baldini M, Cazzaniga M, Schulzky M, Cecconi M, Wittig M, Ciccarelli M, Rodríguez-Gandía M, Bocciolone M, Miozzo M, Montano N, Braun N, Sacchi N, Martínez N, Özer O, Palmieri O, Faverio P, Preatoni P, Bonfanti P, Omodei P, Tentorio P, Castro P, Rodrigues PM, Blandino Ortiz AB, de Cid R, Ferrer R, Gualtierotti R, Nieto R, Goerg S, Badalamenti S, Marsal S, Matullo G, Pelusi S, Juzenas S, Aliberti S, Monzani V, Moreno V, Wesse T, Lenz TL, Pumarola T, Rimoldi V, Bosari S, Albrecht W, Peter W, Romero-Gómez M, D'Amato M, Duga S, Banales JM, Hov JR, Folseraas T, Valenti L, Franke A, Karlsen TH. Genomewide association study of severe Covid-19 with respiratory failure. N Engl J Med, 2020, 383(16): 1522-1534.
doi: 10.1056/NEJMoa2020283 |
| [8] |
Pairo-Castineira E, Clohisey S, Klaric L, Bretherick AD, Rawlik K, Pasko D, Walker S, Parkinson N, Fourman MH, Russell CD, Furniss J, Richmond A, Gountouna E, Wrobel N, Harrison D, Wang B, Wu Y, Meynert A, Griffiths F, Oosthuyzen W, Kousathanas A, Moutsianas L, Yang ZJ, Zhai RR, Zheng CQ, Grimes G, Beale R, Millar J, Shih B, Keating S, Zechner M, Haley C, Porteous DJ, Hayward C, Yang J, Knight J, Summers C, Shankar-Hari M, Klenerman P, Turtle L, Ho A, Moore SC, Hinds C, Horby P, Nichol A, Maslove D, Ling L, McAuley D, Montgomery H, Walsh T, Pereira AC, Renieri A,GenOMICC Investigators, ISARIC4C Investigators, COVID-19 Human Genetics Initiative, 23andMe Investigators, BRACOVID Investigators, Gen-COVID Investigators, Shen X, Ponting CP, Fawkes A, Tenesa A, Caulfield M, Scott R, Rowan K, Murphy L, Openshaw PJM, Semple MG, Law A, Vitart V, Wilson JF, Baillie JK. Genetic mechanisms of critical illness in COVID-19. Nature, 2021, 591(7848): 92-98.
doi: 10.1038/s41586-020-03065-y |
| [9] |
Pairo-Castineira E, Rawlik K, Bretherick AD, Qi T, Wu Y, Nassiri I, McConkey GA, Zechner M, Klaric L, Griffiths F, Oosthuyzen W, Kousathanas A, Richmond A, Millar J, Russell CD, Malinauskas T, Thwaites R, Morrice K, Keating S, Maslove D, Nichol A, Semple MG, Knight J, Shankar-Hari M, Summers C, Hinds C, Horby P, Ling L, McAuley D, Montgomery H, Openshaw PJM, Begg C, Walsh T, Tenesa A, Flores C, Riancho JA, Rojas-Martinez A, Lapunzina P, GenOMICC Investigators, SCOURGE Consortium, ISARICC Investigators, 23andMe COVID-19 Team, Yang J, Ponting CP, Wilson JF, Vitart V, Abedalthagafi M, Luchessi AD, Parra EJ, Cruz R, Carracedo A, Fawkes A, Murphy L, Rowan K, Pereira AC, Law A, Fairfax B, Hendry SC, Baillie JK. GWAS and meta-analysis identifies 49 genetic variants underlying critical COVID-19. Nature, 2023, 617(7962): 764-768.
doi: 10.1038/s41586-023-06034-3 |
| [10] |
Shelton JF, Shastri AJ, Ye C, Weldon CH, Filshtein- Sonmez T, Coker D, Symons A, Esparza-Gordillo J, 23 and Me COVID-19 Team, Aslibekyan S, Auton A. Trans-ancestry analysis reveals genetic and nongenetic associations with COVID-19 susceptibility and severity. Nat Genet, 2021, 53(6): 801-808.
doi: 10.1038/s41588-021-00854-7 pmid: 33888907 |
| [11] |
COVID-19 Host Genetics Initiative.Mapping the human genetic architecture of COVID-19. Nature, 2021, 600(7889): 472-477.
doi: 10.1038/s41586-021-03767-x |
| [12] |
COVID-19 Host Genetics Initiative.A first update on mapping the human genetic architecture of COVID-19. Nature, 2022, 608(7921): E1-E10.
doi: 10.1038/s41586-022-04826-7 |
| [13] |
Roberts GHL, Partha R, Rhead B, Knight SC, Park DS, Coignet MV, Zhang M, Berkowitz N, Turrisini DA, Gaddis M, McCurdy SR, Pavlovic M, Ruiz L, Sass C, AncestryDNA Science Team, Haug Baltzell AK, Guturu H, Girshick AR, Ball CA, Hong EL, Rand KA. Expanded COVID-19 phenotype definitions reveal distinct patterns of genetic association and protective effects. Nat Genet, 2022, 54(4): 374-381.
doi: 10.1038/s41588-022-01042-x pmid: 35410379 |
| [14] |
Shelton JF, Shastri AJ, Fletez-Brant K, 23 and Me COVID- 19 Team, Aslibekyan S, Auton A. The UGT2A1/UGT2A2 locus is associated with COVID-19-related loss of smell or taste. Nat Genet, 2022, 54(2): 121-124.
doi: 10.1038/s41588-021-00986-w pmid: 35039640 |
| [15] |
Wang F, Huang SJ, Gao RS, Zhou YW, Lai CX, Li ZC, Xian WJ, Qian XB, Li ZY, Huang YS, Tang QY, Liu PH, Chen RK, Liu R, Li X, Tong X, Zhou X, Bai Y, Duan G, Zhang T, Xu X, Wang J, Yang HM, Liu SY, He Q, Jin X, Liu L. Initial whole-genome sequencing and analysis of the host genetic contribution to COVID-19 severity and susceptibility. Cell Discov, 2020, 6(1): 83.
doi: 10.1038/s41421-020-00231-4 pmid: 33298875 |
| [16] | Li YF, Ke YH, Xia XY, Wang YH, Cheng FJ, Liu XY, Jin X, Li BA, Xie CY, Liu SY, Chen WJ, Yang CN, Niu YG, Jia RZ, Chen Y, Liu X, Wang ZH, Zheng F, Jin Y, Li Z, Yang N, Cao PB, Chen HX, Ping J, He FC, Wang CJ, Zhou GQ. Genome-wide association study of COVID-19 severity among the Chinese population. Cell Discov, 2021, 7(1): 76. |
| [17] |
Wu P, Ding L, Li XD, Liu SY, Cheng FJ, He Q, Xiao MZ, Wu P, Hou HY, Jiang MH, Long PP, Wang H, Liu LL, Qu MH, Shi X, Jiang Q, Mo TT, Ding WC, Fu Y, Han S, Huo XX, Zeng YC, Zhou YN, Zhang Q, Ke J, Xu X, Ni W, Shao ZY, Wang JZ, Liu PH, Li ZL, Jin Y, Zheng F, Wang F, Liu L, Li WD, Liu K, Peng R, Xu XD, Lin YH, Gao H, Shi LM, Geng ZY, Mu XW, Yan Y, Wang K, Wu DG, Hao XJ, Cheng SS, Qiu GK, Guo H, Li KZ, Chen G, Sun ZY, Lin XH, Jin X, Wang F, Sun CY, Wang CL.Trans-ethnic genome-wide association study of severe COVID-19. Commun Biol, 2021, 4(1): 1034.
doi: 10.1038/s42003-021-02549-5 pmid: 34465887 |
| [18] | Gong B, Huang LL, He YQ, Xie W, Yin Y, Shi Y, Xiao JL, Zhong L, Zhang Y, Jiang ZL, Hao F, Zhou Y, Li H, Jiang L, Yang XX, Song XR, Kang Y, Tuo L, Huang Y, Shuai P, Liu YP, Zheng F, Yang ZL.A genetic variant in IL-6 lowering its expression is protective for critical patients with COVID-19. Signal Transduct Target Ther, 2022, 7(1): 112. |
| [19] |
Namkoong H, Edahiro R, Takano T, Nishihara H, Shirai Y, Sonehara K, Tanaka H, Azekawa S, Mikami Y, Lee H, Hasegawa T, Okudela K, Okuzaki D, Motooka D, Kanai M, Naito T, Yamamoto K, Wang QS, Saiki R, Ishihara R, Matsubara Y, Hamamoto J, Hayashi H, Yoshimura Y, Tachikawa N, Yanagita E, Hyugaji T, Shimizu E, Katayama K, Kato Y, Morita T, Takahashi K, Harada N, Naito T, Hiki M, Matsushita Y, Takagi H, Aoki R, Nakamura A, Harada S, Sasano H, Kabata H, Masaki K, Kamata H, Ikemura S, Chubachi S, Okamori S, Terai H, Morita A, Asakura T, Sasaki J, Morisaki H, Uwamino Y, Nanki K, Uchida S, Uno S, Nishimura T, Ishiguro T, Isono T, Shibata S, Matsui Y, Hosoda C, Takano K, Nishida T, Kobayashi Y, Takaku Y, Takayanagi N, Ueda S, Tada A, Miyawaki M, Yamamoto M, Yoshida E, Hayashi R, Nagasaka T, Arai S, Kaneko Y, Sasaki K, Tagaya E, Kawana M, Arimura K, Takahashi K, Anzai T, Ito S, Endo A, Uchimura Y, Miyazaki Y, Honda T, Tateishi T, Tohda S, Ichimura N, Sonobe K, Sassa CT, Nakajima J, Nakano Y, Nakajima Y, Anan R, Arai R, Kurihara Y, Harada Y, Nishio K, Ueda T, Azuma M, Saito R, Sado T, Miyazaki Y, Sato R, Haruta Y, Nagasaki T, Yasui Y, Hasegawa Y, Mutoh Y, Kimura T, Sato T, Takei R, Hagimoto S, Noguchi Y, Yamano Y, Sasano H, Ota S, Nakamori Y, Yoshiya K, Saito F, Yoshihara T, Wada D, Iwamura H, Kanayama S, Maruyama S, Yoshiyama T, Ohta K, Kokuto H, Ogata H, Tanaka Y, Arakawa K, Shimoda M, Osawa T, Tateno H, Hase I, Yoshida S, Suzuki S, Kawada M, Horinouchi H, Saito F, Mitamura K, Hagihara M, Ochi J, Uchida T, Baba R, Arai D, Ogura T, Takahashi H, Hagiwara S, Nagao G, Konishi S, Nakachi I, Murakami K, Yamada M, Sugiura H, Sano H, Matsumoto S, Kimura N, Ono Y, Baba H, Suzuki Y, Nakayama S, Masuzawa K, Namba S, Suzuki K, Naito Y, Liu YC, Takuwa A, Sugihara F, Wing JB, Sakakibara S, Hizawa N, Shiroyama T, Miyawaki S, Kawamura Y, Nakayama A, Matsuo H, Maeda Y, Nii T, Noda Y, Niitsu T, Adachi Y, Enomoto T, Amiya S, Hara R, Yamaguchi Y, Murakami T, Kuge T, Matsumoto K, Yamamoto Y, Yamamoto M, Yoneda M, Kishikawa T, Yamada S, Kawabata S, Kijima N, Takagaki M, Sasa N, Ueno Y, Suzuki M, Takemoto N, Eguchi H, Fukusumi T, Imai T, Fukushima M, Kishima H, Inohara H, Tomono K, Kato K, Takahashi M, Matsuda F, Hirata H, Takeda Y, Koh H, Manabe T, Funatsu Y, Ito F, Fukui T, Shinozuka K, Kohashi S, Miyazaki M, Shoko T, Kojima M, Adachi T, Ishikawa M, Takahashi K, Inoue T, Hirano T, Kobayashi K, Takaoka H, Watanabe K, Miyazawa N, Kimura Y, Sado R, Sugimoto H, Kamiya A, Kuwahara N, Fujiwara A, Matsunaga T, Sato Y, Okada T, Hirai Y, Kawashima H, Narita A, Niwa K, Sekikawa Y, Nishi K, Nishitsuji M, Tani M, Suzuki J, Nakatsumi H, Ogura T, Kitamura H, Hagiwara E, Murohashi K, Okabayashi H, Mochimaru T, Nukaga S, Satomi R, Oyamada Y, Mori N, Baba T, Fukui Y, Odate M, Mashimo S, Makino Y, Yagi K, Hashiguchi M, Kagyo J, Shiomi T, Fuke S, Saito H, Tsuchida T, Fujitani S, Takita M, Morikawa D, Yoshida T, Izumo T, Inomata M, Kuse N, Awano N, Tone M, Ito A, Nakamura Y, Hoshino K, Maruyama J, Ishikura H, Takata T, Odani T, Amishima M, Hattori T, Shichinohe Y, Kagaya T, Kita T, Ohta K, Sakagami S, Koshida K, Hayashi K, Shimizu T, Kozu Y, Hiranuma H, Gon Y, Izumi N, Nagata K, Ueda K, Taki R, Hanada S, Kawamura K, Ichikado K, Nishiyama K, Muranaka H, Nakamura K, Hashimoto N, Wakahara K, Sakamoto K, Omote N, Ando A, Kodama N, Kaneyama Y, Maeda S, Kuraki T, Matsumoto T, Yokote K, Nakada TA, Abe R, Oshima T, Shimada T, Harada M, Takahashi T, Ono H, Sakurai T, Shibusawa T, Kimizuka Y, Kawana A, Sano T, Watanabe C, Suematsu R, Sageshima H, Yoshifuji A, Ito K, Takahashi S, Ishioka K, Nakamura M, Masuda M, Wakabayashi A, Watanabe H, Ueda S, Nishikawa M, Chihara Y, Takeuchi M, Onoi K, Shinozuka J, Sueyoshi A, Nagasaki Y, Okamoto M, Ishihara S, Shimo M, Tokunaga Y, Kusaka Y, Ohba T, Isogai S, Ogawa A, Inoue T, Fukuyama S, Eriguchi Y, Yonekawa A, Kan-O K, Matsumoto K, Kanaoka K, Ihara S, Komuta K, Inoue Y, Chiba S, Yamagata K, Hiramatsu Y, Kai H, Asano K, Oguma T, Ito Y, Hashimoto S, Yamasaki M, Kasamatsu Y, Komase Y, Hida N, Tsuburai T, Oyama B, Takada M, Kanda H, Kitagawa Y, Fukuta T, Miyake T, Yoshida S, Ogura S, Abe S, Kono Y, Togashi Y, Takoi H, Kikuchi R, Ogawa S, Ogata T, Ishihara S, Kanehiro A, Ozaki S, Fuchimoto Y, Wada S, Fujimoto N, Nishiyama K, Terashima M, Beppu S, Yoshida K, Narumoto O, Nagai H, Ooshima N, Motegi M, Umeda A, Miyagawa K, Shimada H, Endo M, Ohira Y, Watanabe M, Inoue S, Igarashi A, Sato M, Sagara H, Tanaka A, Ohta S, Kimura T, Shibata Y, Tanino Y, Nikaido T, Minemura H, Sato Y, Yamada Y, Hashino T, Shinoki M, Iwagoe H, Takahashi H, Fujii K, Kishi H, Kanai M, Imamura T, Yamashita T, Yatomi M, Maeno T, Hayashi S, Takahashi M, Kuramochi M, Kamimaki I, Tominaga Y, Ishii T, Utsugi M, Ono A, Tanaka T, Kashiwada T, Fujita K, Saito Y, Seike M, Watanabe H, Matsuse H, Kodaka N, Nakano C, Oshio T, Hirouchi T, Makino S, Egi M, Biobank Japan P, Omae Y, Nannya Y, Ueno T, Katayama K, Ai M, Fukui Y, Kumanogoh A, Sato T, Hasegawa N, Tokunaga K, Ishii M, Koike R, Kitagawa Y, Kimura A, Imoto S, Miyano S, Ogawa S, Kanai T, Fukunaga K, Okada Y. DOCK 2 is involved in the host genetics and biology of severe COVID-19. Nature, 2022, 609(7928): 754-760.
doi: 10.1038/s41586-022-05163-5 |
| [20] |
Zhang Q, Bastard P, Liu ZY, Le Pen J, Moncada-Velez M, Chen J, Ogishi M, Sabli IKD, Hodeib S, Korol C, Rosain J, Bilguvar K, Ye JQ, Bolze A, Bigio B, Yang R, Arias AA, Zhou QH, Zhang Y, Onodi F, Korniotis S, Karpf L, Philippot Q, Chbihi M, Bonnet-Madin L, Dorgham K, Smith N, Schneider WM, Razooky BS, Hoffmann HH, Michailidis E, Moens L, Han JE, Lorenzo L, Bizien L, Meade P, Neehus AL, Ugurbil AC, Corneau A, Kerner G, Zhang P, Rapaport F, Seeleuthner Y, Manry J, Masson C, Schmitt Y, Schlüter A, Le Voyer T, Khan T, Li J, Fellay J, Roussel L, Shahrooei M, Alosaimi MF, Mansouri D, Al-Saud H, Al-Mulla F, Almourfi F, Al-Muhsen SZ, Alsohime F, Al Turki S, Hasanato R, van de Beek D, Biondi A, Bettini LR, D'Angio M, Bonfanti P, Imberti L, Sottini A, Paghera S, Quiros-Roldan E, Rossi C, Oler AJ, Tompkins MF, Alba C, Vandernoot I, Goffard JC, Smits G, Migeotte I, Haerynck F, Soler-Palacin P, Martin-Nalda A, Colobran R, Morange PE, Keles S, Çölkesen F, Ozcelik T, Yasar KK, Senoglu S, Karabela ŞN, Rodríguez-Gallego C, Novelli G, Hraiech S, Tandjaoui-Lambiotte Y, Duval X, Laouénan C, COVID-STORM Clinicians, COVID Clinicians, Imagine COVID Group, French COVID Cohort Study Group, CoV-Contact Cohort, Amsterdam UMC Covid-19 Biobank, COVID Human Genetic Effort, NIAID-USUHS/TAGC COVID Immunity Group, Snow AL, Dalgard CL, Milner JD, Vinh DC, Mogensen TH, Marr N, Spaan AN, Boisson B, Boisson-Dupuis S, Bustamante J, Puel A, Ciancanelli MJ, Meyts I, Maniatis T, Soumelis V, Amara A, Nussenzweig M, García-Sastre A, Krammer F, Pujol A, Duffy D, Lifton RP, Zhang SY, Gorochov G, Béziat V, Jouanguy E, Sancho-Shimizu V, Rice CM, Abel L, Notarangelo LD, Cobat A, Su HC, Casanova JL. Inborn errors of type I IFN immunity in patients with life-threatening COVID-19. Science, 2020, 370(6515): eabd4570.
doi: 10.1126/science.abd4570 |
| [21] |
Asano T, Boisson B, Onodi F, Matuozzo D, Moncada- Velez M, Maglorius Renkilaraj MRL, Zhang P, Meertens L, Bolze A, Materna M, Korniotis S, Gervais A, Talouarn E, Bigio B, Seeleuthner Y, Bilguvar K, Zhang Y, Neehus AL, Ogishi M, Pelham SJ, Le Voyer T, Rosain J, Philippot Q, Soler-Palacín P, Colobran R, Martin-Nalda A, Rivière JG, Tandjaoui-Lambiotte Y, Chaïbi K, Shahrooei M, Darazam IA, Olyaei NA, Mansouri D, Hatipoğlu N, Palabiyik F, Ozcelik T, Novelli G, Novelli A, Casari G, Aiuti A, Carrera P, Bondesan S, Barzaghi F, Rovere-Querini P, Tresoldi C, Franco JL, Rojas J, Reyes LF, Bustos IG, Arias AA, Morelle G, Christèle K, Troya J, Planas-Serra L, Schlüter A, Gut M, Pujol A, Allende LM, Rodriguez- Gallego C, Flores C, Cabrera-Marante O, Pleguezuelo DE, de Diego RP, Keles S, Aytekin G, Akcan OM, Bryceson YT, Bergman P, Brodin P, Smole D, Smith CIE, Norlin AC, Campbell TM, Covill LE, Hammarström L, Pan- Hammarström Q, Abolhassani H, Mane S, Marr N, Ata M, Al Ali F, Khan T, Spaan AN, Dalgard CL, Bonfanti P, Biondi A, Tubiana S, Burdet C, Nussbaum R, Kahn- Kirby A, Snow AL, COVID Human Genetic Effort, COVID- STORM Clinicians, COVID Clinicians, Imagine COVID Group, French COVID Cohort Study Group, CoV-Contact Cohort, Amsterdam UMC Covid-, Biobank, NIAID- USUHS COVID Study Group, Bustamante J, Puel A, Boisson-Dupuis S, Zhang SY, Béziat V, Lifton RP, Bastard P, Notarangelo LD, Abel L, Su HC, Jouanguy E, Amara A, Soumelis V, Cobat A, Zhang Q, Casanova JL.X-linked recessive TLR7 deficiency in -1% of men under 60 years old with life-threatening COVID-19. Sci Immunol, 2021, 6(62): eabl4348.
doi: 10.1126/sciimmunol.abl4348 |
| [22] | Kosmicki JA, Horowitz JE, Banerjee N, Lanche R, Marcketta A, Maxwell E, Bai XD, Sun D, Backman JD, Sharma D, Kury FSP, Kang HM, O'Dushlaine C, Yadav A, Mansfield AJ, Li AH, Watanabe K, Gurski L, McCarthy SE, Locke AE, Khalid S, O'Keeffe S, Mbatchou J, Chazara O, Huang YF, Kvikstad E, O'Neill A, Nioi P, Parker MM, Petrovski S, Runz H, Szustakowski JD, Wang QL, Wong E, Cordova-Palomera A, Smith EN, Szalma S, Zheng XW, Esmaeeli S, Davis JW, Lai YP, Chen X, Justice AE, Leader JB, Mirshahi T, Carey DJ, Verma A, Sirugo G, Ritchie MD, Rader DJ, Povysil G, Goldstein DB, Kiryluk K, Pairo-Castineira E, Rawlik K, Pasko D, Walker S, Meynert A, Kousathanas A, Moutsianas L, Tenesa A, Caulfield M, Scott R, Wilson JF, Baillie JK, Butler-Laporte G, Nakanishi T, Lathrop M, Richards JB, Regeneron Genetics Center, UKB Exome Sequencing Consortium, Jones M, Balasubramanian S, Salerno W, Shuldiner AR, Marchini J, Overton JD, Habegger L, Cantor MN, Reid JG, Baras A, Abecasis GR, Ferreira MAR. Pan-ancestry exome-wide association analyses of COVID-19 outcomes in 586,157 individuals. Am J Hum Genet, 2021, 108(7): 1350-1355. |
| [23] |
Liu PH, Fang MY, Luo YX, Zheng F, Jin Y, Cheng FJ, Zhu HH, Jin X. Rare variants in inborn errors of immunity genes associated with COVID-19 severity. Front Cell Infect Microbiol, 2022, 12: 888582.
doi: 10.3389/fcimb.2022.888582 |
| [24] |
Horowitz JE, Kosmicki JA, Damask A, Sharma D, Roberts GHL, Justice AE, Banerjee N, Coignet MV, Yadav A, Leader JB, Marcketta A, Park DS, Lanche R, Maxwell E, Knight SC, Bai XD, Guturu H, Sun D, Baltzell A, Kury FSP, Backman JD, Girshick AR, O'Dushlaine C, McCurdy SR, Partha R, Mansfield AJ, Turissini DA, Li AH, Zhang M, Mbatchou J, Watanabe K, Gurski L, McCarthy SE, Kang HM, Dobbyn L, Stahl E, Verma A, Sirugo G, Regeneron Genetics Center, Ritchie MD, Jones M, Balasubramanian S, Siminovitch K, Salerno WJ, Shuldiner AR, Rader DJ, Mirshahi T, Locke AE, Marchini J, Overton JD, Carey DJ, Habegger L, Cantor MN, Rand KA, Hong EL, Reid JG, Ball CA, Baras A, Abecasis GR, Ferreira MAR. Genome-wide analysis provides genetic evidence that ACE2 influences COVID-19 risk and yields risk scores associated with severe disease. Nat Genet, 2022, 54(4): 382-392.
doi: 10.1038/s41588-021-01006-7 pmid: 35241825 |
| [25] |
Zhao J, Yang Y, Huang HP, Li D, Gu DF, Lu XF, Zhang Z, Liu L, Liu T, Liu YK, He YJ, Sun B, Wei ML, Yang GY, Wang XH, Zhang L, Zhou XY, Xing MZ, Wang PG. Relationship between the ABO blood group and the coronavirus disease 2019(COVID-19) susceptibility. Clin Infect Dis, 2021, 73(2): 328-331.
doi: 10.1093/cid/ciaa1150 |
| [26] | Dendrou CA, Cortes A, Shipman L, Evans HG, Attfield KE, Jostins L, Barber T, Kaur G, Kuttikkatte SB, Leach OA, Desel C, Faergeman SL, Cheeseman J, Neville MJ, Sawcer S, Compston A, Johnson AR, Everett C, Bell JI, Karpe F, Ultsch M, Eigenbrot C, McVean G, Fugger L. Resolving TYK2 locus genotype-to-phenotype differences in autoimmunity. Sci Transl Med, 2016, 8(363): 363ra149. |
| [27] |
Mallen-St Clair J, Pham CTN, Villalta SA, Caughey GH, Wolters PJ. Mast cell dipeptidyl peptidase I mediates survival from sepsis. J Clin Invest, 2004, 113(4): 628-634.
pmid: 14966572 |
| [28] |
Korkmaz B, Lesner A, Marchand-Adam S, Moss C, Jenne DE. Lung protection by cathepsin C inhibition: a new hope for COVID-19 and ARDS? J Med Chem, 2020, 63(22): 13258-13265.
doi: 10.1021/acs.jmedchem.0c00776 |
| [29] |
Mousa M, Vurivi H, Kannout H, Uddin M, Alkaabi N, Mahboub B, Tay GK, Alsafar HS, UAE COVID-19 Collaborative Partnership. Genome-wide association study of hospitalized COVID-19 patients in the United Arab Emirates. EBioMedicine, 2021, 74: 103695.
doi: 10.1016/j.ebiom.2021.103695 |
| [30] |
Pereira AC, Bes TM, Velho M, Marques E, Jannes CE, Valino KR, Dinardo CL, Costa SF, Duarte AJS, Santos AR, Mitne-Neto M, Medina-Pestana J, Krieger JE. Genetic risk factors and COVID-19 severity in Brazil: results from BRACOVID study. Hum Mol Genet, 2022, 31(18): 3021-3031.
doi: 10.1093/hmg/ddac045 pmid: 35368071 |
| [31] |
van der Made CI, Simons A, Schuurs-Hoeijmakers J, van den Heuvel G, Mantere T, Kersten S, van Deuren RC, Steehouwer M, van Reijmersdal SV, Jaeger M, Hofste T, Astuti G, Corominas Galbany J, van der Schoot V, van der Hoeven H, Hagmolen Of Ten Have W, Klijn E, van den Meer C, Fiddelaers J, de Mast Q, Bleeker-Rovers CP, Joosten LAB, Yntema HG, Gilissen C, Nelen M, van der Meer JWM, Brunner HG, Netea MG, van de Veerdonk FL, Hoischen A. Presence of genetic variants among young men with severe COVID-19. JAMA, 2020, 324(7): 663-673.
doi: 10.1001/jama.2020.13719 |
| [32] |
Wainberg M, Sinnott-Armstrong N, Mancuso N, Barbeira AN, Knowles DA, Golan D, Ermel R, Ruusalepp A, Quertermous T, Hao K, Björkegren JLM, Im HK, Pasaniuc B, Rivas MA, Kundaje A. Opportunities and challenges for transcriptome-wide association studies. Nat Genet, 2019, 51(4): 592-599.
doi: 10.1038/s41588-019-0385-z pmid: 30926968 |
| [33] |
Li BL, Ritchie MD. From GWAS to gene: transcriptome- wide association studies and other methods to functionally understand GWAS discoveries. Front Genet, 2021, 12: 713230.
doi: 10.3389/fgene.2021.713230 |
| [34] |
Broekema RV, Bakker OB, Jonkers IH. A practical view of fine-mapping and gene prioritization in the post-genome- wide association era. Open Biol, 2020, 10(1): 190221.
doi: 10.1098/rsob.190221 |
| [35] | Yao Y, Ye F, Li KL, Xu P, Tan WJ, Feng QS, Rao SQ.Genome and epigenome editing identify CCR9 and SLC6A 20 as target genes at the 3p21.31 locus associated with severe COVID-19. Signal Transduct Target Ther, 2021, 6(1): 85. |
| [36] |
Kasela S, Daniloski Z, Bollepalli S, Jordan TX tenOever BR, Sanjana NE, Lappalainen T. Integrative approach identifies SLC6A20 and CXCR6 as putative causal genes for the COVID-19 GWAS signal in the 3p21.31 locus. Genome Biol, 2021, 22(1): 242.
doi: 10.1186/s13059-021-02454-4 pmid: 34425859 |
| [37] | Daniloski Z, Jordan TX, Wessels HH, Hoagland DA, Kasela S, Legut M, Maniatis S, Mimitou EP, Lu L, Geller E, Danziger O, Rosenberg BR, Phatnani H, Smibert P, Lappalainen T tenOever BR, Sanjana NE. Identification of required host factors for SARS-CoV-2 infection in human cells. Cell, 2021, 184(1): 92-105.e16. |
| [38] |
Downes DJ, Cross AR, Hua P, Roberts N, Schwessinger R, Cutler AJ, Munis AM, Brown J, Mielczarek O, de Andrea CE, Melero I, COvid-19 Multi-omics Blood ATlas (COMBAT) Consortium, Gill DR, Hyde SC, Knight JC, Todd JA, Sansom SN, Issa F, Davies JOJ, Hughes JR. Identification of LZTFL1 as a candidate effector gene at a COVID-19 risk locus. Nat Genet, 2021, 53(11): 1606-1615.
doi: 10.1038/s41588-021-00955-3 pmid: 34737427 |
| [39] |
Wei Q, Chen ZH, Wang L, Zhang T, Duan L, Behrens C, Wistuba II, Minna JD, Gao B, Luo JH, Liu ZP. LZTFL1 suppresses lung tumorigenesis by maintaining differentiation of lung epithelial cells. Oncogene, 2016, 35(20): 2655-2663.
doi: 10.1038/onc.2015.328 pmid: 26364604 |
| [40] |
Wang LB, Guo JF, Wang QC, Zhou JC, Xu CP, Teng RY, Chen YX, Wei Q, Liu ZP. LZTFL1 suppresses gastric cancer cell migration and invasion through regulating nuclear translocation of beta-catenin. J Cancer Res Clin Oncol, 2014; 140(12): 1997-2008.
doi: 10.1007/s00432-014-1753-9 |
| [1] | Wenbing Liu, Dan Liu, Jin Yan, Xin Liu, Qianfei Wang. Genetic predisposition in patients with severe COVID-19 [J]. Hereditas(Beijing), 2022, 44(8): 672-681. |
| [2] | Guoqing Qian. Advances in genome-wide association study of chronic obstructive pulmonary disease [J]. Hereditas(Beijing), 2020, 42(9): 832-846. |
| [3] | Li Wang, Yanmei Xu, Zhujun Cheng, Zhaoping Xiong, Libin Deng. The progress of genetics of cholesterol metabolism disorder [J]. HEREDITAS(Beijing), 2014, 36(9): 857-863. |
| [4] | ZHENG Wei JI Lin-Dan XING Wen-Hua TU Wei-Wei XU Jin. Advances in genome-wide association study of tuberculosis [J]. HEREDITAS, 2013, 35(7): 823-829. |
| [5] | YANG Ying-Zhong WANG Ya-Ping MA Lan DU Yang GE Ri-Li. Genome-wide association study of high-altitude pulmonary edema in Han Chinese [J]. HEREDITAS, 2013, 35(11): 1291-1299. |
| [6] | XU Rui-Wei, YAN Wei-Li. Advances in genome-wide association studies on essential hypertension [J]. HEREDITAS, 2012, 34(7): 793-809. |
| [7] | YANG Zhao-Qing, CHU Jia-You. The research progress of human genetic diversity in China [J]. HEREDITAS, 2012, 34(11): 1351-1364. |
| [8] | QUAN Cheng, ZHANG Hua-Jun. Research strategies for the next step of genome-wide association study [J]. HEREDITAS, 2011, 33(2): 100-108. |
| [9] | HAN Jian-Wen, ZHANG Hua-Jun. Current status of genome-wide association study [J]. HEREDITAS, 2011, 33(1): 25-35. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||