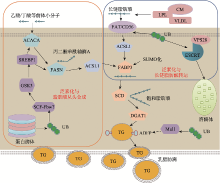
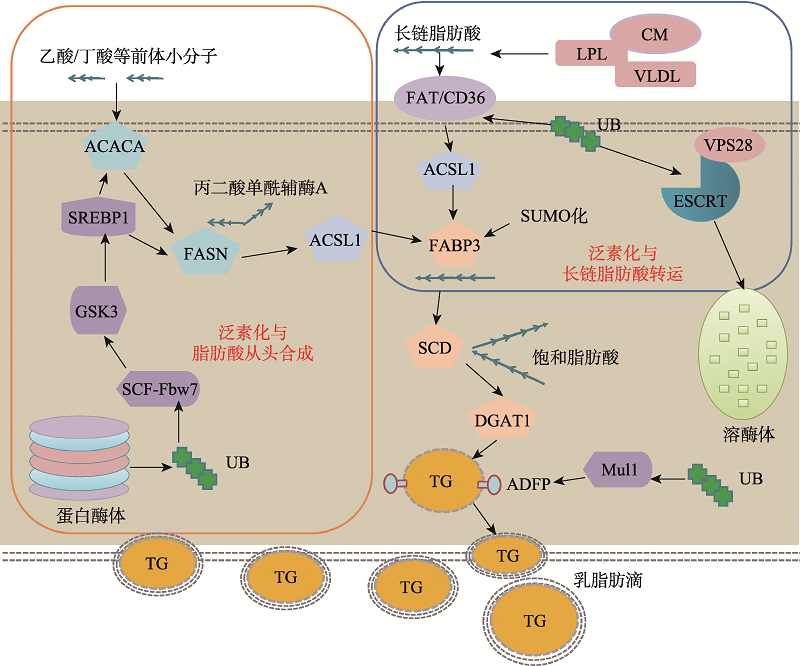
Hereditas(Beijing) ›› 2020, Vol. 42 ›› Issue (6): 548-555.doi: 10.16288/j.yczz.20-037
• Review • Previous Articles Next Articles
The regulation of ubiquitination in milk fat synthesis in bovine
Lili Liu1, Aiwei Guo1, Qingqing Li1,2, Peifu Wu1, Yajin Yang1, Fenfen Chen1, Suhua Li1, Panjiang Guo1, Qin Zhang3
- 1. College of Life Science, Southwest Forestry University, Kunming 650224, China
2. Kunming Xianghao Technology Co. Ltd., Kunming 650204, China
3. College of Animal Science and Technology, Shandong Agricultural University, Tai’an 271000, China
-
Received:2020-02-10Revised:2020-04-15Online:2020-06-20Published:2020-05-07 -
Supported by:Supported by the National Natural Science Foundation of China Nos(31902152);Supported by the National Natural Science Foundation of China Nos(31872327)
Cite this article
Lili Liu, Aiwei Guo, Qingqing Li, Peifu Wu, Yajin Yang, Fenfen Chen, Suhua Li, Panjiang Guo, Qin Zhang. The regulation of ubiquitination in milk fat synthesis in bovine[J]. Hereditas(Beijing), 2020, 42(6): 548-555.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
| [1] | Metin C, Luccardini C . Neuroscience. Ubiquitination inhibits neuronal exit. Science, 2010,330(6012):1754-1755. |
| [2] | Shi Y, Chan DW, Jung SY, Malovannaya A, Wang Y, Qin J. A data set of human endogenous protein ubiquitination sites. Mol Cell Proteomics, 2011, 10(5): M110. 002089. |
| [3] | Liu L, Zhang Q . Identification and functional analysis of candidate gene VPS28 for milk fat in bovine mammary epithelial cells. Biochem Bioph Res Co, 2019,510(4):606-613. |
| [4] | Liu LL, Guo AW, Wu PF, Chen FF, Yang YJ, Zhang Q . Regulation of VPS28 gene knockdown on the milk fat synthesis in Chinese Holstein dairy. Hereditas(Beijing), 2018,40(12):1092-1100. |
| 刘莉莉, 郭爱伟, 吴培福, 陈粉粉, 杨亚晋, 张勤 . 敲降VPS28基因对中国荷斯坦奶牛乳脂合成的调控. 遗传, 2018,40(12):1092-1100. | |
| [5] | Liu L, Jiang L, Ding XD, Liu JF, Zhang. The regulation of glucose on milk fat synthesis is mediated by the ubiquitin- proteasome system in bovine mammary epithelial cells. Biochem Biophys Res Commun, 2015,465(1):59-63. |
| [6] | Yu Y, Hayward GS . The ubiquitin E3 ligase RAUL negatively regulates type i interferon through ubiquitination of the transcription factors IRF7 and IRF3. Immunity, 2010,33(6):863-877. |
| [7] | Strack B, Calistri A, Accola MA, Palù G, Göttlinger HG . A role for ubiquitin ligase recruitment in retrovirus release. Proc Natl Acad Sci USA, 2000,97(24):13063-13068. |
| [8] | Bhoj VG, Chen ZJ . Ubiquitylation in innate and adaptive immunity. Nature, 2009,458(7237):430-437. |
| [9] | Ikeda F, Dikic I . Atypical ubiquitin chains: new molecular signals. ‘Protein Modifications: Beyond the Usual Suspects’ review series. EMBO Rep, 2008,9(6):536-542. |
| [10] | Tanno H, Komada M . The ubiquitin code and its decoding machinery in the endocytic pathway. J Biochem, 2013,153(6):497-504. |
| [11] | Mallette FA, Richard S . K48-linked ubiquitination and protein degradation regulate 53BP1 recruitment at DNA damage sites. Cell Res, 2012,22(8):1221-1223. |
| [12] | Hao ZY, Sheng Y, Duncan GS, Li WY, Dominguez C, Sylvester J, Su YW, Lin GH, Snow BE, Brenner D, You-Ten A, Haight J, Inoue S, Wakeham A, Elford A, Hamilton S, Liang Y, Zúñiga-Pflücker JC, He HH, Ohashi PS, Mak TW . K48-linked KLF4 ubiquitination by E3 ligase Mule controls T-cell proliferation and cell cycle progression. Nat Commun, 2017,8:14003. |
| [13] | He S, Zhang LQ . Research progress in linear ubiquitin modification. Hereditas(Beijing), 2015,37(9):911-917. |
| 何珊, 张令强 . 线性泛素化修饰研究进展. 遗传, 2015,37(9):911-917. | |
| [14] | Tang Y, Tu HL, Zhang J, Zhao XQ, Wang YN, Qin J, Lin X . K63-linked ubiquitination regulates RIPK1 kinase activity to prevent cell death during embryogenesis and inflammation. Nat Commun, 2019,10(1):4157. |
| [15] | Liu CS, Yang-Yen HF, Suen CS, Hwang MJ, Yen JJY . Cbl-mediated K63-linked ubiquitination of JAK2 enhances JAK2 phosphorylation and signal transduction. Sci Rep, 2017,7(1):4613. |
| [16] | Zhang QY, Zhang YZ, Shen K, Zhang SY, Cao JP . Histone ubiquitylation and its roles in DNA damage response. Hereditas (Beijing), 2019,41(1):29-40. |
| 张卿义, 张樱子, 沈凯, 张舒羽, 曹建平 . 组蛋白泛素化修饰及其在DNA损伤应答中的作用. 遗传, 2019,41(1):29-40. | |
| [17] | Ray DM, Rogers BA, Sunman JA, Akiyama SK, Olden K, Roberts JD . Lysine 63-linked ubiquitination is important for arachidonic acid-induced cellular adhesion and migration. Biochem Cell Biol, 2010,88(6):947-956. |
| [18] | Békés M, Okamoto K, Crist SB, Jones MJ, Chapman JR, Brasher BB, Melandri FD, Ueberheide BM, Denchi EL, Huang TT . DUB-resistant ubiquitin to survey ubiquitination switches in mammalian cells. Cell Rep, 2013,5(3):826-838. |
| [19] | Clague MJ . Biochemistry: Oxidation controls the DUB step. Nature, 2013,497(7447):49-50. |
| [20] | Wright MH, Berlin I, Nash PD . Regulation of endocytic sorting by ESCRT-DUB-mediated deubiquitination. Cell Biochem Biophys, 2011,60(1-2):39-46. |
| [21] | Rathaus M, Lerrer B, Cohen HY . DeubiKuitylation: a novel DUB enzymatic activity for the DNA repair protein, Ku70. Cell Cycle, 2009,8(12):1843-1852. |
| [22] | Zhu Y, Carroll M, Papa FR, Hochstrasser M, D'Andrea AD. DUB-1, a deubiquitinating enzyme with growth-suppressing activity. Proc Natl Acad Sci USA, 1996,93(8):3275-3279. |
| [23] | Leestemaker Y, Ovaa H . Tools to investigate the ubiquitin proteasome system. Drug Discov Today Technol, 2017,26:25-31. |
| [24] | Hao YH, Doyle JM, Ramanathan S, Gomez TS, Jia D, Xu M, Chen ZJ, Billadeau DD, Rosen MK, Potts PR . Regulation of WASH-dependent actin polymerization and protein trafficking by ubiquitination. Cell, 2013,152(5):1051-1064. |
| [25] | Nakayama KI, Nakayama K . Ubiquitin ligases: cell-cycle control and cancer. Nat Rev Cancer, 2006,6(5):369-381. |
| [26] | Malynn BA, Ma A . Ubiquitin makes its mark on immune regulation. Immunity, 2010,33(6):843-852. |
| [27] | Chen K, Cheng HH, Zhou RJ . Molecular mechanisms and functions of autophagy and the ubiq-uitin-proteasome pathway. Hereditas(Beijing), 2012,34(1):5-18. |
| 陈科, 程汉华, 周荣家 . 自噬与泛素化蛋白降解途径的分子机制及其功能. 遗传, 2012,34(1):5-18. | |
| [28] | Pickart CM . Mechanisms underlying ubiquitination. Annu Rev Biochem, 2001,70:503-533. |
| [29] | Villarroya-Beltri C, Baixauli F, Mittelbrunn M, Fernández- Delgado I, Torralba D, Moreno-Gonzalo O, Baldanta S, Enrich C, Guerra S, Sánchez-Madrid F . ISGylation controls exosome secretion by promoting lysosomal degradation of MVB proteins. Nat Commun, 2016,7:13588. |
| [30] | Pohl C, Dikic I . Cellular quality control by the ubiquitin- proteasome system and autophagy. Science, 2019,366(6467):818-822. |
| [31] | Smalle J, Vierstra RD . The ubiquitin 26S proteasome proteolytic pathway. Annu Rev Plant Biol, 2004,55:555-590. |
| [32] | Bard JAM, Goodall EA, Greene ER, Jonsson E, Dong KC, Martin A . Structure and Function of the 26S Proteasome. Annu Rev Biochem, 2018,87:697-724. |
| [33] | Chen YJ, Wu H, Shen XZ . The ubiquitin-proteasome system and its potential application in hepatocellular carcinoma therapy. Cancer Lett, 2016,379(2):245-252. |
| [34] | Dong LH, Ran ML, Li Z, Peng FZ, Chen B . The role of ubiquitin-proteasome pathway in spermatogenesis. Hereditas (Beijing), 2016,38(9):791-800. |
| 董莲花, 冉茂良, 李智, 彭馥芝, 陈斌 . 泛素-蛋白酶体途径在精子生成中的作用. 遗传, 2016,38(9):791-800. | |
| [35] | Ross JM, Olson L, Coppotelli G . Mitochondrial and ubiquitin proteasome system dysfunction in ageing and disease: two sides of the same coin. Int J Mol Sci, 2015,16(8):19458-19476. |
| [36] | Garrido C, Brunet M, Didelot C, Zermati Y, Schmitt E, Kroemer G . Heat shock proteins 27 and 70: anti-apoptotic proteins with tumorigenic properties. Cell Cycle, 2006,5(22):2592-2601. |
| [37] | Saksena S, Sun J, Chu T, Emr SD . ESCRTing proteins in the endocytic pathway. Trends Biochem Sci, 2007,32(12):561-573. |
| [38] | Odorizzi G . ESCRTs take on a job in surveillance. Cell, 2014,159(2):240-241. |
| [39] | Shields SB, Piper RC . How ubiquitin functions with ESCRTs. Traffic, 2011,12(10):1306-1317. |
| [40] | Gutierrez MG, Carlton JG . ESCRTs offer repair service. Science, 2018,360(6384):33-34. |
| [41] | Majumder P, Chakrabarti O . ESCRTs and associated proteins in lysosomal fusion with endosomes and autophagosomes. Biochem Cell Biol, 2016,94(5):443-450. |
| [42] | Radulovic M, Stenmark H . ESCRTs in membrane sealing. Biochem Soc Trans, 2018,46(4):773-778. |
| [43] | Zhang S, Yu CM, Yin Y, Chen W . Research progress of the role of autophagy in infectious diseases. Bull Acad Mil Med Sci, 2009,33(5):469-472. |
| 章晟, 于长明, 殷瑛, 陈薇 . 细胞自噬在病原体感染过程中的作用研究进展. 军事医学科学院院刊, 2009,33(5):469-472. | |
| [44] | Espert L, Codogno P, Biard-Piechaczyk M . Involvement of autophagy in viral infections: antiviral function and subversion by viruses. J Mol Med, 2007,85(8):811-823. |
| [45] | Walther TC, Farese RV . The life of lipid droplets. Biochim Biophys Acta, 2009,1791(6):459-466. |
| [46] | Bionaz M, Loor JJ . Gene networks driving bovine milk fat synthesis during the lactation cycle. BMC Genomics, 2008,9:366. |
| [47] | Reinhardt TA, Lippolis JD . Bovine milk fat globule membrane proteome. J Dairy Res, 2006,73(4):406-416. |
| [48] | Doege H, Stahl A . Protein-mediated fatty acid uptake: novel insights from in vivo models. Physiology, 2006,21:259-268. |
| [49] | Bionaz M, Loor JJ . ACSL1, AGPAT6, FABP3, LPIN1, and SLC27A6 are the most abundant isoforms in bovine mammary tissue and their expression is affected by stage of lactation. J Nutr, 2008,138(6):1019-1024. |
| [50] | Smith J, Su X, El-Maghrabi R, Stahl PD, Abumrad NA . Opposite regulation of CD36 ubiquitination by fatty acids and insulin: effects on fatty acid uptake. J Biol Chem, 2008,283(20):13578-13585. |
| [51] | Kan CF, Singh AB, Stafforini DM, Azhar S, Liu J . Arachidonic acid downregulates acyl-CoA synthetase 4 expression by promoting its ubiquitination and proteasomal degradation. J Lipid Res, 2014,55(8):1657-1667. |
| [52] | Chung SS, Ahn BY, Kim M, Kho JH, Jung HS, Park KS . SUMO modification selectively regulates transcriptional activity of peroxisome-proliferator-activated receptor γ in C2C12 myotubes. Biochem J, 2011,433(1):155-161. |
| [53] | Bernard L, Leroux C, Chilliard Y . Expression and nutritional regulation of lipogenic genes in the ruminant lactating mammary gland. Adv Exp Med Biol, 2008,606:67-108. |
| [54] | Roy R, Ordovas L, Zaragoza P, Romero A, Moreno C, Altarriba J, Rodellar C . Association of polymorphisms in the bovine FASN gene with milk-fat content. Anim Genet, 2006,37(3):215-218. |
| [55] | Tao H, Chang GJ, Xu TL, Zhao HJ, Zhang K, Shen XZ . Feeding a high concentrate diet down-regulates expression of ACACA, LPL and SCD and modifies milk composition in lactating goats. PLoS One, 2015,10(6):e0130525. |
| [56] | McManaman JL, Zabaronick W, Schaack J, Orlicky DJ . Lipid droplet targeting domains of adipophilin. J Lipid Res, 2003,44(4):668-673. |
| [57] | Heid HW, Moll R, Schwetlick I, Rackwitz HR, Keenan TW . Adipophilin is a specific marker of lipid accumulation in diverse cell types and diseases. Cell Tissue Res, 1998,294(2):309-321. |
| [58] | Heid HW, Keenan TW . Intracellular origin and secretion of milk fat globules. Eur J Cell Biol, 2005,84(2-3):245-258. |
| [59] | McManaman JL, Russell TD, Schaack J, Orlicky DJ, Robenek H . Molecular determinants of milk lipid secretion. J Mammary Gland Biol Neoplasia, 2007,12(4):259-268. |
| [60] | Lopez JM, Bennett MK, Sanchez HB, Rosenfeld JM, Osborne TF . Sterol regulation of acetyl coenzyme A carboxylase: a mechanism for coordinate control of cellular lipid. Proc Natl Acad Sci USA, 1996,93(3):1049-1053. |
| [61] | Rudolph MC, Monks J, Burns V, Phistry M, Marians R, Foote MR, Bauman DE, Anderson SM, Neville MC . Sterol regulatory element binding protein and dietary lipid regulation of fatty acid synthesis in the mammary epithelium. Am J Physiol Endocrinol Metab, 2010,299(6):E918-E927. |
| [62] | Punga T, Bengoechea-Alonso MT, Ericsson J . Phosphorylation and ubiquitination of the transcription factor sterol regulatory element-binding protein-1 in response to DNA binding. J Biol Chem, 2006,281(35):25278-25286. |
| [63] | Bengoechea-Alonso MT, Ericsson J . A phosphorylation cascade controls the degradation of active SREBP1. J Biol Chem, 2009,284(9):5885-5895. |
| [64] | Han YM, Hu ZM, Cui AY, Liu ZS, Ma FG, Xue YQ, Liu YX, Zhang FF, Zhao ZH, Yu YY, Gao J, Wei C, Li JY, Fang J, Li J, Fan JG, Song BL, Li Y . Post-translational regulation of lipogenesis via AMPK-dependent phosphorylation of insulin-induced gene. Nat commun, 2019,10(1):623. |
| [65] | Yuan YJ, Li XZ, Xu YY, Zhao HB, Su ZM, Lai DH, Yang WQ, Chen SX, He YZ, Li X, Liu LY, Xu GB . Mitochondrial E3 ubiquitin ligase 1 promotes autophagy flux to suppress the development of clear cell renal cell carcinomas. Cancer Sci, 2019,110(11):3533-3542. |
| [1] | Lianhua Dong, Maoliang Ran, Zhi Li, Fuzhi Peng, Bin Chen. The role of ubiquitin-proteasome pathway in spermatogenesis [J]. Hereditas(Beijing), 2016, 38(9): 791-800. |
| [2] | WU Shen LU Tao-Feng WU Shuang GUAN Wei-Jun. Research progress in G protein-coupled receptor 40 [J]. HEREDITAS, 2013, 35(1): 27-34. |
| [3] | LU Liang LI Dong HE Fu-Chu. Bioinformatics advances in protein ubiquitination [J]. HEREDITAS, 2013, 35(1): 17-26. |
| [4] | YIN Yan-Hui, SUN Min, CHEN Ting-Feng, ZHANG Ya-Ni, ZHU Cai-Ye, LI Wei, LI Bi-Chun. Generation of goat H-FABP overexpression transgenic mice by testicular injection [J]. HEREDITAS, 2012, 34(6): 727-735. |
| [5] | FU Ai-Sai, LIU Ran, SHU Jing, LIU Tian-Gang. Genetic engineering of microbial metabolic pathway for production of advanced biodiesel [J]. HEREDITAS, 2011, 33(10): 1121-1133. |
| [6] | CAO Yang-Ping, DAN Jin-Lei, LI Zhong, MENG Feng. Isolation of OsFAD2, OsFAD6 and FAD family members response to abiotic stresses in Oryza sativa L. [J]. HEREDITAS, 2010, 32(8): 839-847. |
| [7] | NI Yu, GUO Yan-Jun. Progress in the study on genes encoding enzymes involved in bio-synthesis of very long chain fatty acids and cuticular wax in plants [J]. HEREDITAS, 2008, 30(5): 561-567. |
| [8] | ZHU Gui-Ming, CHEN Hong, LU Jian-Shen, ZHOU Yan-Rong, WU Xiao-Jie, CHEN Hong-Xing, DENG Ji-Xian. Synthesis of Total sFat-1 Gene by PCR-Restrict Enzyme Ligation Method [J]. HEREDITAS, 2006, 28(9): 1123-1128. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||