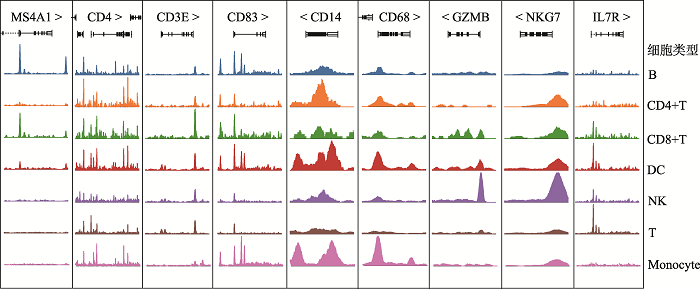
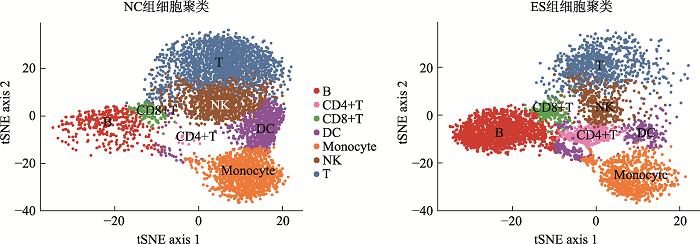
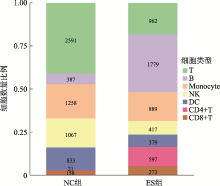
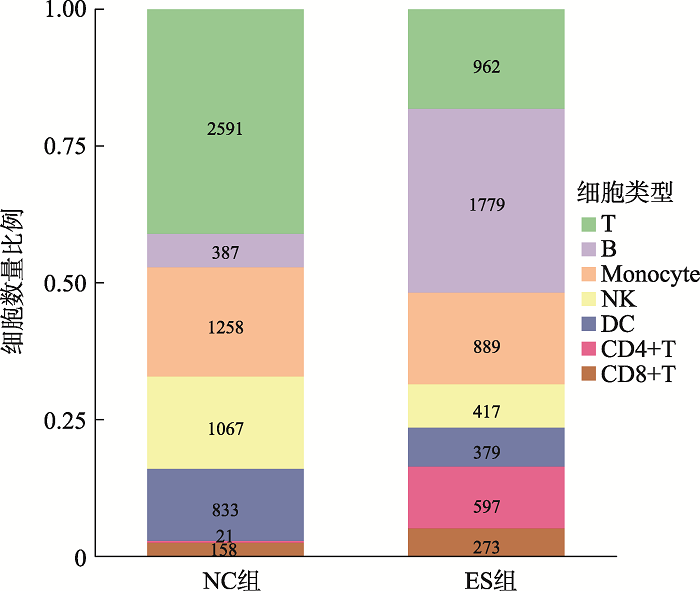
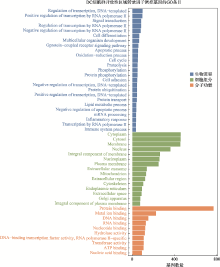
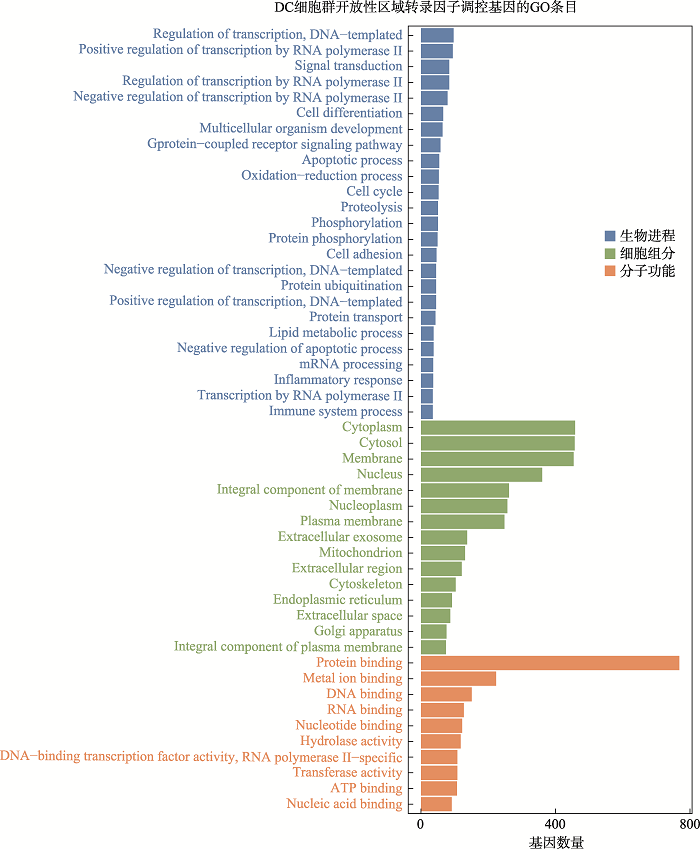
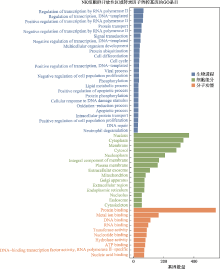
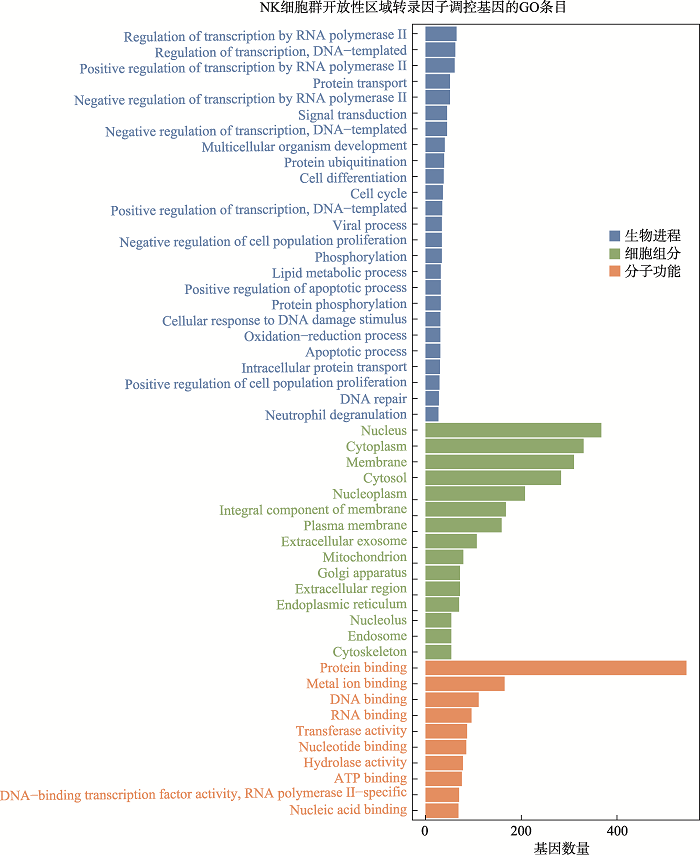
Hereditas(Beijing) ›› 2021, Vol. 43 ›› Issue (1): 74-83.doi: 10.16288/j.yczz.20-283
• Research Article • Previous Articles Next Articles
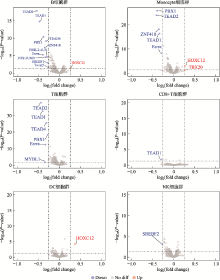
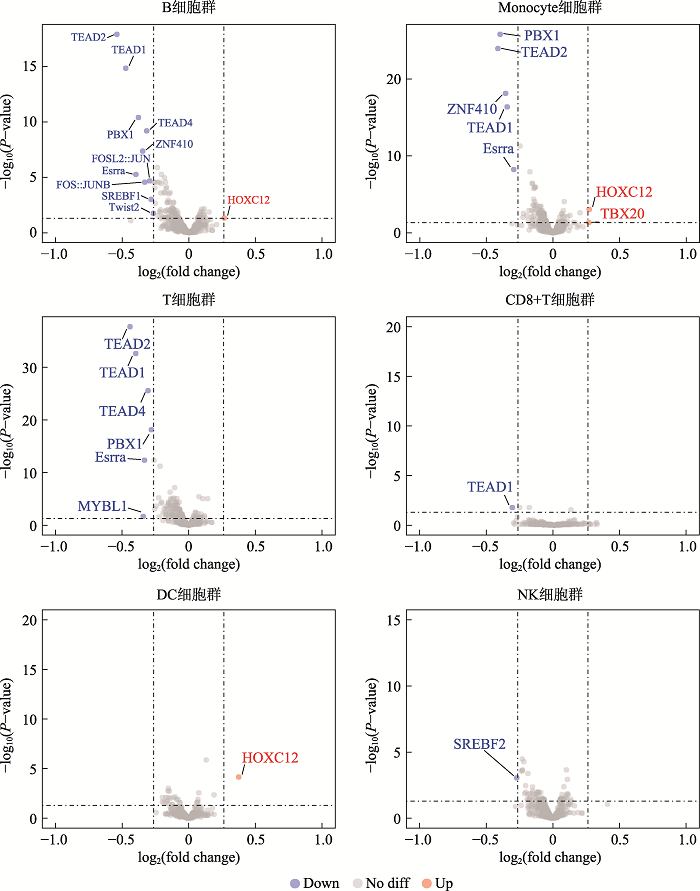
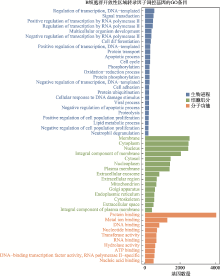
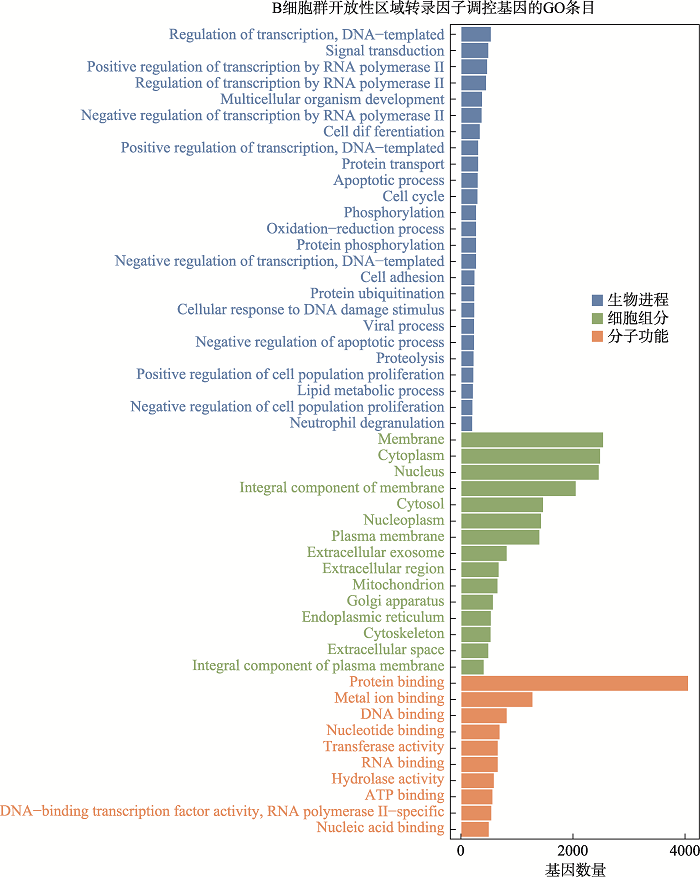
Analysis of transcription factors in accessible open chromatin in the 18-trisomy syndrome based on single cell ATAC sequencing technique
Xiaofen Qiu1,2,3, Dong’e Tang3, Haiyan Yu3, Qiuyan Liao3, Zhiyang Hu3, Jun Zhou3, Xin Zhao3, Huiyan He3, Zhuojian Liang3, Chengming Xu2, Ming Yang1,2( ), Yong Dai3(
), Yong Dai3( )
)
- 1. College of Life Science, Guangxi Normal University, Guilin 541006, China
2. Guangxi Key Laboratory of Metabolic Diseases Research, Department of Clinical Laboratory of Guilin No.924 Hospital, Guilin 541002, China
3. Clinical Medical Research Center, Guangdong Provincial Engineering Research Center of Autoimmune Disease Precision Medicine, Shenzhen Engineering Research Center of Autoimmune Disease, The Second Clinical Medical College of Jinan University, The First Affiliated Hospital of Southern University of Science and Technology, Shenzhen People’s Hospital, Shenzhen 518020, China;
-
Received:2020-11-08Revised:2020-12-24Online:2021-01-20Published:2021-01-04 -
Contact:Yang Ming,Dai Yong E-mail:yangming181@yeah.net;daiyong22@aliyun.com -
Supported by:Supported by Science and Technology Planning Project of Guangdong Province, China(2017B020209001);the Science and Technology Plan of Shenzhen No(GJHZ20180413181714541);Guangxi Key Laboratory of Metabolic Diseases Research No(20-065-76);the Scientific Research and Technology Development Planning Project of Guilin No(2016012702-1)
Cite this article
Xiaofen Qiu, Dong’e Tang, Haiyan Yu, Qiuyan Liao, Zhiyang Hu, Jun Zhou, Xin Zhao, Huiyan He, Zhuojian Liang, Chengming Xu, Ming Yang, Yong Dai. Analysis of transcription factors in accessible open chromatin in the 18-trisomy syndrome based on single cell ATAC sequencing technique[J]. Hereditas(Beijing), 2021, 43(1): 74-83.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
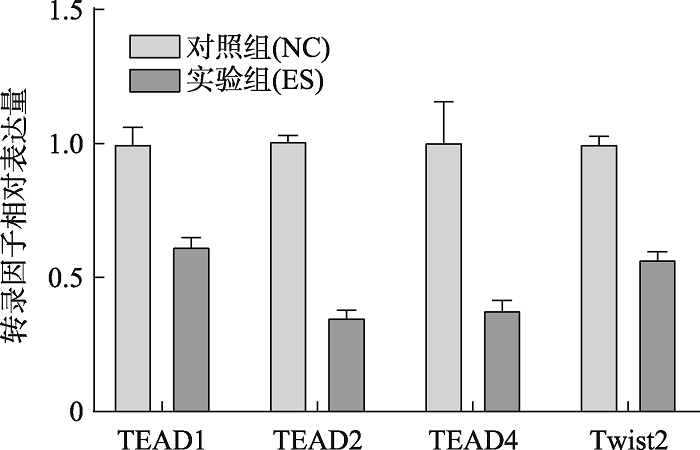
Table 2
Primer sequences of qPCR"
| 名称 | 序列(5'→ 3') | 碱基数 |
|---|---|---|
| H-TEAD1-F | GCCACTGCCATTCATAACAAGC | 22 |
| H-TEAD1-R | CCTGGCTGCCCTGTTTGAATC | 21 |
| H-TEAD2-F | CCATTCTCACAGACACCGTTCAC | 23 |
| H-TEAD2-R | TCCACGAAGGCTGAGAACTCTAC | 23 |
| H-TEAD4-F | GACACGTACAACAAGCACCTG | 21 |
| H-TEAD4-R | CCGTTCGAAGAGATCCTTGAGTC | 23 |
| H-Twist2-F | CCCTCTGACAAGCTGAGCAAG | 21 |
| H-Twist2-R | CATGCGCCACACGGAGAAG | 19 |
| [1] |
Edwards JH, Harnden DG, Cameron AH, Crosse VM, Wolff OH . A new trisomic syndrome. Lancet, 1960,1(7128):787-790.
doi: 10.1016/s0140-6736(60)90675-9 pmid: 13819419 |
| [2] |
Smith DW, Patau K, Therman E, Inhorn SL . A new autosomal trisomy syndrome: multiple congenital anomalies caused by an extra chromosome. J Pediatr, 1960,57:338-345.
doi: 10.1016/s0022-3476(60)80241-7 pmid: 13831938 |
| [3] |
Cereda A, Carey JC . The trisomy 18 syndrome. Orphanet J Rare Dis, 2012,7:81.
doi: 10.1186/1750-1172-7-81 pmid: 23088440 |
| [4] |
Cavadino A, Morris JK . Revised estimates of the risk of fetal loss following a prenatal diagnosis of trisomy 13 or trisomy 18. Am J Med Genet A, 2017,173(4):953-958.
doi: 10.1002/ajmg.a.38123 pmid: 28328132 |
| [5] |
Yamanaka M, Setoyama T, Igarashi Y, Kurosawa K, Itani Y, Hashimoto S, Saitoh K, Takei M, Hirabuki T . Pregnancy outcome of fetuses with trisomy 18 identified by prenatal sonography and chromosomal analysis in a perinatal center. Am J Med Genet A, 2006,140(11):1177-1182.
doi: 10.1002/ajmg.a.31241 pmid: 16652360 |
| [6] |
Rasmussen SA, Wong LYC, Yang QH, May KM, Friedman JM . Population-based analyses of mortality in trisomy 13 and trisomy 18. Pediatrics, 2003,111(4 Pt 1):777-784.
doi: 10.1542/peds.111.4.777 pmid: 12671111 |
| [7] |
Nelson KE, Rosella LC, Mahant S, Guttmann A . Survival and surgical interventions for children with trisomy 13 and 18. Jama, 2016,316(4):420-428.
doi: 10.1001/jama.2016.9819 pmid: 27458947 |
| [8] |
FitzPatrick DR, Ramsay J, McGill NI, Shade M, Carothers AD, Hastie ND . Transcriptome analysis of human autosomal trisomy. Hum Mol Genet, 2002,11(26):3249-3256.
doi: 10.1093/hmg/11.26.3249 pmid: 12471051 |
| [9] |
Buenrostro JD, Wu BJ, Litzenburger UM, Ruff D, Gonzales ML, Snyder MP, Chang HY, Greenleaf WJ . Single-cell chromatin accessibility reveals principles of regulatory variation. Nature, 2015,523(7561):486-490.
doi: 10.1038/nature14590 pmid: 26083756 |
| [10] |
Schueler MG, Sullivan BA . Structural and functional dynamics of human centromeric chromatin. Annu Rev Genomics Hum Genet, 2006,7:301-313.
doi: 10.1146/annurev.genom.7.080505.115613 pmid: 16756479 |
| [11] |
Hsiung CCS, Morrissey CS, Udugama M, Frank CL, Keller CA, Baek S, Giardine B, Crawford GE, Sung MH, Hardison RC, Blobel GA . Genome accessibility is widely preserved and locally modulated during mitosis. Genome Res, 2015,25(2):213-225.
doi: 10.1101/gr.180646.114 pmid: 25373146 |
| [12] |
González J, Muñoz A, Martos G . Asymmetric latent semantic indexing for gene expression experiments visualization. J Bioinform Comput Biol, 2016,14(4):1650023.
doi: 10.1142/S0219720016500232 pmid: 27427382 |
| [13] |
Sandelin A, Alkema W, Engström P, Wasserman WW, Lenhard B . JASPAR: an open-access database for eukaryotic transcription factor binding profiles. Nucleic Acids Res, 2004,32(Database issue):D91-94.
doi: 10.1093/nar/gkh012 pmid: 14681366 |
| [14] |
Schelker M, Feau S, Du JY, Ranu N, Klipp E, MacBeath G, Schoeberl B, Raue A. Estimation of immune cell content in tumour tissue using single-cell RNA-seq data. Nat Commun, 2017,8(1):2032.
doi: 10.1038/s41467-017-02289-3 pmid: 29230012 |
| [15] |
Sinha D, Kumar A, Kumar H, Bandyopadhyay S, Sengupta D. dropClust: efficient clustering of ultra-large scRNA-seq data. Nucleic Acids Res, 2018,46(6):e36.
doi: 10.1093/nar/gky007 pmid: 29361178 |
| [16] |
Makrydimas G, Plachouras N, Thilaganathan B, Nicolaides KH . Abnormal immunological development in fetuses with trisomy 18. Prenat Diagn, 1994,14(4):239-241.
doi: 10.1002/pd.1970140403 pmid: 8066033 |
| [17] |
Lin KC, Park HW, Guan KL . Regulation of the Hippo pathway transcription factor TEAD. Trends Biochem Sci, 2017,42(11):862-872.
doi: 10.1016/j.tibs.2017.09.003 pmid: 28964625 |
| [18] |
Chan P, Han X, Zheng BH, DeRan M, Yu JZ, Jarugumilli GK, Deng H, Pan DJ, Luo XL, Wu X. Autopalmitoylation of TEAD proteins regulates transcriptional output of the Hippo pathway. Nat Chem Biol, 2016,12(4):282-289.
doi: 10.1038/nchembio.2036 pmid: 26900866 |
| [19] |
Noland CL, Gierke S, Schnier PD, Murray J, Sandoval WN, Sagolla M, Dey A, Hannoush RN, Fairbrother WJ, Cunningham CN . Palmitoylation of tead transcription factors is required for their stability and function in hippo pathway signaling. Structure, 2016,24(1):179-186.
doi: 10.1016/j.str.2015.11.005 pmid: 26724994 |
| [20] |
Akerberg BN, Gu F, VanDusen NJ, Zhang XR, Dong R, Li K, Zhang B, Zhou B, Sethi I, Ma Q, Wasson L, Wen T, Liu JH, Dong KZ, Conlon FL, Zhou JL, Yuan GC, Zhou PZ, Pu WT. A reference map of murine cardiac transcription factor chromatin occupancy identifies dynamic and conserved enhancers. Nat Commun, 2019,10(1):4907.
doi: 10.1038/s41467-019-12812-3 pmid: 31659164 |
| [21] |
Joshi S, Davidson G, Le Gras S, Watanabe S, Braun T, Mengus G, Davidson I . TEAD transcription factors are required for normal primary myoblast differentiation in vitro and muscle regeneration in vivo. PLoS Genet, 2017,13(2):e1006600.
doi: 10.1371/journal.pgen.1006600 pmid: 28178271 |
| [22] |
Wen T, Liu JH, He XQ, Dong KZ, Hu GQ, Yu LY, Yin Q, Osman I, Peng JT, Zheng ZQ, Xin HB, Fulton D, Du QS, Zhang W, Zhou JL . Transcription factor TEAD1 is essential for vascular development by promoting vascular smooth muscle differentiation. Cell Death Differ, 2019,26(12):2790-2806.
doi: 10.1038/s41418-019-0335-4 pmid: 31024075 |
| [23] |
Osman I, He XQ, Liu JH, Dong KZ, Wen T, Zhang FZ, Yu LY, Hu GQ, Xin HB, Zhang W, Zhou JL . TEAD1 (TEA domain transcription factor 1) promotes smooth muscle cell proliferation through upregulating slc1a5 (solute carrier family 1 member 5)-mediated glutamine uptake. Circ Res, 2019,124(9):1309-1322.
doi: 10.1161/CIRCRESAHA.118.314187 pmid: 30801233 |
| [24] |
Liu RY, Lee J, Kim BS, Wang QL, Buxton SK, Balasubramanyam N, Kim JJ, Dong JR, Zhang AJ, Li SM, Gupte AA, Hamilton DJ, Martin JF, Rodney GG, Coarfa C, Wehrens XH, Yechoor VK, Moulik M . Tead1 is required for maintaining adult cardiomyocyte function, and its loss results in lethal dilated cardiomyopathy. JCI insight, 2017,2(17):e93343.
doi: 10.1172/jci.insight.93343 |
| [25] |
Franco HL, Casasnovas J, Rodríguez-Medina JR, Cadilla CL . Redundant or separate entities?--roles of Twist1 and Twist2 as molecular switches during gene transcription. Nucleic Acids Res, 2011,39(4):1177-1186.
doi: 10.1093/nar/gkq890 pmid: 20935057 |
| [26] | Zhao CX, Ze Y . Biological function and molecular mechanism of Twist2. Hereditas(Beijing), 2015,37(1):17-24. |
| [27] |
Liu N, Garry GA, Li S, Bezprozvannaya S, Sanchez-Ortiz E, Chen BB, Shelton JM, Jaichander P, Bassel-Duby R, Olson EN . A Twist2-dependent progenitor cell contributes to adult skeletal muscle. Nat Cell Biol, 2017,19(3):202-213.
doi: 10.1038/ncb3477 pmid: 28218909 |
| [28] |
Albizua I, Chopra P, Sherman SL, Gambello MJ, Warren ST . Analysis of the genomic expression profile in trisomy 18: insight into possible genes involved in the associated phenotypes. Hum Mol Genet, 2020,29(2):238-247.
doi: 10.1093/hmg/ddz279 pmid: 31813999 |
| [29] |
Koide K, Slonim DK, Johnson KL, Tantravahi U, Cowan JM, Bianchi DW . Transcriptomic analysis of cell-free fetal RNA suggests a specific molecular phenotype in trisomy 18. Hum Genet, 2011,129(3):295-305.
doi: 10.1007/s00439-010-0923-3 pmid: 21152935 |
| [1] | Dandan Wu, Mingkun Zhu, Zhongyan Fang, Wei Ma. Progress on molecular composition and genetic mechanism of plant B chromosomes [J]. Hereditas(Beijing), 2022, 44(9): 772-782. |
| [2] | Yuan Zhang, Yuting Zhao, Lenan Zhuang, Jin He. Transcriptional regulation of transcriptional Mediator complexes in cardiovascular development and disease [J]. Hereditas(Beijing), 2022, 44(5): 383-397. |
| [3] | Guofang Liu, Peidong Ren, Wenxin Ye, Guangtao Lu. Analysis of transcriptional regulators HpaR1 and Clp regulating the expression of glycoside hydrolase-encoding gene in the Xanthomonas campestris pv. campestris [J]. Hereditas(Beijing), 2021, 43(9): 910-920. |
| [4] | Tianyi Wang, Yingxiang Wang, Chenjiang You. Structural and functional characteristics of plant PHD domain-containing proteins [J]. Hereditas(Beijing), 2021, 43(4): 323-339. |
| [5] | Menggang Lv, Aijia Liu, Qingwei Li, Peng Su. Progress on the origin, function and evolutionary mechanism of RHR transcription factor family [J]. Hereditas(Beijing), 2021, 43(3): 215-225. |
| [6] | Yuanyuan Hao, Xiangqian Zhao, Fudeng Huang, Chunshou Li. The role of PPR proteins in posttranscriptional regulation of organelle components in plants [J]. Hereditas(Beijing), 2021, 43(11): 1050-1065. |
| [7] | Xiaomeng Gao, Zhihua Zhang. Three-dimensional structure and function of chromatin regulated by “liquid-liquid phase separation” of biological macromolecules. [J]. Hereditas(Beijing), 2020, 42(1): 45-56. |
| [8] | Yu Zhang, Yuda Fang. Progresses on the structure and function of cohesin [J]. Hereditas(Beijing), 2020, 42(1): 57-72. |
| [9] | Xiaofei Zheng,Haiyan Huang,Qiang Wu. Chromatin architectural protein CTCF regulates gene expression of the UGT1 cluster [J]. Hereditas(Beijing), 2019, 41(6): 509-523. |
| [10] | Haoqiang Yu,Fuai Sun,Wenqi Feng,Fengzhong Lu,Wanchen Li,Fengling Fu. The BES1/BZR1 transcription factors regulate growth, development and stress resistance in plants [J]. Hereditas(Beijing), 2019, 41(3): 206-214. |
| [11] | Chunyou Ning,Mengnan He,Qianzi Tang,Qing Zhu,Mingzhou Li,Diyan Li. Advances in mammalian three-dimensional genome by using Hi-C technology approach [J]. Hereditas(Beijing), 2019, 41(3): 215-233. |
| [12] | Junxia Zou, Ke Chen. Roles and molecular mechanisms of hypoxia-inducible factors in renal cell carcinoma [J]. Hereditas(Beijing), 2018, 40(5): 341-356. |
| [13] | Lili Liu, Aiwei Guo, Peifu Wu, Fenfen Chen, Yajin Yang, Qin Zhang. Regulation of VPS28 gene knockdown on the milk fat synthesis in Chinese Holstein dairy [J]. Hereditas(Beijing), 2018, 40(12): 1092-1100. |
| [14] | Lan Ren,Rudan Xiao,Qian Zhang,Xiaomin Lou,Zhaojun Zhang,Xiangdong Fang. Synergistic regulation of the erythroid differentiation of K562 cells by KLF1 and KLF9 [J]. Hereditas(Beijing), 2018, 40(11): 998-1006. |
| [15] | Min Yue, Yu Yang, Gaili Guo, Ximing Qin. Genetic and epigeneticregulations of mammalian circadian rhythms [J]. Hereditas(Beijing), 2017, 39(12): 1122-1137. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||