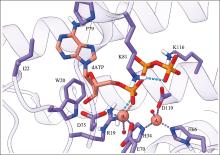
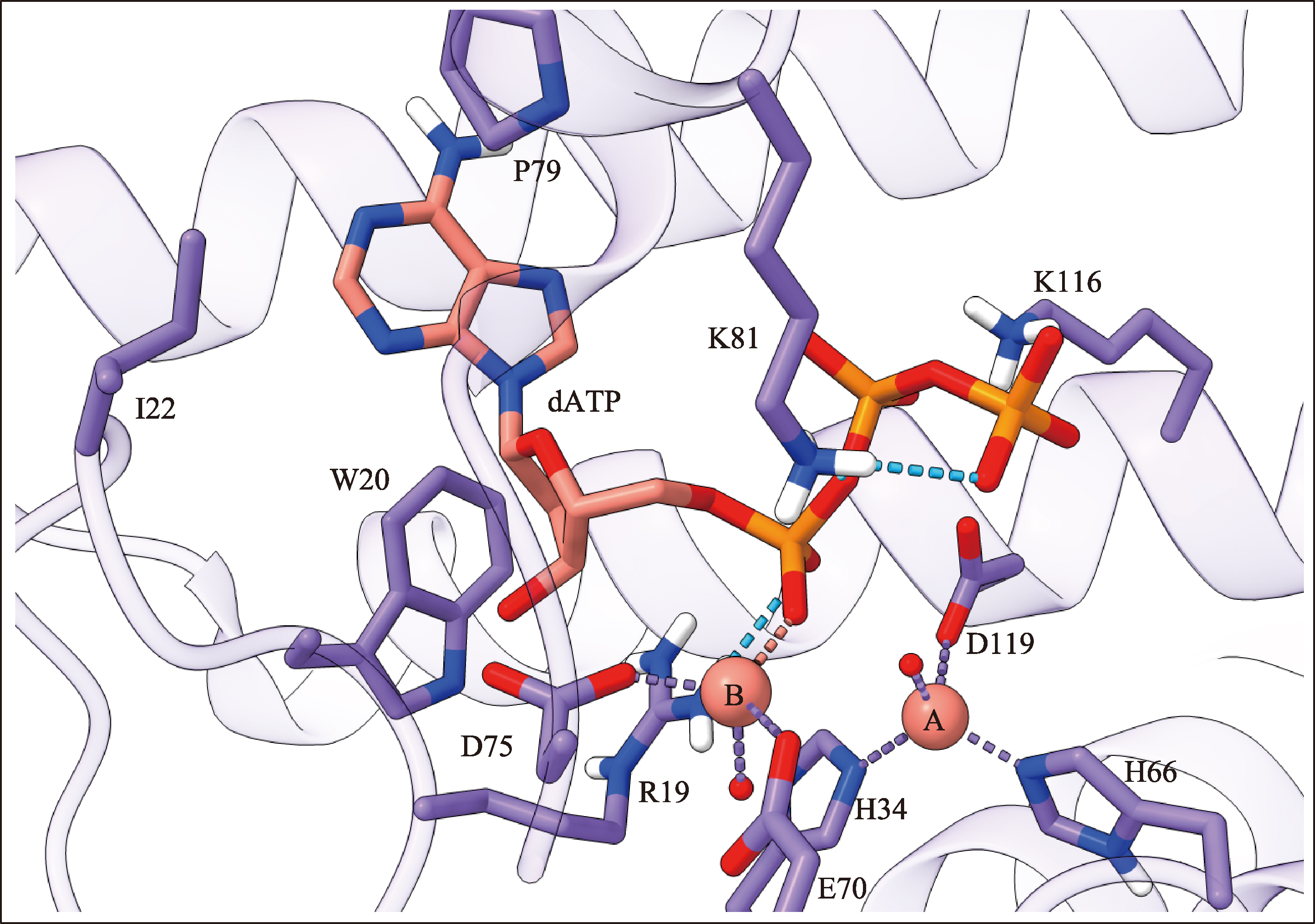
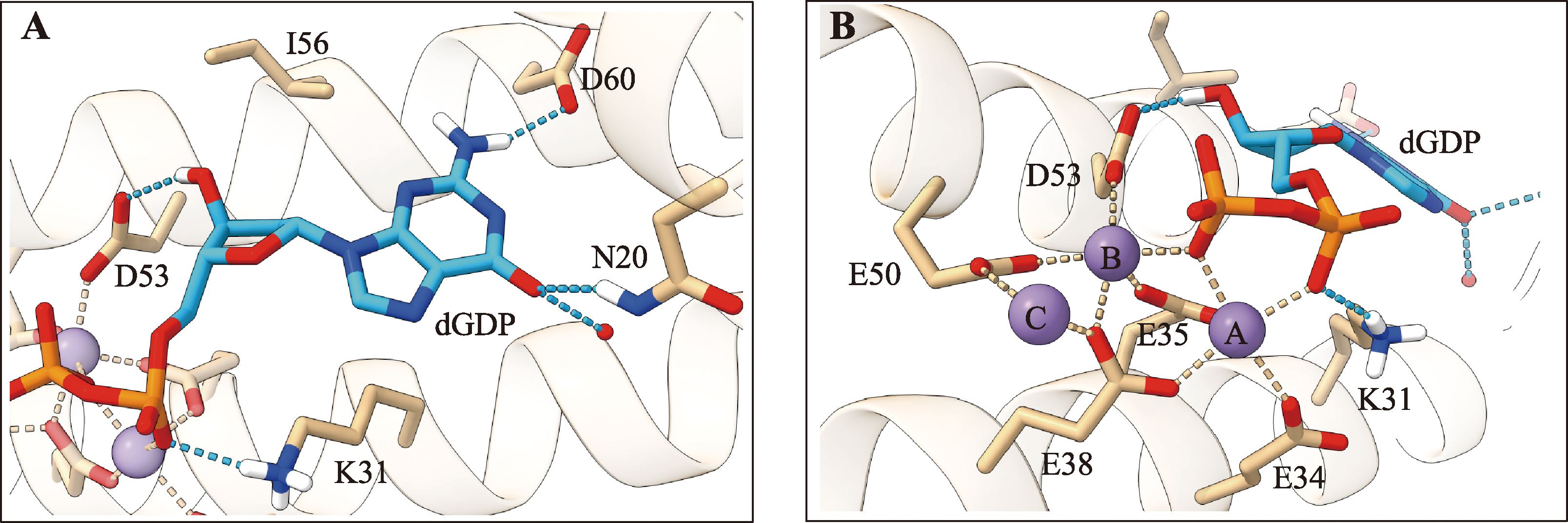
Hereditas(Beijing) ›› 2023, Vol. 45 ›› Issue (10): 887-903.doi: 10.16288/j.yczz.23-059
• Review • Previous Articles Next Articles
Progress on Z genome biosynthetic pathway of bacteriophage
- iHuman Institute and School of Life Science and Technology, ShanghaiTech University, Shanghai 201210, China
-
Received:2023-05-26Revised:2023-08-17Online:2023-10-20Published:2023-08-18 -
Contact:Suwen Zhao E-mail:chenhy6@shanghaitech.edu.cn;zhaosw@shanghaitech.edu.cn -
Supported by:National Natural Science Foundation of China(32122024);Shanghai Frontiers Science Center for Bomacromolecules and Precision Medicine
Cite this article
Huiyu Chen, Suwen Zhao. Progress on Z genome biosynthetic pathway of bacteriophage[J]. Hereditas(Beijing), 2023, 45(10): 887-903.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
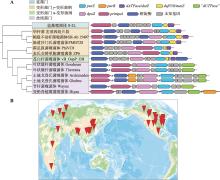
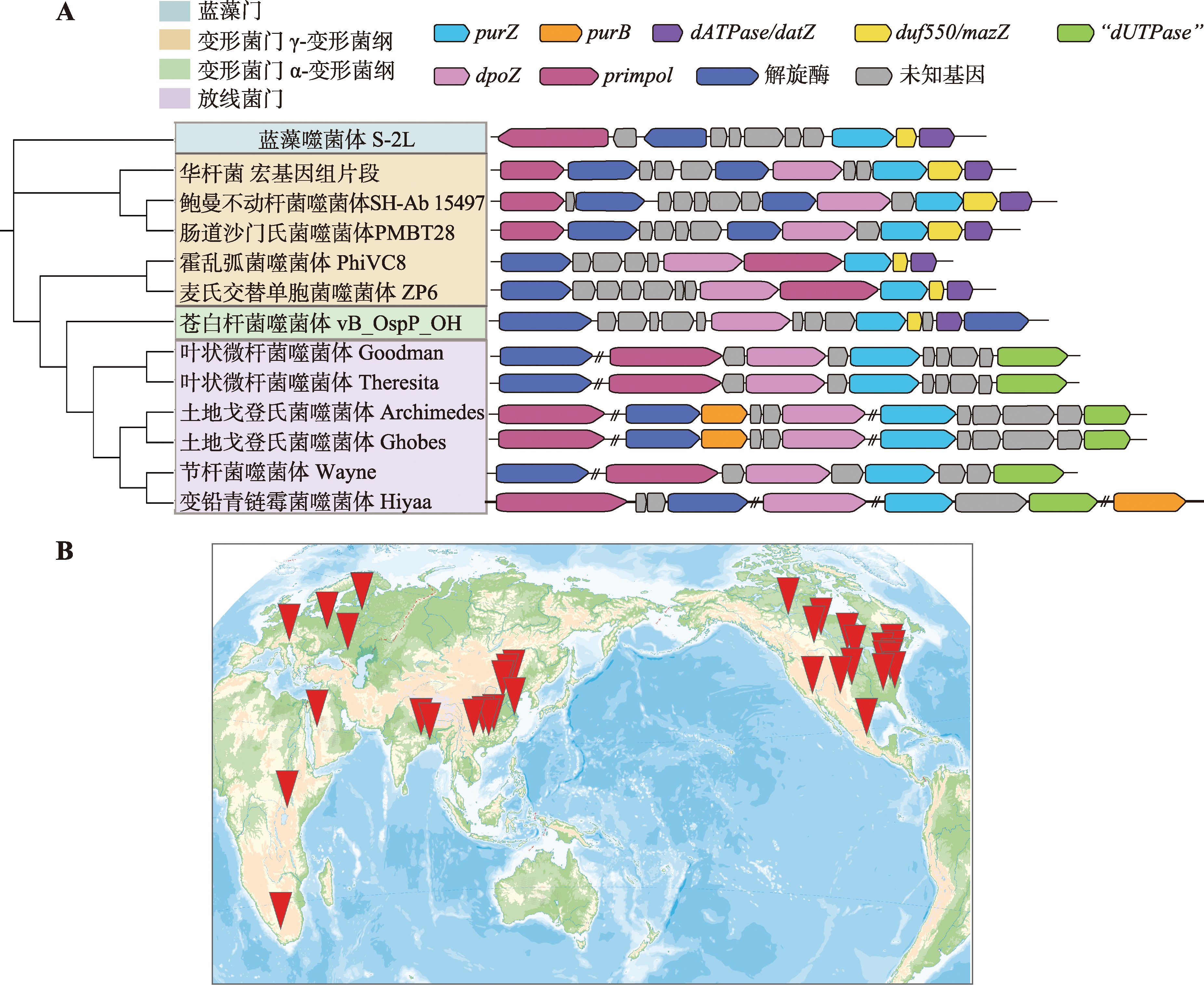
Table 1
List of isolated and sequenced Z-genome phages"
| 物种名 | 基因组编号 | 基因组大小(kb) | GC含量(%) | 宿主的物种分类 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ochrobactrum phage vBOspPOH | MT028492.1 | 41.23 | 55.2 | 变形菌门-α变形菌纲-根瘤菌目-布鲁氏菌科-苍白杆菌属 | ||||||||
| Burkholderia phage BCSR129 | MW460247.1 | 66.15 | 58.4 | 变形菌门-β变形菌纲-伯克氏菌目-伯克氏菌科-伯克氏菌属 | ||||||||
| Salmonella phage PMBT28 | MG641885.1 | 48.11 | 58.6 | 变形菌门-γ变形菌纲-肠杆菌目-肠杆菌科-沙门氏菌属 | ||||||||
| Providencia phage Kokobel2 | MW145138.1 | 45.88 | 47.5 | 变形菌门-γ变形菌纲-肠杆菌目-摩根菌科-普罗威登斯菌属 | ||||||||
| Vibrio phage 23E28 1 | MW824387.1 | 40.73 | 49.9 | 变形菌门-γ变形菌纲-弧菌目-弧菌科-弧菌属 | ||||||||
| Vibrio phage H1 | KM612261.1 | 39.53 | 50.5 | 变形菌门-γ变形菌纲-弧菌目-弧菌科-弧菌属 | ||||||||
| Vibrio phage H2 SGB-2014 | KM612262.1 | 39.53 | 50.5 | 变形菌门-γ变形菌纲-弧菌目-弧菌科-弧菌属 | ||||||||
| Vibrio phage H3 | KM612263.1 | 39.53 | 50.5 | 变形菌门-γ变形菌纲-弧菌目-弧菌科-弧菌属 | ||||||||
| Vibrio phage J3 | KM612265.1 | 39.78 | 50.5 | 变形菌门-γ变形菌纲-弧菌目-弧菌科-弧菌属 | ||||||||
| Vibrio phage QH | KM612259.1 | 39.73 | 50.5 | 变形菌门-γ变形菌纲-弧菌目-弧菌科-弧菌属 | ||||||||
| Vibrio phage VP5 | AY510084.1 | 39.78 | 50.5 | 变形菌门-γ变形菌纲-弧菌目-弧菌科-弧菌属 | ||||||||
| Vibrio phage CJY | KM612260.1 | 39.54 | 50.6 | 变形菌门-γ变形菌纲-弧菌目-弧菌科-弧菌属 | ||||||||
| Vibrio phage J2 | KM612264.1 | 39.53 | 50.6 | 变形菌门-γ变形菌纲-弧菌目-弧菌科-弧菌属 | ||||||||
| Vibrio phage JSF33 | KY883647.1 | 39.66 | 50.6 | 变形菌门-γ变形菌纲-弧菌目-弧菌科-弧菌属 | ||||||||
| Vibrio phage JSF9 | KY883656.1 | 40.01 | 50.6 | 变形菌门-γ变形菌纲-弧菌目-弧菌科-弧菌属 | ||||||||
| Vibrio phage VP2 | AY5505112 | 39.85 | 50.6 | 变形菌门-γ变形菌纲-弧菌目-弧菌科-弧菌属 | ||||||||
| Vibrio phage JSF15 | KY883642.1 | 39.64 | 50.7 | 变形菌门-γ变形菌纲-弧菌目-弧菌科-弧菌属 | ||||||||
| Vibrio phage Rostov 6 | MH105773.1 | 39.93 | 50.7 | 变形菌门-γ变形菌纲-弧菌目-弧菌科-弧菌属 | ||||||||
| Vibrio phage phiVC8 | JF712866.1 | 39.42 | 50.8 | 变形菌门-γ变形菌纲-弧菌目-弧菌科-弧菌属 | ||||||||
| Pseudomonas phage PSA28 | MZ089737.1 | 48.44 | 58.3 | 变形菌门-γ变形菌纲-假单胞菌目-假单胞菌科-假单胞菌属 | ||||||||
| Acinetobacter phage Barton | MW176032.1 | 43.04 | 47.7 | 变形菌门-γ变形菌纲-假单胞菌目-莫拉氏菌科-不动杆菌属 | ||||||||
| Acinetobacter phage DMU1 | MT992243.1 | 43.48 | 47.8 | 变形菌门-γ变形菌纲-假单胞菌目-莫拉氏菌科-不动杆菌属 | ||||||||
| Acinetobacter phage JeffCo | MW176034.1 | 43.29 | 47.8 | 变形菌门-γ变形菌纲-假单胞菌目-莫拉氏菌科-不动杆菌属 | ||||||||
| Acinetobacter phage SH-Ab 15497 | MG674163.1 | 43.42 | 47.9 | 变形菌门-γ变形菌纲-假单胞菌目-莫拉氏菌科-不动杆菌属 | ||||||||
| Alteromonas phage ZP6[ | MK203850.1 | 37.74 | 50.1 | 变形菌门-γ变形菌纲-交替单胞菌目-交替单胞菌科-交替单胞菌属 | ||||||||
| Gordonia phage Archimedes | MT771339.1 | 44.94 | 62.8 | 放线菌门-放线菌纲-放线菌目-分枝杆菌科-戈登氏菌属 | ||||||||
| Gordonia phage Ghobes | KX557278.1 | 45.29 | 65.2 | 放线菌门-放线菌纲-放线菌目-分枝杆菌科-戈登氏菌属 | ||||||||
| Streptomyces phage Hiyaa | MK279841.1 | 83.22 | 66.7 | 放线菌门-放线菌纲-链霉菌目-链霉菌科-链霉菌属 | ||||||||
| Streptomyces phage Keanu | MT723933.1 | 82.1 | 66.8 | 放线菌门-放线菌纲-链霉菌目-链霉菌科-链霉菌属 | ||||||||
| Streptomyces phage Galactica | MT316461.1 | 81.22 | 66.9 | 放线菌门-放线菌纲-链霉菌目-链霉菌科-链霉菌属 | ||||||||
| Curtobacterium phage Parvaparticeps | MZ357096.1 | 40.29 | 65.9 | 放线菌门-放线菌纲-微球菌目-微杆菌科-短小杆菌属 | ||||||||
| Microbacterium phage FireCastle | Not Found | 43 | 64.8 | 放线菌门-放线菌纲-微球菌目-微杆菌科-微杆菌属 | ||||||||
| Microbacterium phage Rasovi | MT310855.1 | 42.92 | 65.2 | 放线菌门-放线菌纲-微球菌目-微杆菌科-微杆菌属 | ||||||||
| Microbacterium phage PermaG | MZ150782.1 | 42.28 | 65.5 | 放线菌门-放线菌纲-微球菌目-微杆菌科-微杆菌属 | ||||||||
| Microbacterium phage Theresita | MK660713.1 | 40.23 | 65.9 | 放线菌门-放线菌纲-微球菌目-微杆菌科-微杆菌属 | ||||||||
| Microbacterium phage Sucha | MK737942.1 | 41.83 | 66.1 | 放线菌门-放线菌纲-微球菌目-微杆菌科-微杆菌属 | ||||||||
| Microbacterium phage Goodman | MK016495.1 | 42.36 | 66.3 | 放线菌门-放线菌纲-微球菌目-微杆菌科-微杆菌属 | ||||||||
| Microbacterium phage Johann | MK016497.1 | 42.36 | 66.3 | 放线菌门-放线菌纲-微球菌目-微杆菌科-微杆菌属 | ||||||||
| Microbacterium phage Cicada | MT498057.1 | 42.35 | 66.5 | 放线菌门-放线菌纲-微球菌目-微杆菌科-微杆菌属 | ||||||||
| Microbacterium phage SBlackberry | MZ747515.1 | 42.05 | 66.9 | 放线菌门-放线菌纲-微球菌目-微杆菌科-微杆菌属 | ||||||||
| Microbacterium phage TurboVicky | ON970583.1 | 42.3 | 67 | 放线菌门-放线菌纲-微球菌目-微杆菌科-微杆菌属 | ||||||||
| Microbacterium phage Typher | ON260821.1 | 41.86 | 67 | 放线菌门-放线菌纲-微球菌目-微杆菌科-微杆菌属 | ||||||||
| Microbacterium phage Zanella | MN369765.1 | 42.11 | 67 | 放线菌门-放线菌纲-微球菌目-微杆菌科-微杆菌属 | ||||||||
| Arthrobacter phage DrRobert | KU160643.1 | 42.6 | 60.6 | 放线菌门-放线菌纲-微球菌目-微球菌科-节杆菌属 | ||||||||
| Arthrobacter phage Bodacious | MK112531.1 | 43.24 | 60.7 | 放线菌门-放线菌纲-微球菌目-微球菌科-节杆菌属 | ||||||||
| Arthrobacter phage ChewChew | MK279844.1 | 43.44 | 60.7 | 放线菌门-放线菌纲-微球菌目-微球菌科-节杆菌属 | ||||||||
| Arthrobacter phage Christian | MF140404.1 | 43.08 | 60.7 | 放线菌门-放线菌纲-微球菌目-微球菌科-节杆菌属 | ||||||||
| Arthrobacter phage CristinaYang | MK279847.1 | 43.37 | 60.7 | 放线菌门-放线菌纲-微球菌目-微球菌科-节杆菌属 | ||||||||
| Arthrobacter phage DreamTeam | MK919484.1 | 43.09 | 60.7 | 放线菌门-放线菌纲-微球菌目-微球菌科-节杆菌属 | ||||||||
| Arthrobacter phage Gisselle | MT498050.1 | 43.09 | 60.7 | 放线菌门-放线菌纲-微球菌目-微球菌科-节杆菌属 | ||||||||
| Arthrobacter phage Huntingdon | MG210949.1 | 43.89 | 60.7 | 放线菌门-放线菌纲-微球菌目-微球菌科-节杆菌属 | ||||||||
| Arthrobacter phage Joann | KU160652.1 | 44.18 | 60.7 | 放线菌门-放线菌纲-微球菌目-微球菌科-节杆菌属 | ||||||||
| Arthrobacter phage Kittykat | MW314848.1 | 43.6 | 60.7 | 放线菌门-放线菌纲-微球菌目-微球菌科-节杆菌属 | ||||||||
| Arthrobacter phage Lennox | MK279862.1 | 43.09 | 60.7 | 放线菌门-放线菌纲-微球菌目-微球菌科-节杆菌属 | ||||||||
| Arthrobacter phage LilStuart | MN813680.1 | 42.95 | 60.7 | 放线菌门-放线菌纲-微球菌目-微球菌科-节杆菌属 | ||||||||
| Arthrobacter phage Lucy | KX576641.1 | 42.94 | 60.7 | 放线菌门-放线菌纲-微球菌目-微球菌科-节杆菌属 | ||||||||
| Arthrobacter phage Nancia | MK279867.1 | 43.24 | 60.7 | 放线菌门-放线菌纲-微球菌目-微球菌科-节杆菌属 | ||||||||
| Arthrobacter phage Nubia | MF140424.1 | 44.05 | 60.7 | 放线菌门-放线菌纲-微球菌目-微球菌科-节杆菌属 | ||||||||
| Arthrobacter phage OurGirlNessie | MK279869.1 | 42.56 | 60.7 | 放线菌门-放线菌纲-微球菌目-微球菌科-节杆菌属 | ||||||||
| Arthrobacter phage PitaDog | MF140425.1 | 42.96 | 60.7 | 放线菌门-放线菌纲-微球菌目-微球菌科-节杆菌属 | ||||||||
| Arthrobacter phage Preamble | KU160659.1 | 43.37 | 60.7 | 放线菌门-放线菌纲-微球菌目-微球菌科-节杆菌属 | ||||||||
| Arthrobacter phage RcigaStruga | KX576640.1 | 43.89 | 60.7 | 放线菌门-放线菌纲-微球菌目-微球菌科-节杆菌属 | ||||||||
| Arthrobacter phage Albanese | OP434444.1 | 44.11 | 60.8 | 放线菌门-放线菌纲-微球菌目-微球菌科-节杆菌属 | ||||||||
| Arthrobacter phage Glenn | KU160645.1 | 44.39 | 60.8 | 放线菌门-放线菌纲-微球菌目-微球菌科-节杆菌属 | ||||||||
| Arthrobacter phage GreenHearts | MK279854.1 | 44.25 | 60.8 | 放线菌门-放线菌纲-微球菌目-微球菌科-节杆菌属 | ||||||||
| Arthrobacter phage Greenhouse | KX688103.1 | 43.98 | 60.8 | 放线菌门-放线菌纲-微球菌目-微球菌科-节杆菌属 | ||||||||
| Arthrobacter phage Lasagna | MK279860.1 | 42.6 | 60.8 | 放线菌门-放线菌纲-微球菌目-微球菌科-节杆菌属 | ||||||||
| Arthrobacter phage Oxynfrius | KX688102.1 | 44.16 | 60.8 | 放线菌门-放线菌纲-微球菌目-微球菌科-节杆菌属 | ||||||||
| Arthrobacter phage Pterodactyl | MK279872.1 | 43.27 | 60.8 | 放线菌门-放线菌纲-微球菌目-微球菌科-节杆菌属 | ||||||||
| Arthrobacter phage WonderBoy | Not Found | 42.74 | 60.8 | 放线菌门-放线菌纲-微球菌目-微球菌科-节杆菌属 | ||||||||
| Arthrobacter phage Fluke | MG198781.1 | 43.81 | 60.9 | 放线菌门-放线菌纲-微球菌目-微球菌科-节杆菌属 | ||||||||
| Arthrobacter phage MeganNoll | MG198782.1 | 44.26 | 60.9 | 放线菌门-放线菌纲-微球菌目-微球菌科-节杆菌属 | ||||||||
| Arthrobacter phage MrGloopy | MK660714.1 | 43.7 | 60.9 | 放线菌门-放线菌纲-微球菌目-微球菌科-节杆菌属 | ||||||||
| Arthrobacter phage RAP15 | KU160662.1 | 44.26 | 60.9 | 放线菌门-放线菌纲-微球菌目-微球菌科-节杆菌属 | ||||||||
| Arthrobacter phage Wawa | MK279892.1 | 43.85 | 60.9 | 放线菌门-放线菌纲-微球菌目-微球菌科-节杆菌属 | ||||||||
| Arthrobacter phage Beethoven | MK112528.1 | 43.93 | 61 | 放线菌门-放线菌纲-微球菌目-微球菌科-节杆菌属 | ||||||||
| Arthrobacter phage Carpal | MK112538.1 | 43.71 | 61 | 放线菌门-放线菌纲-微球菌目-微球菌科-节杆菌属 | ||||||||
| Arthrobacter phage Cholula | MK279845.1 | 43.66 | 61 | 放线菌门-放线菌纲-微球菌目-微球菌科-节杆菌属 | ||||||||
| Arthrobacter phage Immaculata | KU160649.1 | 43.66 | 61 | 放线菌门-放线菌纲-微球菌目-微球菌科-节杆菌属 | ||||||||
| Arthrobacter phage Jumboset | OP820465.1 | 43.78 | 61 | 放线菌门-放线菌纲-微球菌目-微球菌科-节杆菌属 | ||||||||
| Arthrobacter phage Potatoes | MK279901.1 | 43.66 | 61 | 放线菌门-放线菌纲-微球菌目-微球菌科-节杆菌属 | ||||||||
| Arthrobacter phage Riverdale | MK279875.1 | 43.39 | 61 | 放线菌门-放线菌纲-微球菌目-微球菌科-节杆菌属 | ||||||||
| Arthrobacter phage Rozby | MK279876.1 | 43.2 | 61 | 放线菌门-放线菌纲-微球菌目-微球菌科-节杆菌属 | ||||||||
| Arthrobacter phage Savage2526 | MK279880.1 | 43.42 | 61 | 放线菌门-放线菌纲-微球菌目-微球菌科-节杆菌属 | ||||||||
| Arthrobacter phage TattModd | MK279886.1 | 43.55 | 61 | 放线菌门-放线菌纲-微球菌目-微球菌科-节杆菌属 | ||||||||
| Arthrobacter phage Vallejo | KX621005.1 | 43.61 | 61 | 放线菌门-放线菌纲-微球菌目-微球菌科-节杆菌属 | ||||||||
| Arthrobacter phage AppleCider | MN735429.1 | 43.7 | 61.1 | 放线菌门-放线菌纲-微球菌目-微球菌科-节杆菌属 | ||||||||
| Arthrobacter phage Dino | MF140407.1 | 43.56 | 61.1 | 放线菌门-放线菌纲-微球菌目-微球菌科-节杆菌属 | ||||||||
| Arthrobacter phage Korra | KU160653.2 | 43.71 | 61.1 | 放线菌门-放线菌纲-微球菌目-微球菌科-节杆菌属 | ||||||||
| Arthrobacter phage Litotes | MK279863.1 | 43.5 | 61.1 | 放线菌门-放线菌纲-微球菌目-微球菌科-节杆菌属 | ||||||||
| Arthrobacter phage Scuttle | MK814749.1 | 43.73 | 61.1 | 放线菌门-放线菌纲-微球菌目-微球菌科-节杆菌属 | ||||||||
| Arthrobacter phage Wayne | KU160672.2 | 44.37 | 61.1 | 放线菌门-放线菌纲-微球菌目-微球菌科-节杆菌属 | ||||||||
| Arthrobacter phage Zorro | MK279896.1 | 43.56 | 61.1 | 放线菌门-放线菌纲-微球菌目-微球菌科-节杆菌属 | ||||||||
| Arthrobacter phage CallieOMalley | MK112535.1 | 43.86 | 61.2 | 放线菌门-放线菌纲-微球菌目-微球菌科-节杆菌属 | ||||||||
| Arthrobacter phage Canowicakte | MF140400.1 | 43.91 | 61.2 | 放线菌门-放线菌纲-微球菌目-微球菌科-节杆菌属 | ||||||||
| Arthrobacter phage Suppi | KX621004.1 | 43.91 | 61.2 | 放线菌门-放线菌纲-微球菌目-微球菌科-节杆菌属 | ||||||||
| Arthrobacter phage Urla | MG198779.1 | 43.94 | 61.2 | 放线菌门-放线菌纲-微球菌目-微球菌科-节杆菌属 | ||||||||
| Arthrobacter phage BigMack | MK112529.1 | 43.36 | 61.3 | 放线菌门-放线菌纲-微球菌目-微球菌科-节杆菌属 | ||||||||
| Arthrobacter phage EstebanJulior | MK919476.1 | 43.75 | 61.3 | 放线菌门-放线菌纲-微球菌目-微球菌科-节杆菌属 | ||||||||
| Arthrobacter phage MamaPearl | MT498045.1 | 43.75 | 61.3 | 放线菌门-放线菌纲-微球菌目-微球菌科-节杆菌属 | ||||||||
| Arthrobacter phage Moki | MH744421.1 | 43.16 | 61.3 | 放线菌门-放线菌纲-微球菌目-微球菌科-节杆菌属 | ||||||||
| Arthrobacter phage Bennie | KU160640.2 | 43.08 | 61.4 | 放线菌门-放线菌纲-微球菌目-微球菌科-节杆菌属 | ||||||||
| Arthrobacter phage HeadNerd | MK279907.1 | 42.97 | 61.4 | 放线菌门-放线菌纲-微球菌目-微球菌科-节杆菌属 | ||||||||
| Arthrobacter phage Huckleberry | MK279856.1 | 42.97 | 61.4 | 放线菌门-放线菌纲-微球菌目-微球菌科-节杆菌属 | ||||||||
| Stenotrophomonas phage BUCT555 | MW291508.1 | 39.44 | 61.4 | 放线菌门-放线菌纲-微球菌目-微球菌科-节杆菌属 | ||||||||
| Arthrobacter phage Chridison | OP751151.1 | 43.02 | 61.5 | 放线菌门-放线菌纲-微球菌目-微球菌科-节杆菌属 | ||||||||
| Arthrobacter phage Eunoia | MK279851.1 | 44.42 | 61.5 | 放线菌门-放线菌纲-微球菌目-微球菌科-节杆菌属 | ||||||||
| Arthrobacter phage Aledel | MK112526.1 | 43.74 | 61.6 | 放线菌门-放线菌纲-微球菌目-微球菌科-节杆菌属 | ||||||||
| Arthrobacter phage HunterDalle | KU160648.1 | 43.34 | 61.6 | 放线菌门-放线菌纲-微球菌目-微球菌科-节杆菌属 | ||||||||
| Arthrobacter phage OMalley | MK279868.1 | 43.74 | 61.6 | 放线菌门-放线菌纲-微球菌目-微球菌科-节杆菌属 | ||||||||
| Arthrobacter phage Riovina | MK279874.1 | 43.74 | 61.6 | 放线菌门-放线菌纲-微球菌目-微球菌科-节杆菌属 | ||||||||
| Arthrobacter phage Vulture | KU160671.1 | 43.34 | 61.6 | 放线菌门-放线菌纲-微球菌目-微球菌科-节杆菌属 | ||||||||
| Arthrobacter phage Pumancara | KU160661.1 | 42.83 | 61.7 | 放线菌门-放线菌纲-微球菌目-微球菌科-节杆菌属 | ||||||||
| Arthrobacter phage Supakev | MK279884.1 | 43.76 | 61.7 | 放线菌门-放线菌纲-微球菌目-微球菌科-节杆菌属 | ||||||||
| Arthrobacter phage Daiboju | MH450117.1 | 43.95 | 61.9 | 放线菌门-放线菌纲-微球菌目-微球菌科-节杆菌属 | ||||||||
| Arthrobacter phage Herb | MH450118.1 | 43.95 | 61.9 | 放线菌门-放线菌纲-微球菌目-微球菌科-节杆菌属 | ||||||||
| Arthrobacter phage KingBob | MH450121.1 | 43.95 | 61.9 | 放线菌门-放线菌纲-微球菌目-微球菌科-节杆菌属 | ||||||||
| Arthrobacter phage Maria1952 | MN586061.1 | 43.95 | 61.9 | 放线菌门-放线菌纲-微球菌目-微球菌科-节杆菌属 | ||||||||
| Arthrobacter phage Sergei | MH450131.1 | 43.95 | 61.9 | 放线菌门-放线菌纲-微球菌目-微球菌科-节杆菌属 | ||||||||
| Arthrobacter phage Temper16 | MF668285.1 | 43.95 | 61.9 | 放线菌门-放线菌纲-微球菌目-微球菌科-节杆菌属 | ||||||||
| Phage 66_12 | MN304821.1 | 40.56 | 66.1 | 宏基因组 | ||||||||
| Bacillus phage vB_BpsS-140 | MH884512.1 | 55.09 | 39.8 | 厚壁菌门-芽孢杆菌纲-芽孢杆菌目-芽孢杆菌科-芽孢杆菌属 | ||||||||
| Cyanophage S-2L | AX955019.1 | 45.57 | 69.4 | 蓝藻门 | ||||||||
Table 1
| [1] | Liyanage VRB, Jarmasz JS, Murugeshan N, Del Bigio MR, Rastegar M, Davie JR. DNA modifications: function and applications in normal and disease States. Biology (Basel), 2014, 3(4): 670-723. |
| [2] | Weigele P, Raleigh EA. Biosynthesis and function of modified bases in bacteria and their Viruses. Chem Rev, 2016, 116(20): 12655-12687. |
| [3] | Gommers-Ampt JH, Borst P. Hypermodified bases in DNA. FASEB J, 1995, 9(11): 1034-1042. |
| [4] | Wyatt GR, Cohen SS. The bases of the nucleic acids of some bacterial and animal viruses: the occurrence of 5-hydroxymethylcytosine. Biochem J, 1953, 55(5): 774-782. |
| [5] | Bryson AL, Hwang Y, Sherrill-Mix S, Wu GD, Lewis JD, Black L, Clark TA, Bushman FD. Covalent modification of bacteriophage T 4 DNA inhibits CRISPR-Cas9. mBio, 2015, 6(3): e00648. |
| [6] | Kuo TT, Huang TC, Teng MH. 5-Methylcytosine replacing cytosine in the deoxyribonucleic acid of a bacteriophage for Xanthomonas oryzae. J Mol Biol, 1968, 34(2): 373-375. |
| [7] | Swinton D, Hattman S, Benzinger R, Buchanan-Wollaston V, Beringer J. Replacement of the deoxycytidine residues in Rhizobium bacteriophage RL38JI DNA. FEBS Lett, 1985, 184(2): 294-298. |
| [8] | Kallen RG, Simon M, Marmur J. The new occurrence of a new pyrimidine base replacing thymine in a bacteriophage DNA:5-hydroxymethyl uracil. J Mol Biol, 1962, 5: 248-250. |
| [9] | Lee YJ, Dai N, Walsh SE, Müller S, Fraser ME, Kauffman KM, Guan CD, Corrêa IR, Weigele PR. Identification and biosynthesis of thymidine hypermodifications in the genomic DNA of widespread bacterial viruses. Proc Natl Acad Sci USA, 2018, 115(14): E3116-E3125. |
| [10] | Warren RA. Modified bases in bacteriophage DNAs. Annu Rev Microbiol, 1980, 34: 137-158. |
| [11] | Kropinski AM, Bose RJ, Warren RA. 5-(4- Aminobutylaminomethyl)uracil, an unusual pyrimidine from the deoxyribonucleic acid of bacteriophage phiW-14. Biochemistry, 1973, 12(1): 151-157. |
| [12] | Swinton D, Hattman S, Crain PF, Cheng CS, Smith DL, McCloskey JA. Purification and characterization of the unusual deoxynucleoside, alpha-N-(9-beta-D-2′- deoxyribofuranosylpurin-6-yl)glycinamide, specified by the phage Mu modification function. Proc Natl Acad Sci USA, 1983, 80(24): 7400-7404. |
| [13] | Thiaville JJ, Kellner SM, Yuan YF, Hutinet G, Thiaville PC, Jumpathong W, Mohapatra S, Brochier-Armanet C, Letarov AV, Hillebrand R, Malik CK, Rizzo CJ, Dedon PC, de Crécy-Lagard V. Novel genomic island modifies DNA with 7-deazaguanine derivatives. Proc Natl Acad Sci USA, 2016, 113(11): E1452-E1459. |
| [14] | Tsai R, Corrêa IR, Xu MY, Xu SY. Restriction and modification of deoxyarchaeosine (dG+)-containing phage 9 g DNA. Sci Rep, 2017, 7(1): 8348. |
| [15] | Hutinet G, Kot W, Cui L, Hillebrand R, Balamkundu S, Gnanakalai S, Neelakandan R, Carstens AB, Fa Lui C, Tremblay D, Jacobs-Sera D, Sassanfar M, Lee YJ, Weigele P, Moineau S, Hatfull GF, Dedon PC, Hansen LH, de Crécy-Lagard V.. 7-Deazaguanine modifications protect phage DNA from host restriction systems. Nat Commun, 2019, 10(1): 5442. |
| [16] | Kirnos MD, Khudyakov IY, Alexandrushkina NI, Vanyushin BF. 2-Aminoadenine is an adenine substituting for a base in S-2L cyanophage DNA. Nature, 1977, 270(5635): 369-370. |
| [17] | Khudyakov IY, Kirnos MD, Alexandrushkina NI, Vanyushin BF. Cyanophage S-2L contains DNA with 2,6-diaminopurine substituted for adenine. Virology, 1978, 88(1): 8-18. |
| [18] | Cheong C, Tinoco I, Chollet A. Thermodynamic studies of base pairing involving 2,6-diaminopurine. Nucleic Acids Res, 1988, 16(11): 5115-5122. |
| [19] | Sági J, Szakonyi E, Vorlícková M, Kypr J. Unusual contribution of 2-aminoadenine to the thermostability of DNA. J Biomol Struct Dyn, 1996, 13(6): 1035-1041. |
| [20] | Cristofalo M, Kovari D, Corti R, Salerno D, Cassina V, Dunlap D, Mantegazza F. Nanomechanics of diaminopurine- substituted DNA. Biophys J, 2019, 116(5): 760-771. |
| [21] | Szekeres M, Matveyev AV. Cleavage and sequence recognition of 2,6-diaminopurine-containing DNA by site- specific endonucleases. FEBS Lett, 1987, 222(1): 89-94. |
| [22] | Chollet A, Kawashima E. DNA containing the base analogue 2-aminoadenine: preparation, use as hybridization probes and cleavage by restriction endonucleases. Nucleic Acids Res, 1988, 16(1): 305-317. |
| [23] | Tan Y, You CJ, Park J, Kim HS, Guo S, Schärer OD, Wang YS. Transcriptional perturbations of 2,6-diaminopurine and 2-aminopurine. ACS Chem Biol, 2022, 17(7): 1672-1676. |
| [24] | Philippe Marliere, Pierre-Alexandre Kaminski, Frédérique GALISSON, Madeleine BouzonSylvie, PochetJean Weissenbach, William Saurin, Catherine Robert, Virginie Vico. Banque genomique du cyanophage s-2l et analyse fonctionnelle. WO2003093461A2. 2003-11-13. |
| [25] | Zhou Y, Xu XX, Wei YF, Cheng Y, Guo Y, Khudyakov I, Liu FL, He P, Song ZY, Li Z, Gao Y, Ang EL, Zhao HM, Zhang Y, Zhao SW. A widespread pathway for substitution of adenine by diaminopurine in phage genomes. Science, 2021, 372(6541): 512-516. |
| [26] | Sleiman D, Garcia PS, Lagune M, Loc'h J, Haouz A, Taib N, Röthlisberger P, Gribaldo S, Marlière P, Kaminski PA. A third purine biosynthetic pathway encoded by aminoadenine-based viral DNA genomes. Science, 2021, 372(6541): 516-520. |
| [27] | Pezo V, Jaziri F, Bourguignon PY, Louis D, Jacobs-Sera D, Rozenski J, Pochet S, Herdewijn P, Hatfull GF, Kaminski PA, Marliere P. Noncanonical DNA polymerization by aminoadenine-based siphoviruses. Science, 2021, 372(6541): 520-524 |
| [28] | Czernecki D, Bonhomme F, Kaminski PA, Delarue M. Characterization of a triad of genes in cyanophage S-2L sufficient to replace adenine by 2-aminoadenine in bacterial DNA. Nat Commun, 2021, 12(1): 4710. |
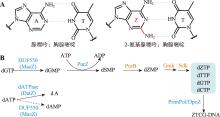
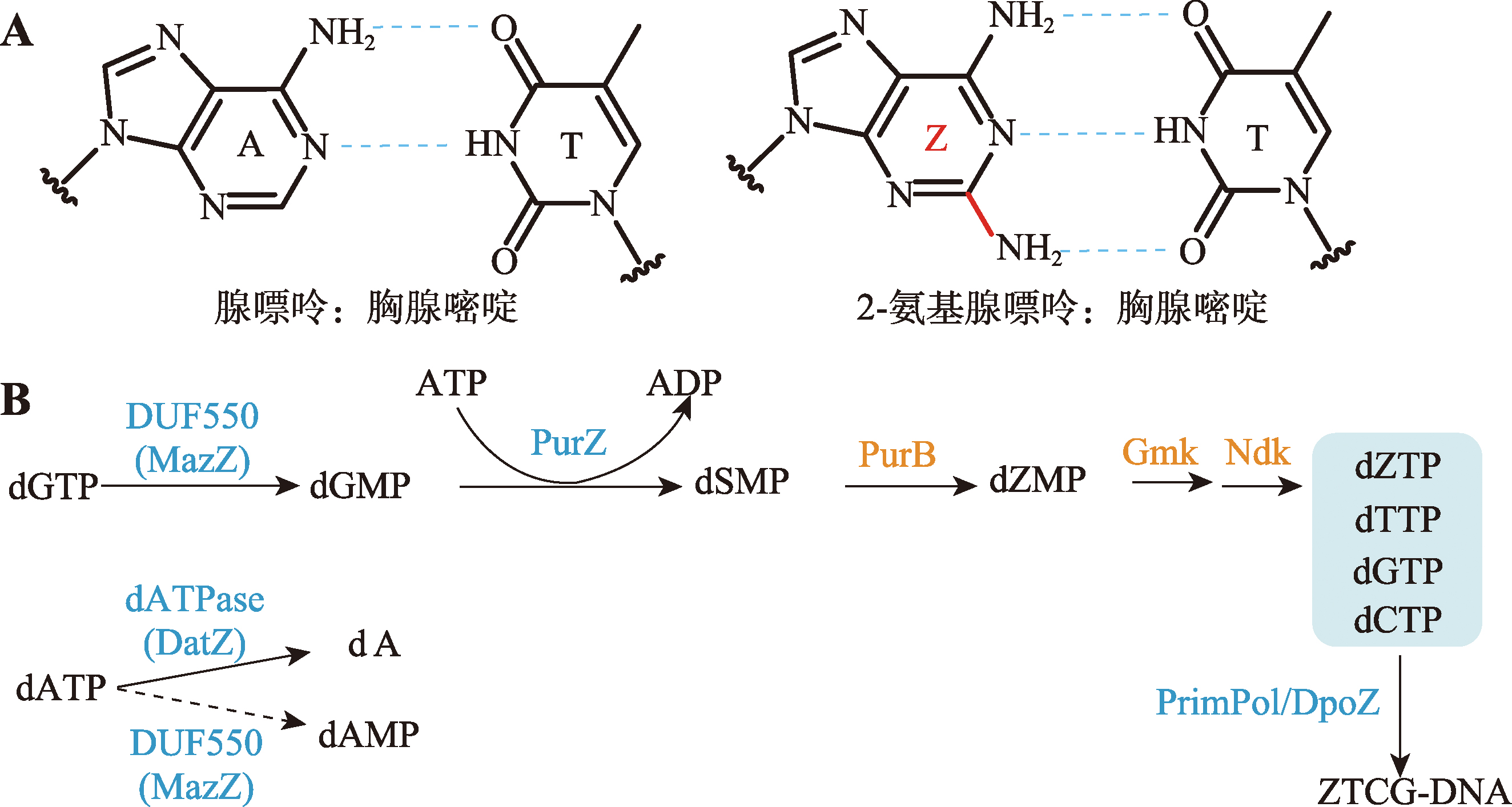
| [29] | Jin JY, Zhou JH. The mystery of Z-genome biosynthesis has been elucidated. Synth Biol J, 2022, 3(1): 1-5. |
| 金交羽, 周佳海. Z-基因组的生物合成奥秘被揭示. 合成生物学, 2022, 3(1): 1-5. | |
| [30] | Callahan MP, Smith KE, Cleaves HJ, Ruzicka J, Stern JC, Glavin DP, House CH, Dworkin JP. Carbonaceous meteorites contain a wide range of extraterrestrial nucleobases. Proc Natl Acad Sci USA, 2011, 108(34): 13995-13998. |
| [31] | Wang ZY, Zhang F, Liang YT, Zheng KY, Gu CX, Zhang WJ, Liu YD, Zhang XR, Shao HB, Jiang Y, Guo C, He H, Wang HL, Sung YY, Mok WJ, Wong LL, He JF, McMinn A, Wang M. Genome and ecology of a novel alteromonas podovirus, ZP6, representing a new viral genus, Mareflavirus. Microbiol Spectr, 2021, 9(2): e0046321. |
| [32] | Honzatko RB, Fromm HJ. Structure-function studies of adenylosuccinate synthetase from Escherichia coli. Arch Biochem Biophys, 1999, 370(1): 1-8. |
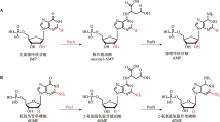
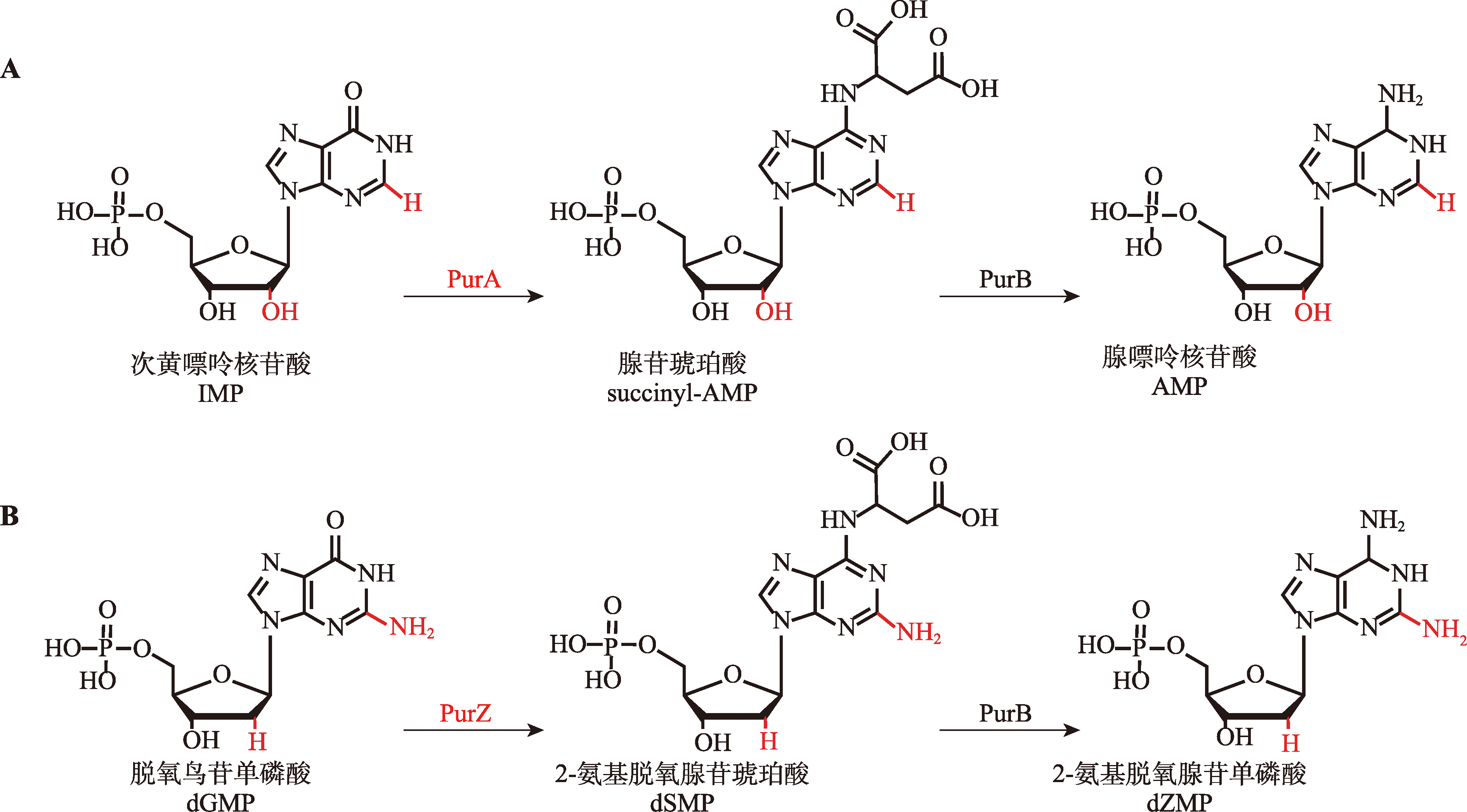
| [33] | Tong Y, Wu XY, Liu Y, Chen HY, Zhou Y, Jiang L, Li M, Zhao SW, Zhang Y. Alternative Z-genome biosynthesis pathway shows evolutionary progression from Archaea to phage. Nat Microbiol, 2023, 8(7): 1330-1338. |
| [34] | Wang W, Gorrell A, Honzatko RB, Fromm HJ. A study of Escherichia coli adenylosuccinate synthetase association states and the interface residues of the homodimer. J Biol Chem, 1997, 272(11): 7078-7084. |
| [35] | Mehrotra S, Balaram H. Kinetic characterization of adenylosuccinate synthetase from the thermophilic archaea Methanocaldococcus jannaschii. Biochemistry, 2007, 46(44): 12821-12832. |
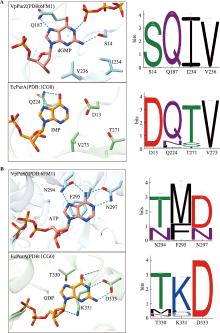
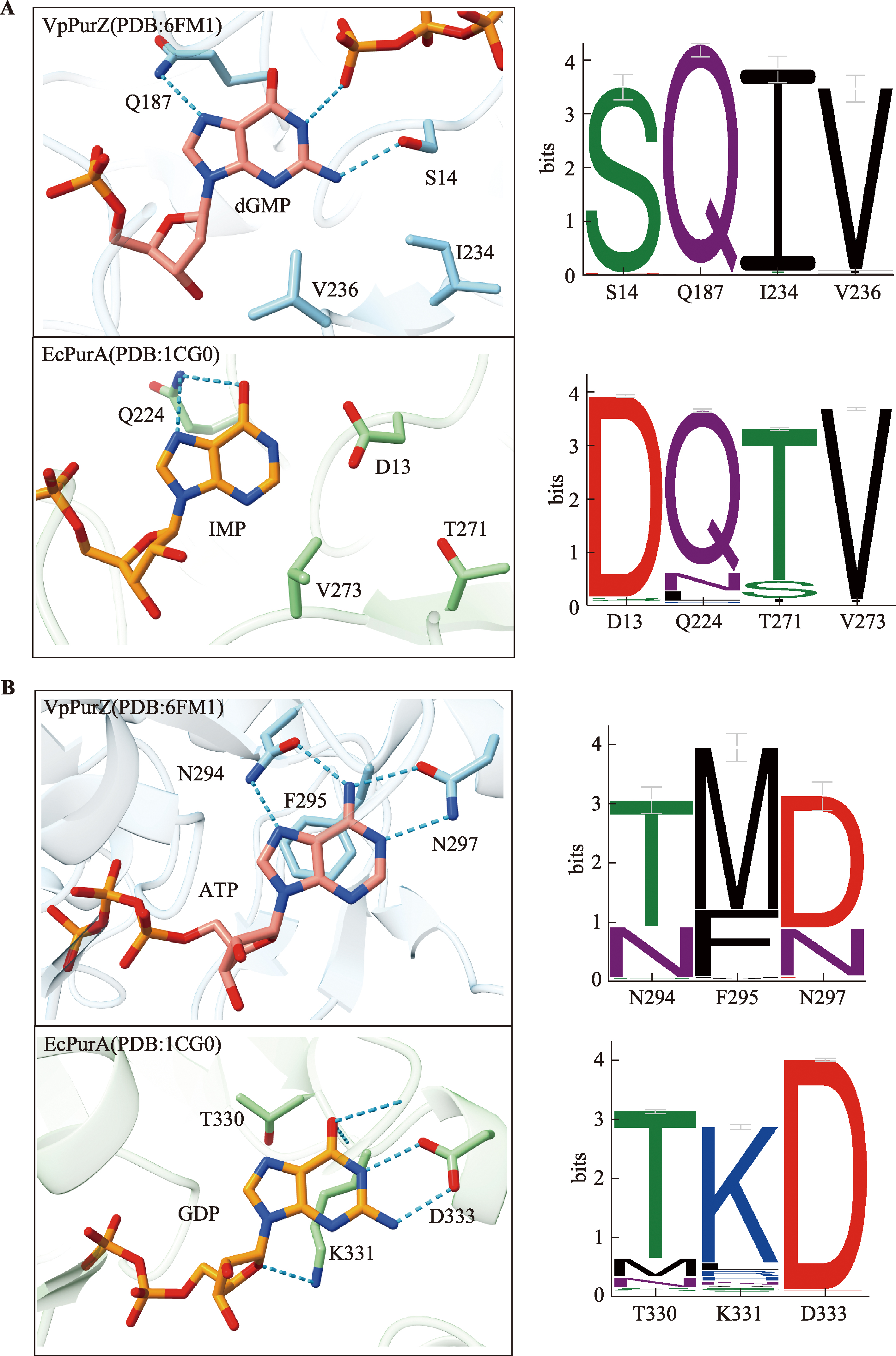
| [36] | Czernecki D, Legrand P, Tekpinar M, Rosario S, Kaminski PA, Delarue M. How cyanophage S-2L rejects adenine and incorporates 2-aminoadenine to saturate hydrogen bonding in its DNA. Nat Commun, 2021, 12(1): 2420. |
| [37] | Bochner BR, Ames BN. Complete analysis of cellular nucleotides by two-dimensional thin layer chromatography. J Biol Chem, 1982, 257(16): 9759-9769. |
| [38] | Kim EE, Wyckoff HW. Reaction mechanism of alkaline phosphatase based on crystal structures: Two-metal ion catalysis. J Mol Biol, 1991, 218(2): 449-464. |
| [39] | Moroz OV, Harkiolaki M, Galperin MY, Vagin AA, González-Pacanowska D, Wilson KS. The crystal structure of a complex of Campylobacter jejuni dUTPase with substrate analogue sheds light on the mechanism and suggests the "basic module" for dimeric d(C/U)TPases. J Mol Biol, 2004, 342(5): 1583-1597. |
| [40] | Guilliam TA, Keen BA, Brissett NC, Doherty AJ. Primase-polymerases are a functionally diverse superfamily of replication and repair enzymes. Nucleic Acids Res, 2015, 43(14): 6651-6664. |
| [41] | Braithwaite DK, Ito J. Compilation, alignment, and phylogenetic relationships of DNA polymerases. Nucleic Acids Res, 1993, 21(4): 787-802. |
| [42] | Czernecki D, Hu HD, Romoli F, Delarue M.Structural dynamics and determinants of 2-aminoadenine specificity in DNA polymerase DpoZ of Vibriophage ϕVC8. Nucleic Acids Res, 2021, 49(20): 11974-11985. |
| [43] | Li Y, Korolev S, Waksman G. Crystal structures of open and closed forms of binary and ternary complexes of the large fragment of Thermus aquaticus DNA polymerase I: structural basis for nucleotide incorporation. EMBO J, 1998, 17(24): 7514-7525. |
| [44] | Wu EY, Beese LS. The structure of a high fidelity DNA polymerase bound to a mismatched nucleotide reveals an “Ajar” intermediate conformation in the nucleotide selection mechanism. J Biol Chem, 2011, 286(22): 19758-19767. |
| [45] | Ceze L, Nivala J, Strauss K. Molecular digital data storage using DNA. Nat Rev Genet, 2019, 20(8): 456-466. |
| [46] | Zhao FP, Wei YF, Wang XY, Zhou Y, Tong Y, Ang EL, Liu SN, Zhao HM, Zhang Y. Enzymatic synthesis of the unnatural nucleotide 2′-deoxyisoguanosine 5′-monophosphate. ChemBioChem, 2022, 23(21): e202200295. |
| [47] | Rothemund PWK. Folding DNA to create nanoscale shapes and patterns. Nature, 2006, 440(7082): 297-302. |
| [48] | Zhang DL, Ge Q, Feng YB, Chen WG. Comparison and analysis on scientific research programs on DNA data storage. China Biotechnol, 2022, 42(6): 116-129. |
| 张大璐, 葛奇, 冯一博, 陈为刚. DNA数据存储的科研概况国际对比与分析. 中国生物工程杂志, 2022, 42(6): 116-129. | |
| [49] | Zhang LQ, Yang ZY, Sefah K, Bradley KM, Hoshika S, Kim MJ, Kim HJ, Zhu GZ, Jiménez E, Cansiz S, Teng IT, Champanhac C, McLendon C, Liu C, Zhang W, Gerloff DL, Huang Z, Tan WH, Benner SA. Evolution of functional six-nucleotide DNA. J Am Chem Soc, 2015, 137(21): 6734-6737. |
| [50] | Tian EK, Wang Y, Wu ZX, Wan ZQH, Cheng W. Bacteriophage therapy: retrospective review and future prospects. J Sichuan Univ (Med Sci Ed), 2021, 52(2): 170-175. |
| 田而慷, 王玥, 吴卓轩, 万紫千红, 程伟. 噬菌体疗法:回顾与展望. 四川大学学报(医学版), 2021, 52(2): 170-175. |
| [1] | Caijiao Liang, Fanmei Meng, Yuncan Ai. CRISPR/Cas systems in genome engineering of bacteriophages [J]. Hereditas(Beijing), 2018, 40(5): 378-389. |
| [2] | Xiangyu Fan, Ying He, Jianping Xie. Undergraduate teaching in life science exemplified by mycobacteriophages [J]. HEREDITAS(Beijing), 2014, 36(8): 842-846. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||