Hereditas(Beijing) ›› 2021, Vol. 43 ›› Issue (1): 66-73.doi: 10.16288/j.yczz.20-245
• Review • Previous Articles Next Articles
Progress on the GntR family transcription regulators in bacteria
Guofang Liu1, Xinxin Wang2, Huizhao Su2, Guangtao Lu2( )
)
- 1. Guangxi Key Laboratory for Polysaccharide Materials and Modifications,School of Marine Sciences and Biotechnology, Guangxi University for Nationalities, Nanning 530008, China
2. College of Life Science and Technology, Guangxi University, Nanning 530004, China
-
Received:2020-09-18Revised:2020-12-07Online:2021-01-20Published:2020-12-10 -
Contact:Lu Guangtao E-mail:lugt@gxu.edu.cn -
Supported by:Supported by Guangxi University for Nationalities Scientific Research Fund No(2019KJQN004);Science and Technology Major Project of Guangxi No(AA18242026)
Cite this article
Guofang Liu, Xinxin Wang, Huizhao Su, Guangtao Lu. Progress on the GntR family transcription regulators in bacteria[J]. Hereditas(Beijing), 2021, 43(1): 66-73.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
| [1] |
Majidian P, Kuse J, Tanaka K, Najafi H, Zeinalabedini M, Takenaka S, Yoshida KI . Bacillus subtilis GntR regulation modified to devise artificial transient induction systems. J Gen Appl Microbiol, 2017,62(6):277-285.
doi: 10.2323/jgam.2016.05.004 pmid: 27829583 |
| [2] |
Pabo CO, Sauer RT . Transcription factors: structural families and principles of DNA recognition. Annu Rev Biochem, 1992,61:1053-1095.
doi: 10.1146/annurev.bi.61.070192.005201 pmid: 1497306 |
| [3] |
Haydon DJ, Guest JR . A new family of bacterial regulatory proteins. FEMS Microbiol Lett, 1991,63(2-3):291-295.
doi: 10.1016/0378-1097(91)90101-f pmid: 2060763 |
| [4] |
Fujita Y, Fujita R . New diagnostic systems--technics, efficiency and limitations. Cholangioscopy. a) Peroral cholangioscopy. Nihon Rinsho, 1987,45(7):1466-1471.
pmid: 3669339 |
| [5] |
Hoskisson PA, Rigali S . Chapter 1: Variation in form and function the helix-turn-helix regulators of the GntR superfamily. Adv Appl Microbiol, 2009,69:1-22.
doi: 10.1016/S0065-2164(09)69001-8 pmid: 19729089 |
| [6] |
Suvorova IA, Korostelev YD, Gelfand MS . GntR family of bacterial transcription factors and their DNA binding motifs: structure, positioning and co-evolution. PLoS One, 2015,10(7):e0132618.
doi: 10.1371/journal.pone.0132618 pmid: 26151451 |
| [7] |
Vindal V, Suma K, Ranjan A . GntR family of regulators in Mycobacterium smegmatis: a sequence and structure based characterization. BMC Genomics, 2007,8:289.
doi: 10.1186/1471-2164-8-289 pmid: 17714599 |
| [8] |
Zheng MY, Cooper DR, Grossoehme NE, Yu MM, Hung LW, Cieslik M, Derewenda U, Lesley SA, Wilson IA, Giedroc DP, Derewenda ZS . Structure of Thermotoga maritima TM0439: implications for the mechanism of bacterial GntR transcription regulators with Zn 2+-binding FCD domains. Acta Crystallogr D Biol Crystallogr , 2009,65(Pt 4):356-365.
doi: 10.1107/S0907444909004727 pmid: 19307717 |
| [9] |
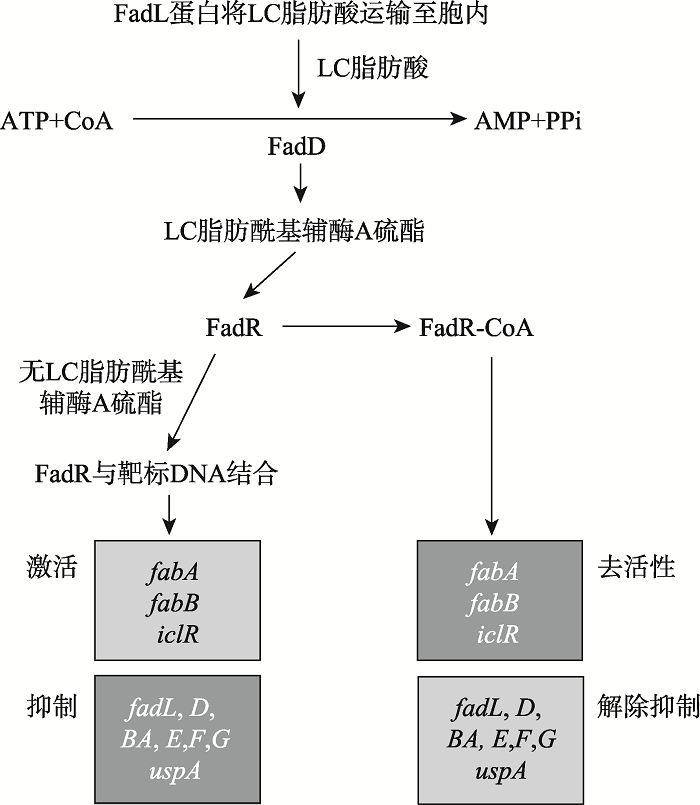
van Aalten DM, DiRusso CC, Knudsen J, Wierenga RK . Crystal structure of FadR, a fatty acid-responsive transcription factor with a novel acyl coenzyme A-binding fold. EMBO J, 2000,19(19):5167-5177.
doi: 10.1093/emboj/19.19.5167 pmid: 11013219 |
| [10] |
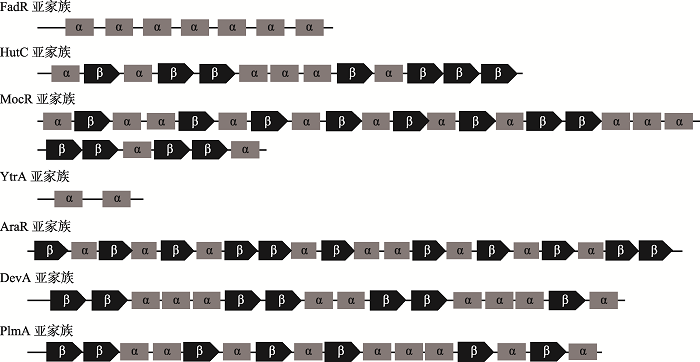
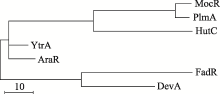
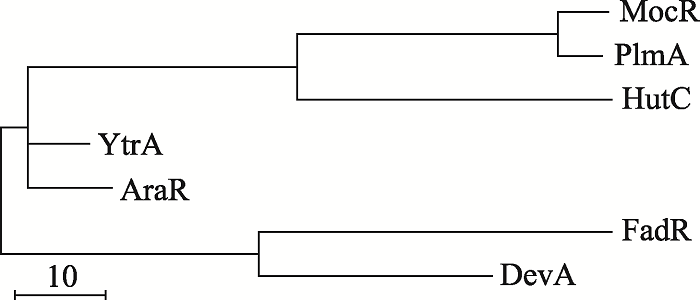
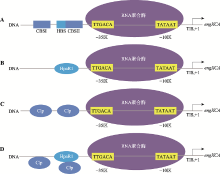
Rigali S, Derouaux A, Giannotta F, Dusart J . Subdivision of the helix-turn-helix GntR family of bacterial regulators in the FadR, HutC, MocR, and YtrA subfamilies. J Biol Chem, 2002,277(15):12507-12515.
doi: 10.1074/jbc.M110968200 pmid: 11756427 |
| [11] |
Allison SL, Phillips AT . Nucleotide sequence of the gene encoding the repressor for the histidine utilization genes of Pseudomonas putida. J Bacteriol, 1990,172(9):5470-5476.
doi: 10.1128/jb.172.9.5470-5476.1990 pmid: 2203753 |
| [12] |
Fillenberg SB, Friess MD, Körner S, Böckmann RA, Muller YA . Crystal structures of the global regulator DasR from Streptomyces coelicolor: implications for the allosteric regulation of GntR/HutC repressors. PLoS One, 2016,11(6):e0157691.
doi: 10.1371/journal.pone.0157691 pmid: 27337024 |
| [13] |
Milano T, Angelaccio S, Tramonti A, Di Salvo ML, Contestabile R, Pascarella S . Structural properties of the linkers connecting the N- and C- terminal domains in the MocR bacterial transcriptional regulators. Biochim Open, 2016,3:8-18.
doi: 10.1016/j.biopen.2016.07.002 pmid: 29450126 |
| [14] |
Gao YG, Yao M, Itou H, Zhou Y, Tanaka I . The structures of transcription factor CGL2947 from Corynebacterium glutamicum in two crystal forms: A novel homodimer assembling and the implication for effector-binding mode. Protein Sci, 2007,16(9):1878-1886.
doi: 10.1110/ps.072976907 pmid: 17766384 |
| [15] |
Yoshida KI, Fujita Y, Ehrlich SD . An operon for a putative ATP-binding cassette transport system involved in acetoin utilization of Bacillus subtilis. J Bacteriol, 2000,182(19):5454-5461.
doi: 10.1128/jb.182.19.5454-5461.2000 pmid: 10986249 |
| [16] |
Gu D, Meng HM, Li Y, Ge HJ, Jiao XN . A GntR family transcription factor (VPA1701) for swarming motility and colonization of Vibrio parahaemolyticus. Pathogens, 2019,8(4):235.
doi: 10.3390/pathogens8040235 |
| [17] |
Hoskisson PA, Rigali S, Fowler K, Findlay KC, Buttner MJ . DevA, a GntR-like transcriptional regulator required for development in Streptomyces coelicolor. J Bacteriol, 2006,188(14):5014-5023.
doi: 10.1128/JB.00307-06 pmid: 16816174 |
| [18] |
Daddaoua A, Corral-Lugo A, Ramos JL, Krell T . Identification of GntR as regulator of the glucose metabolism in Pseudomonas aeruginosa. Environ Microbiol, 2017,19(9):3721-3733.
doi: 10.1111/1462-2920.13871 pmid: 28752954 |
| [19] |
Truong-Bolduc QC, Hooper DC . The transcriptional regulators NorG and MgrA modulate resistance to both quinolones and beta-lactams in Staphylococcus aureus. J Bacteriol, 2007,189(8):2996-3005.
doi: 10.1128/JB.01819-06 pmid: 17277059 |
| [20] |
Li ZQ, Wang SL, Zhang H, Zhang JL, Xi L, Zhang JB, Chen CF . Transcriptional regulator GntR of Brucella abortus regulates cytotoxicity, induces the secretion of inflammatory cytokines and affects expression of the type IV secretion system and quorum sensing system in macrophages. World J Microbiol Biotechnol, 2017,33(3):60.
doi: 10.1007/s11274-017-2230-9 pmid: 28243986 |
| [21] |
Zhou XF, Yan Q, Wang N . Deciphering the regulon of a GntR family regulator via transcriptome and ChIP-exo analyses and its contribution to virulence in Xanthomonas citri. Mol Plant Pathol, 2017,18(2):249-262.
doi: 10.1111/mpp.12397 pmid: 26972728 |
| [22] |
Zhou Y, Nie RN, Liu XY, Kong JH, Wang XH, Li JQ . GntR is involved in the expression of virulence in strain Streptococcus suis P1/7. FEMS Microbiol Lett, 2018,365(14).
doi: 10.1093/femsle/fny052 pmid: 29514248 |
| [23] |
Gao RS, Li DF, Lin Y, Lin JX, Xia XY, Wang H, Bi LJ, Zhu J, Hassan B, Wang SH, Feng YJ . Structural and functional characterization of the FadR regulatory protein from Vibrio alginolyticus. Front Cell Infect Microbiol, 2017,7:513.
doi: 10.3389/fcimb.2017.00513 pmid: 29312893 |
| [24] |
Huang WL, Wilks A . A rapid seamless method for gene knockout in Pseudomonas aeruginosa. BMC Microbiol, 2017,17(1):199.
doi: 10.1186/s12866-017-1112-5 pmid: 28927382 |
| [25] |
Wang TT, Qi YH, Wang ZH, Zhao JR, Ji LX, Li J, Cai Z, Yang L, Wu M, Liang HH . Coordinated regulation of anthranilate metabolism and bacterial virulence by the GntR family regulator MpaR in Pseudomonas aeruginosa. Mol Microbiol, 2020,114(5):857-869.
doi: 10.1111/mmi.14584 pmid: 32748556 |
| [26] |
Li ZB, Xiang ZT, Zeng JM, Li YQ, Li JY . A GntR family transcription factor in Streptococcus mutans regulates biofilm formation and expression of multiple sugar transporter genes. Front Microbiol, 2019,9:3224.
doi: 10.3389/fmicb.2018.03224 pmid: 30692967 |
| [27] |
Wu KF, Xu HM, Zheng YQ, Wang LB, Zhang XM, Yin YB . CpsR, a GntR family regulator, transcriptionally regulates capsular polysaccharide biosynthesis and governs bacterial virulence in Streptococcus pneumoniae. Sci Rep, 2016,6:29255.
doi: 10.1038/srep29255 pmid: 27386955 |
| [28] |
Truong-Bolduc QC, Dunman PM, Eidem T, Hooper DC . Transcriptional profiling analysis of the global regulator NorG, a GntR-Like protein of Staphylococcus aureus. J Bacteriol, 2011,193(22):6207-6214.
doi: 10.1128/JB.05847-11 |
| [29] |
An SQ, Lu GT, Su HZ, Li RF, He YQ, Jiang BL, Tang DJ, Tang JL . Systematic mutagenesis of all predicted gntR genes in Xanthomonas campestris pv. campestris reveals a GntR family transcriptional regulator controlling hypersensitive response and virulence. Mol Plant Microbe Interact, 2011,24(9):1027-1039.
doi: 10.1094/MPMI-08-10-0180 pmid: 21615202 |
| [30] |
Su HZ, Wu L, Qi YH, Liu GF, Lu GT, Tang JL . Characterization of the GntR family regulator HpaR1 of the crucifer black rot pathogen Xanthomonas campestris pathovar campestris. Sci Rep, 2016,6:19862.
doi: 10.1038/srep19862 pmid: 26818230 |
| [31] |
Liu GF, Su HZ, Sun HY, Lu GT, Tang JL . Competitive control of endoglucanase gene engXCA expression in the plant pathogen Xanthomonas campestris by the global transcriptional regulators HpaR1 and Clp. Mol Plant Pathol, 2019,20(1):51-68.
doi: 10.1111/mpp.12739 pmid: 30091270 |
| [32] |
Zhou XF, Yan Q, Wang N . Deciphering the regulon of a GntR family regulator via transcriptome and ChIP-exo analyses and its contribution to virulence in Xanthomonas citri. Mol Plant Pathol, 2017,18(2):249-262.
doi: 10.1111/mpp.12397 pmid: 26972728 |
| [33] |
Chi WJ . Retracted: DasR, a GntR-family global regulator, regulates N-acetylglucosamine metabolism in Streptomyces griseus. J Microbiol Biotechnol, 2017.
doi: 10.4014/jmb.2011.11029 pmid: 33263336 |
| [34] |
Taw MN, Lee HI, Lee SH, Chang WS . Characterization of MocR, a GntR-like transcriptional regulator, in Bradyrhizobium japonicum: its impact on motility, biofilm formation, and soybean nodulation. J Microbiol, 2015,53(8):518-525.
doi: 10.1007/s12275-015-5313-z pmid: 26224454 |
| [35] |
Browning DF, Busby SJ . The regulation of bacterial transcription initiation. Nat Rev Microbiol, 2004,2(1):57-65.
doi: 10.1038/nrmicro787 pmid: 15035009 |
| [1] | Chang Lu, Yinhua Huang. Progress in long non-coding RNAs in animals [J]. Hereditas(Beijing), 2017, 39(11): 1054-1065. |
| [2] | Huanping Zhang, Tongming Yin. Advances in lineage-specific genes [J]. HEREDITAS(Beijing), 2015, 37(6): 544-553. |
| [3] | Xiaoqing Huang,Dandan Li,Juan Wu. Long non-coding RNAs in plants [J]. HEREDITAS(Beijing), 2015, 37(4): 344-359. |
| [4] | Yanli Yu, Yanjiao Li, Kaiyuan Pang, Fajun Zhang, Qi Sun, Wencai Li, Zhaodong Meng. Structure and biological functions of plant FKBP family [J]. HEREDITAS(Beijing), 2014, 36(6): 536-546. |
| [5] | WANG Hong LI Gang-Bo ZHANG Da-Yong LIN Jing SHENG Bao-Long CHANG You-Hong. Biological functions of HD-Zip transcription factors [J]. HEREDITAS, 2013, 35(10): 1179-1188. |
| [6] | ZHANG Ji-Yu, WANG Qing-Ju, GUO Zhong-Ren. Progresses on plant AP2/ERF transcription factors [J]. HEREDITAS, 2012, 34(7): 835-847. |
| [7] | SHI Ya-Li, ZHANG Rui, LIN Qin, GUO San-Dui. Biological function of the Somatic embryogenesis receptor-like kinases in plant [J]. HEREDITAS, 2012, 34(5): 551-559. |
| [8] | LUO Jun-Ling ZHAO Na LU Chang-Ming. Plant Trihelix transcription factors family [J]. HEREDITAS, 2012, 34(12): 1551-1560. |
| [9] | TANG Qing-Qiu, JIANG Jing-Yan, YANG Chu-Fen, JU Xiao-Ting, DONG Xiao-Yang. Research and development of Lipin family [J]. HEREDITAS, 2010, 32(10): 981-993. |
| [10] | FENG Hua, CHEN Chen, WANG Yi-Qin, QIU Jin-Long, CHU Cheng-Cai, DU Xi-Hua. Plant SNAREs and their biological functions [J]. HEREDITAS, 2009, 31(5): 471-478. |
| [11] | E Zhi-Guo, Wang Lei. Advance on the cloning and functional analysis of disease resistance genes in rice [J]. HEREDITAS, 2009, 31(10): 999-1005. |
| [12] | YU Wen-Bo, JIANG Song-Min, YU Long. Research progresses on septin family [J]. HEREDITAS, 2008, 30(9): 1107-1107. |
| [13] | WANG Yan, XU Heng-Yong, ZHU Qing. Progress in the study on mammalian diacylgycerol acyltransgerase (DGAT) gene and its biological function [J]. HEREDITAS, 2007, 29(10): 1167-1167―1172. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||