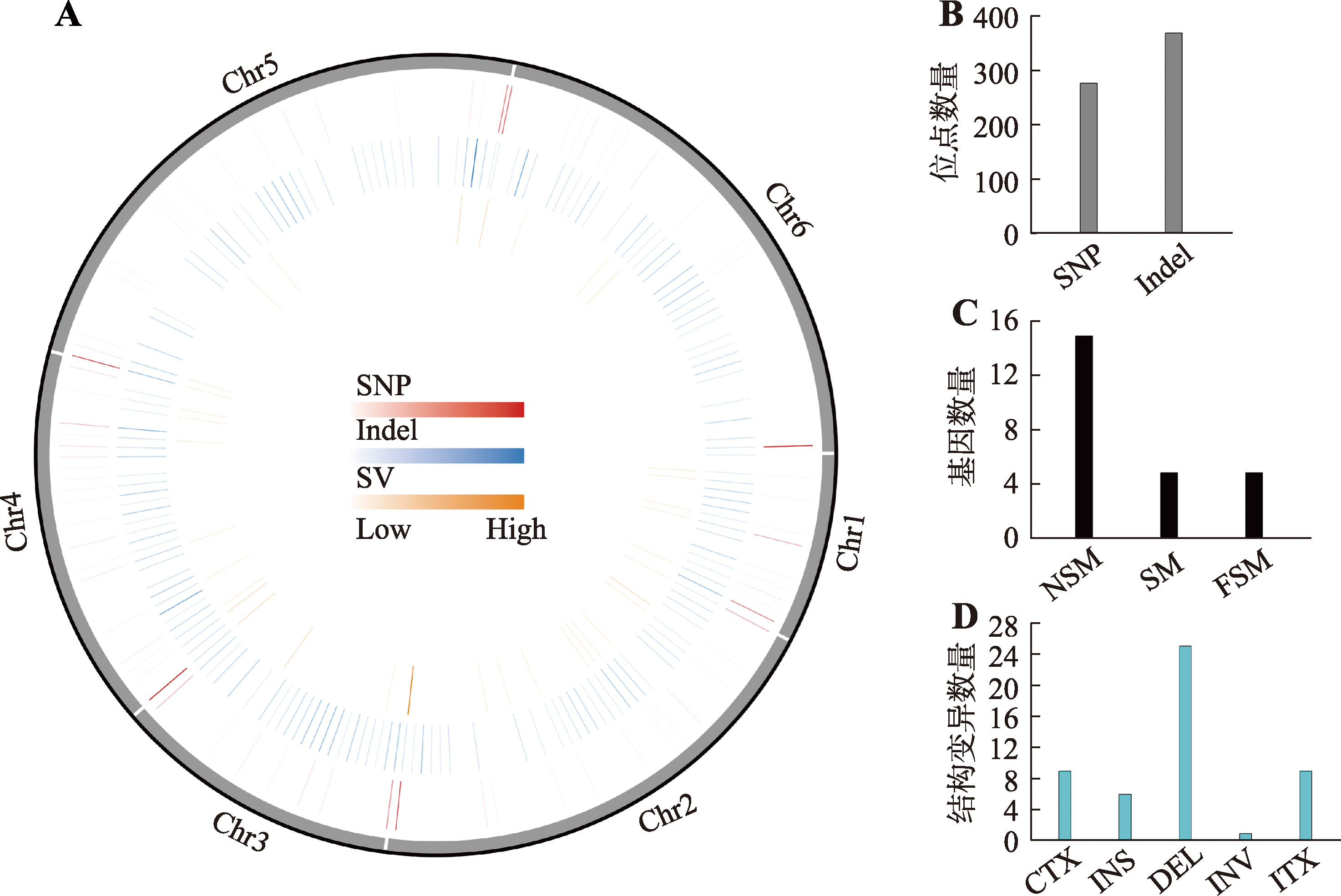
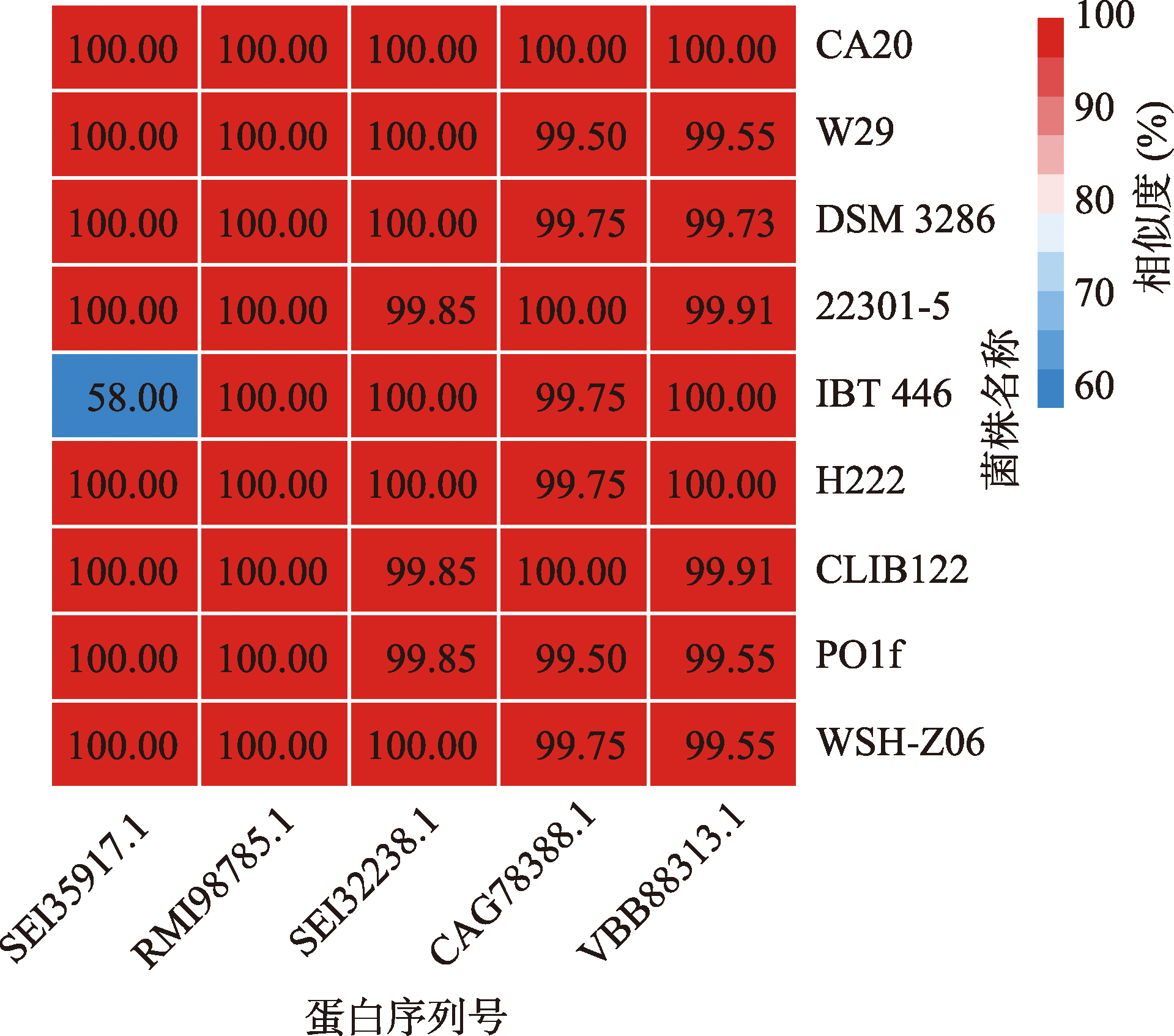
Hereditas(Beijing) ›› 2023, Vol. 45 ›› Issue (10): 904-921.doi: 10.16288/j.yczz.23-139
• Research Article • Previous Articles Next Articles
Mechanism and evolutionary analysis of Yarrowia lipolytica CA20 capable of producing erythritol with a high yield based on comparative genomics
Kai Xia( ), Fangmei Liu, Yuqing Chen, Shanshan Chen, Chunying Huang, Xuequn Zhao, Ruyi Sha, Jun Huang(
), Fangmei Liu, Yuqing Chen, Shanshan Chen, Chunying Huang, Xuequn Zhao, Ruyi Sha, Jun Huang( )
)
- Zhejiang Provincial Collaborative Innovation Center of Agricultural Biological Resources Biochemical Manufacturing, Key Laboratory of Chemical and Biological Processing Technology for Farm Products of Zhejiang Province, School of Biological and Chemical Engineering, Zhejiang University of Science and Technology, Hangzhou 310023, China
-
Received:2023-06-08Revised:2023-08-11Online:2023-10-20Published:2023-08-23 -
Contact:Jun Huang E-mail:xiakai05@zust.edu.cn;huangjun@zust.edu.cn -
Supported by:Research Start-Up Foundation of Zhejiang University of Science and Technology(F701103L11);Research Innovation Foundation of Postgraduate(2021yjskc11);National Training Program for College Students’ Innovation(202211057026)
Cite this article
Kai Xia, Fangmei Liu, Yuqing Chen, Shanshan Chen, Chunying Huang, Xuequn Zhao, Ruyi Sha, Jun Huang. Mechanism and evolutionary analysis of Yarrowia lipolytica CA20 capable of producing erythritol with a high yield based on comparative genomics[J]. Hereditas(Beijing), 2023, 45(10): 904-921.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
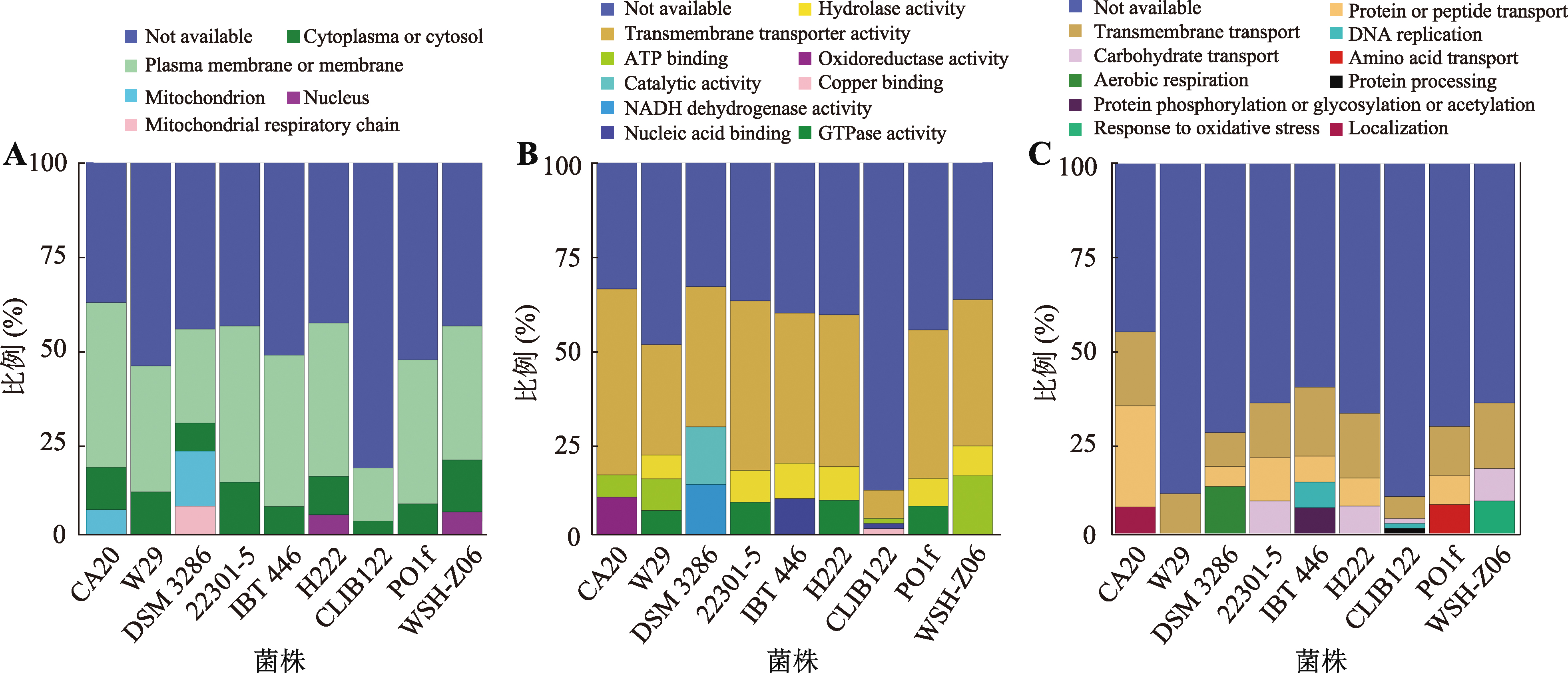
Table 1
Genomic information"
| 菌株 | 数据来源 | 大小(Mb) | GC% | 染色体数量 | 分离环境 |
|---|---|---|---|---|---|
| Y. lipolytica CA20 | PRJNA957569 | 20.42 | 48.97 | 6 | 水果、蜂蜜和糖蜜混合物 |
| Y. lipolytica W29 | PRJNA437435 | 20.50 | 48.98 | 6 | 废水 |
| Y. lipolytica DSM 3286 | PRJNA641784 | 21.22 | 48.91 | 6 | 未记录 |
| Y. lipolytica 22301-5 | PRJNA802556 | 20.95 | 48.96 | 6 | 废水 |
| Y. lipolytica IBT 446 | PRJNA437435 | 19.86 | 49.09 | 6 | 奶酪 |
| Y. lipolytica H222 | PRJNA437435 | 19.72 | 49.11 | 6 | 泥土 |
| Y. lipolytica CLIB122 | PRJNA13837 | 20.55 | 48.98 | 6 | 未记录 |
| Y. lipolytica Po1f | PRJNA233955 | 20.62 | 49.01 | 6 | 废水 |
| Y. lipolytica WSH-Z06 | PRJEB5051 | 20.09 | 49.03 | 6 | 泥土 |
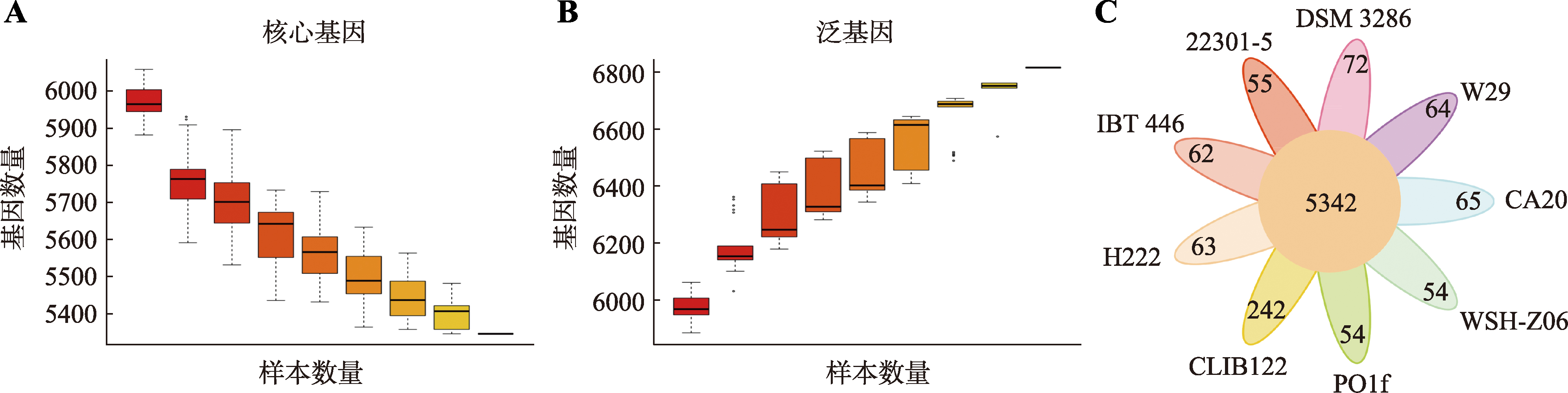
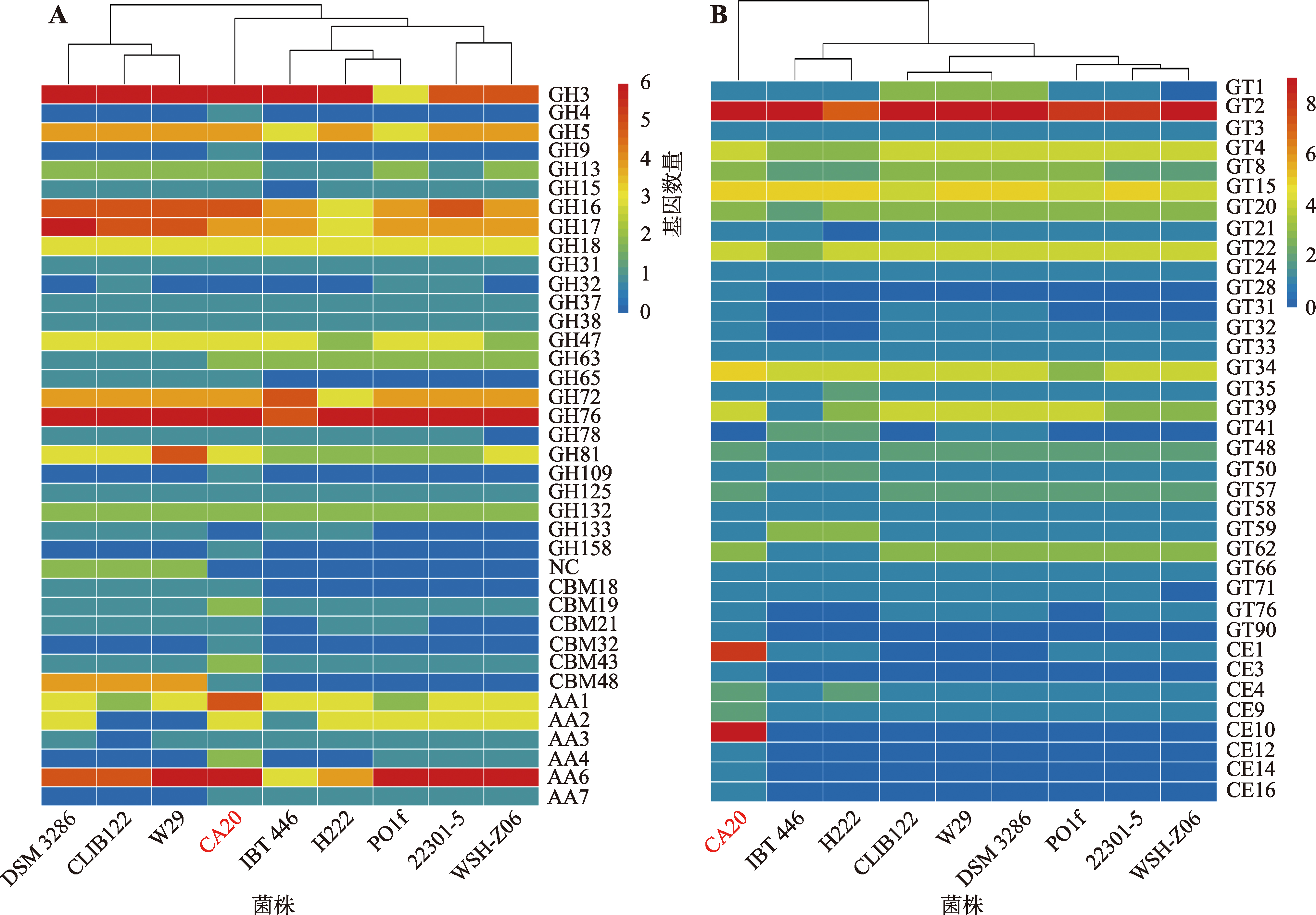
Table 3
Number of carbohydrate-active enzymes in different Y. lipolytica"
| 菌株 | GHs | GTs | CBMs | CEs | AAs | 总和 |
|---|---|---|---|---|---|---|
| Y. lipolytica CA20 | 55 | 60 | 8 | 25 | 18 | 166 |
| Y. lipolytica W29 | 56 | 60 | 8 | 2 | 10 | 136 |
| Y. lipolytica DSM 3286 | 55 | 60 | 8 | 2 | 12 | 137 |
| Y. lipolytica 22301-5 | 48 | 53 | 2 | 3 | 15 | 121 |
| Y. lipolytica IBT 446 | 46 | 48 | 2 | 3 | 9 | 108 |
| Y. lipolytica H222 | 44 | 50 | 3 | 4 | 12 | 113 |
| Y. lipolytica CLIB122 | 55 | 58 | 8 | 2 | 7 | 130 |
| Y. lipolytica PO1f | 45 | 52 | 3 | 3 | 14 | 117 |
| Y. lipolytica WSH-Z06 | 46 | 51 | 2 | 3 | 15 | 117 |
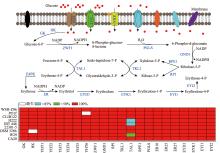
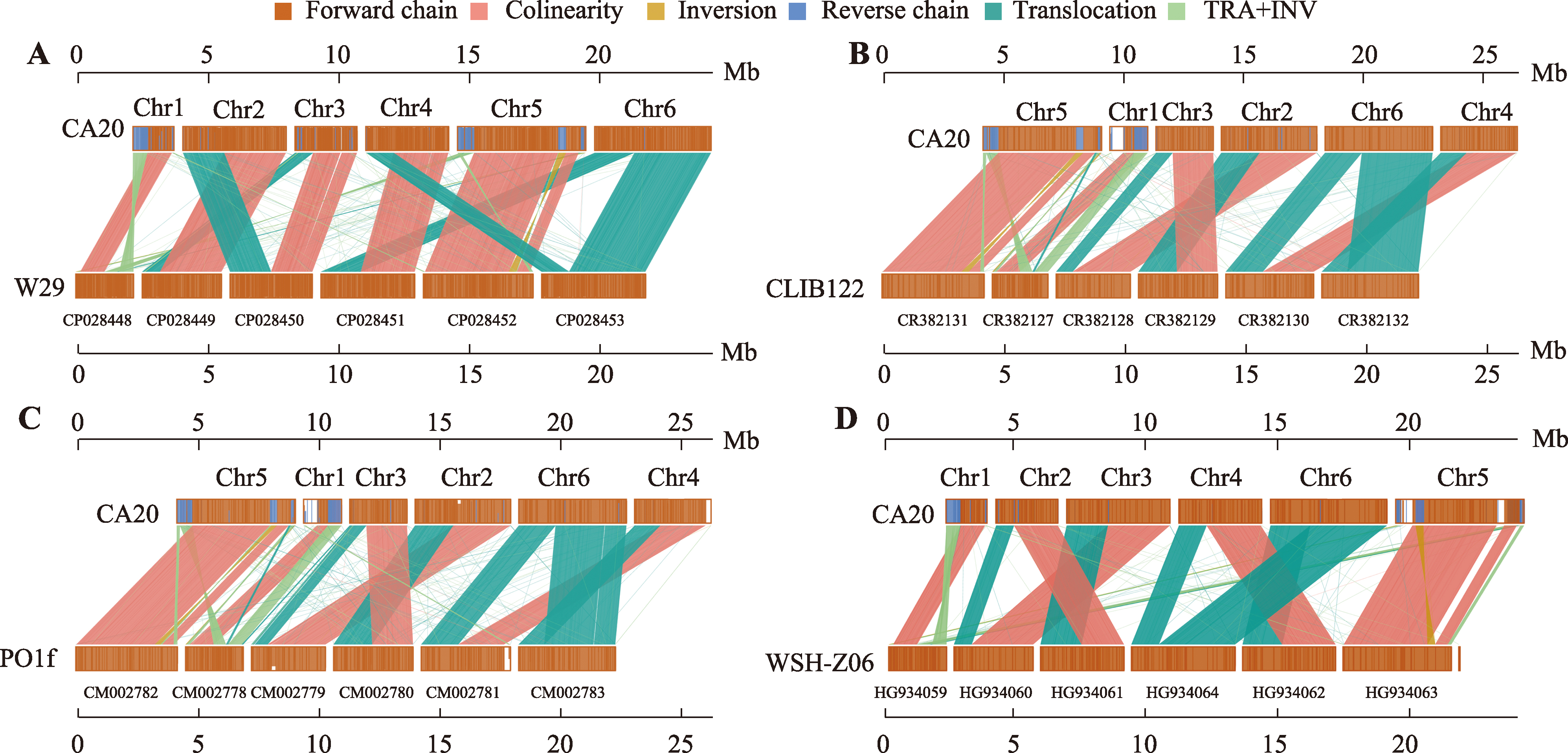
Fig. 9
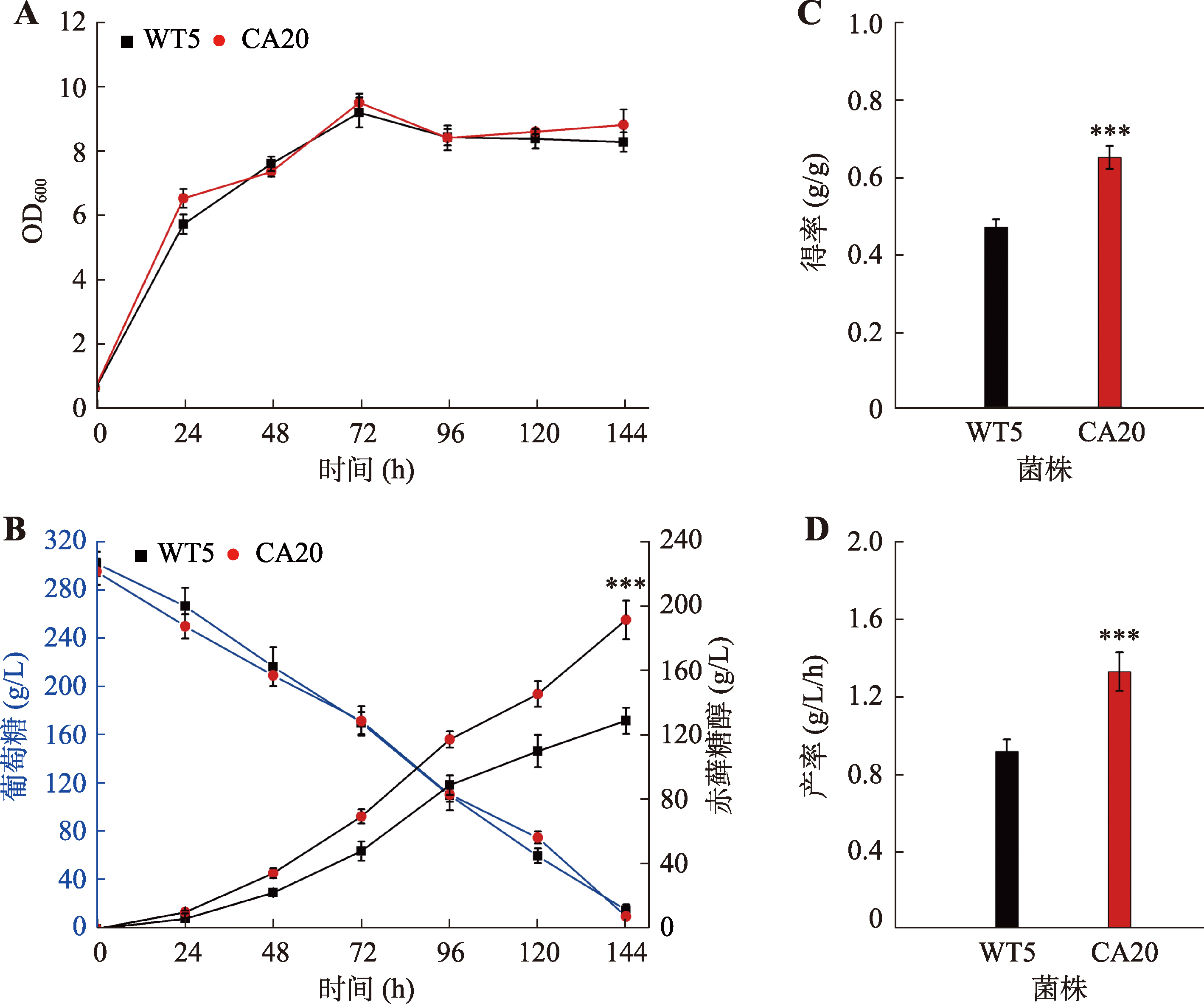
Comparative analysis of the key enzymes involved in the erythritol synthesis YHT1-YHT6:glucose transporter (XP_501516.1,XP_501621.1,XP_505610.1,XP_504302.1,XP_500382.1,and XP_500566.1); GK:glucokinase (VBB78679.1);HK:hexokinase (XP_501216.1);ZWF1:glucose-6-phosphate dehydrogenase (XP_504275.1);PGLS:6-phosphogluconolactonase (VBB78832.1);GND1:phosphogluconate dehydrogenase (XP_500938.1); RPE1:ribulose phosphate 3-epimerase (QNP96251.1);RPI:ribulose-5-phosphate isomerase (XP_500588.1);TKL1:transketolase (XP_503628.1);TAL1:transaldolase (XP_505460.1);E4PK:erythrose-4-phosphate kinase;ER:erythrose reductase [ER10 (XP_502540.1),ER25 (XP_501796.1),and ER27 (XP_505585.1)];EYD1:erythritol dehydrogenase (XP_504870.1);EYK1: erythrulose kinase (XP_504868.1);EYI1:erythrulose-1-phosphate isomerase (XP_504867.1);EYI2:erythrulose-4-phosphate isomerase (XP_504869.1);P:phosphate。"
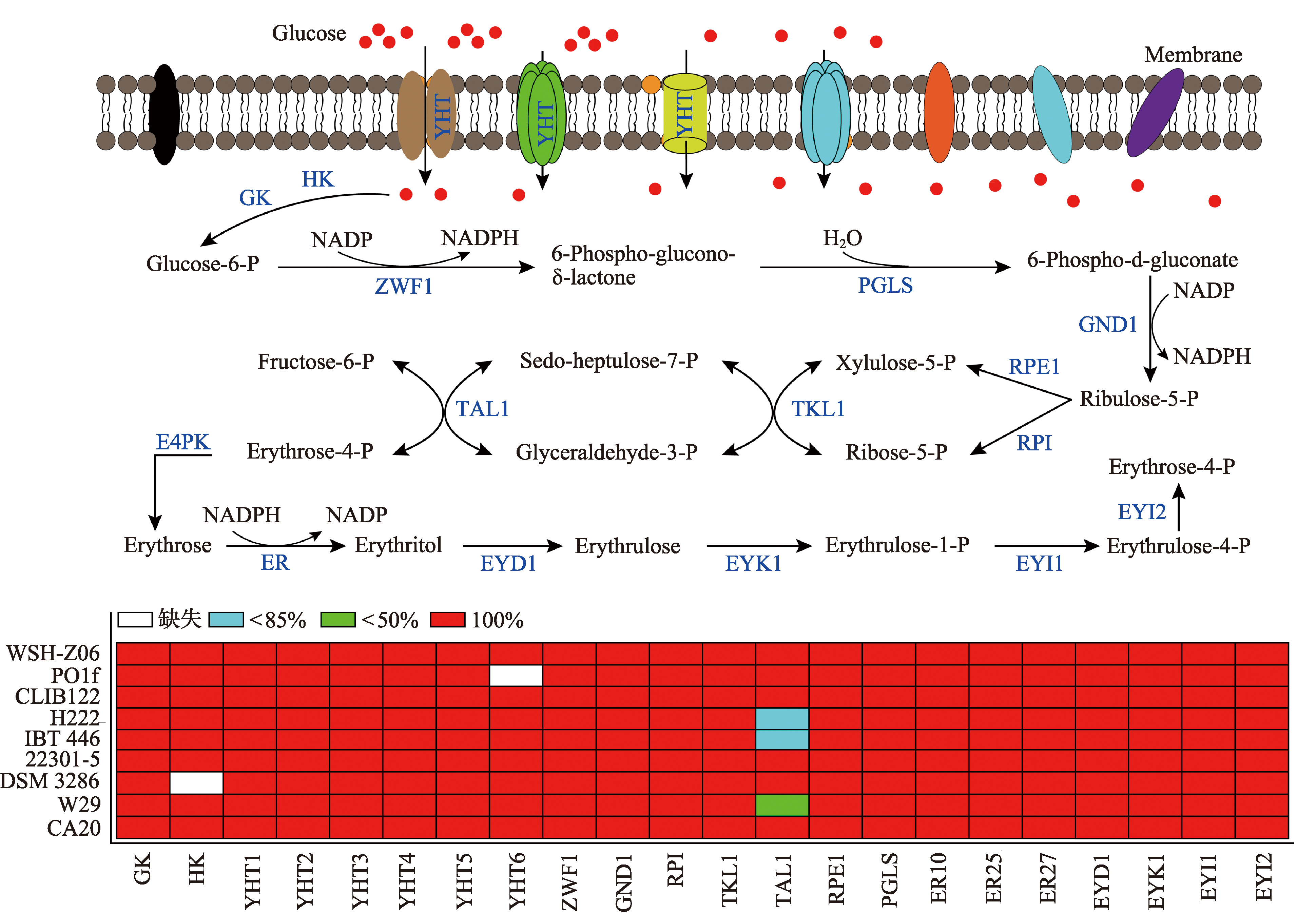
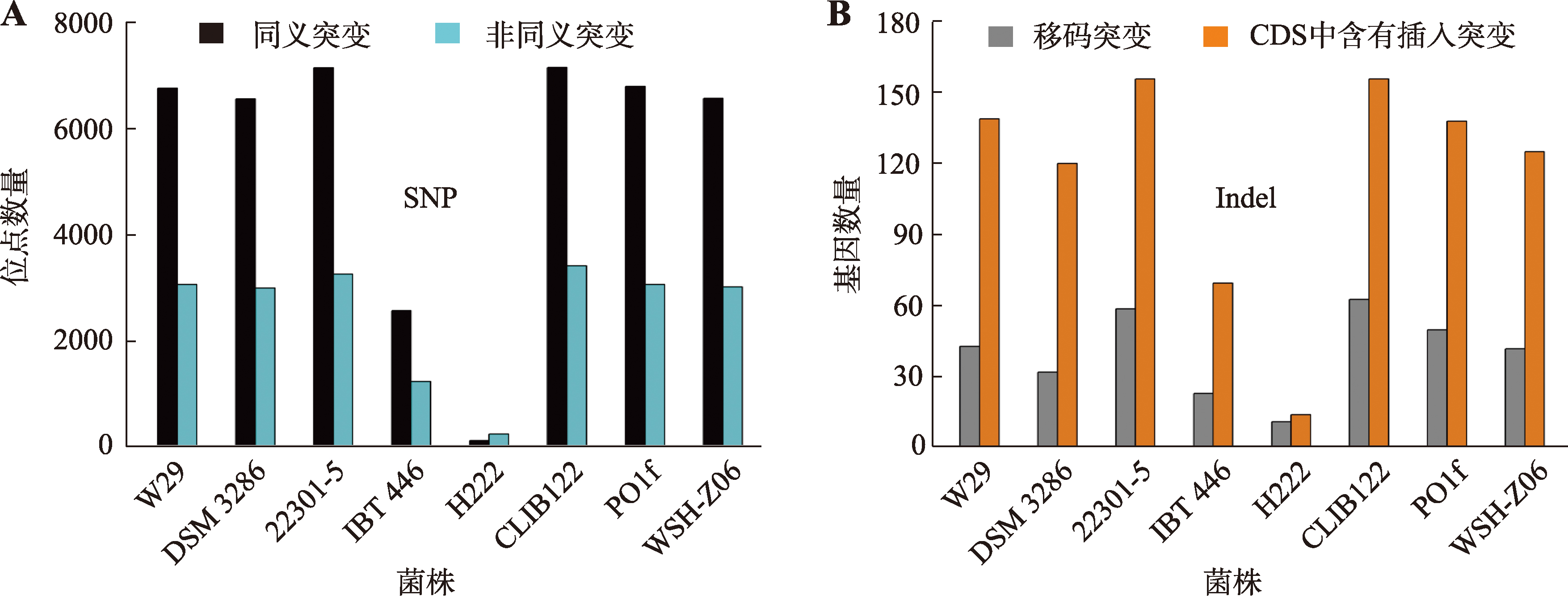
The mutation of CDS resulted from SNP and Indel in CA20 compared with that in WT5"
| 序列号 | 功能 | 突变类型 | 突变位点 |
|---|---|---|---|
| VBB85012.1 | Scaffold protein | 非同义突变 | E1378G |
| RDW32605.1 | Putative methyltransferase | 非同义突变 | L11P |
| VBB85253.1 | Chitin transglycosylase | 非同义突变 | F308S |
| VBB85575.1 | Histone deacetylation protein Rxt3 | 非同义突变 | P230H |
| VBB85808.1 | Hypothetical protein | 非同义突变 | I609N |
| RDW34848.1 | Aspartate/glutamate/uridylate kinase | 非同义突变 | S502P |
| RDW40799.1 | LsmAD domain-domain-containing protein | 非同义突变 | R19G |
| RDW36538.1 | Cytochrome c/c1 heme-lyase | 非同义突变 | S50P |
| RDW34205.1 | Hypothetical protein B0I72DRAFT_135086 | 非同义突变 | L148I |
| VBB88378.1 | Vacuolar alpha mannosidase | 非同义突变 | R1042C |
| VBB88398.1 | Extracellular chitinase | 非同义突变 | L564P |
| VBB79189.1 | Disulfide isomerase precursor | 非同义突变 | S126P |
| SEI34914.1 | YALIA101S05e13674g1_1 | 非同义突变 | F64S |
| VBB87964.1 | Histone transcription regulator 3 | 非同义突变 | F367S |
| RDW50006.1 | Hypothetical protein B0I75DRAFT_142202 | 非同义突变 | P185A |
| SEI35917.1 | YALIA101S09e01662g1_1 | 移码变异 | 基因序列764位插入碱基T |
| RMI98785.1 | Hypothetical protein | 移码变异 | 基因序列1092位插入碱基T |
| SEI32238.1 | Protein kinases | 移码变异 | 基因序列1200位插入碱基A |
| CAG78388.1 | YALI0F18458p | 移码变异 | 基因序列564位插入碱基C |
| VBB88313.1 | Hypothetical protein | 移码变异 | 基因序列3030位A碱基变G碱基 |
| [1] | Liang PX, Cao MF, Li J, Wang QH, Dai ZJ. Expanding sugar alcohol industry: microbial production of sugar alcohols and associated chemocatalytic derivatives. Biotechnol Adv, 2023, 64: 108105. |
| [2] | Liu FM, Zhao XQ, Sha RY, Xia K, Huang J. Advances in microbial production of erythritol. Chin J Bioprocess Eng, 2022, 20(2): 195-205. |
| 刘芳美, 赵学群, 沙如意, 夏凯, 黄俊. 赤藓糖醇微生物合成研究进展. 生物加工过程, 2022, 20(2): 195-205. | |
| [3] | Erian AM, Sauer M. Utilizing yeasts for the conversion of renewable feedstocks to sugar alcohols-a review. Bioresour Technol, 2022, 346: 126296. |
| [4] | Daza-Serna L, Serna-Loaiza S, Masi A, Mach RL, Mach-Aigner AR, Friedl A. From the culture broth to the erythritol crystals: an opportunity for circular economy. Appl Microbiol Biotechnol, 2021, 105(11): 4467-4486. |
| [5] | Chinese Institute of Food Science and Technology. Scientific consensus on erythritol. J Chin Inst Food Sci Technol, 2022, 22(12): 405-412. |
| 中国食品科学技术学会. 赤藓糖醇的科学共识. 中国食品学报, 2022, 22(12): 405-412. | |
| [6] | Liu XY, Yu XJ, He AY, Xia J, He JL, Deng YF, Xu N, Qiu ZY, Wang XY, Zhao PS. One-pot fermentation for erythritol production from distillers grains by the co-cultivation of Yarrowia lipolytica and Trichoderma reesei. Bioresour Technol, 2022, 351: 127053. |
| [7] | Zhang Y, Xu S, Wang N, Chi P, Zhang XY, Cheng HR. Construction and characterization of Yarrowia lipolytica strain with reduced foaming ability. Acta Microbiol Sin, 2022, 62(11): 4165-4175. |
| 张悦, 徐硕, 王楠, 池萍, 张馨月, 程海荣. 发酵产泡性能降低的解脂耶氏酵母菌株的获得及其评价. 微生物学报, 2022, 62(11): 4165-4175. | |
| [8] | Zhang L, Nie MY, Liu F, Chen J, Wei LJ, Hua Q. Multiple gene integration to promote erythritol production on glycerol in Yarrowia lipolytica. Biotechnol Lett, 2021, 43(7): 1277-1287. |
| [9] | Mirończuk AM, Biegalska A, Dobrowolski A. Functional overexpression of genes involved in erythritol synthesis in the yeast Yarrowia lipolytica. Biotechnol Biofuels, 2017, 10: 77. |
| [10] | Liang PX, Li J, Wang QH, Dai ZJ. Enhancing the thermotolerance and erythritol production of Yarrowia lipolytica by introducing heat-resistant devices. Front Bioeng Biotechnol, 2023, 11: 1108653. |
| [11] | Wang N, Chi P, Zou YW, Xu YR, Xu S, Bilal M, Fickers P, Cheng HR. Metabolic engineering of Yarrowia lipolytica for thermoresistance and enhanced erythritol productivity. Biotechnol Biofuels, 2020, 13: 176. |
| [12] | Mirończuk AM, Rakicka M, Biegalska A, Rymowicz W, Dobrowolski A. A two-stage fermentation process of erythritol production by yeast Y. lipolytica from molasses and glycerol. Bioresour Technol, 2015, 198: 445-455. |
| [13] | Qiu XL, Xu P, Zhao XR, Du GC, Zhang J, Li JH. Combining genetically-encoded biosensors with high throughput strain screening to maximize erythritol production in Yarrowia lipolytica. Metab Eng, 2020, 60: 66-76. |
| [14] | Liu XY, Yu XJ, Lv JS, Xu JX, Xia J, Wu Z, Zhang T, Deng YF. A cost-effective process for the coproduction of erythritol and lipase with Yarrowia lipolytica M53 from waste cooking oil. Food Bioprod Process, 2017, 103: 86-94. |
| [15] | Ghezelbash GR, Nahvi I, Emamzadeh R. Improvement of erythrose reductase activity, deletion of by-products and statistical media optimization for enhanced erythritol production from Yarrowia lipolytica mutant 49. Curr Microbiol, 2014, 69(2): 149-157 |
| [16] | Song B, Li XF, Shi ZY, Li CT, Fu YP, Li Y. Breeding of new Pleurotus ostreatus strain by mutation with60Co-γ irradiation. Acta Microbiol Sin, 2019, 59(11): 2155-2164. |
| 宋冰, 李雪飞, 史泽宇, 李长田, 付永平, 李玉. 钴60诱变选育平菇新菌株的研究. 微生物学报, 2019, 59(11): 2155-2164. | |
| [17] | Liu FM, Xia K, Peng YT, Zhao XQ, Sha RY, Huang J. Breeding of Yarrowia lipolytica strains with improved erythritol production by combined mutation and optimization of fermentation process. J Nucl Agric Sci, 2023, 37(5): 907-916. |
| 刘芳美, 夏凯, 彭艳婷, 赵学群, 沙如意, 黄俊. 复合诱变选育高产赤藓糖醇解脂亚罗酵母及其发酵工艺优化. 核农学报, 2023, 37(5): 907-916. | |
| [18] | Feng JY, He JK, Sun JY, Fu XZ, Yuan JA, Guo HP. Genome sequencing and comparative genome analysis of Pseudomonas boreopolis GO2, a strain producing bioflocculant with lignocellulosic biomass. Acta Microbiol Sin, 2023, 63(2): 786-804. |
| 冯嘉茵, 何继堃, 孙嘉怡, 符学志, 袁佳骜, 郭海朋. 利用木质纤维素类生物质产微生物絮凝剂Pseudomonas boreopolis GO2的全基因组测序及比较基因组学分析. 微生物学报, 2023, 63(2): 786-804. | |
| [19] | Xia K, Han CC, Xu J, Liang XL. Toxin-antitoxin HicAB regulates the formation of persister cells responsible for the acid stress resistance in Acetobacter pasteurianus. Appl Microbiol Biotechnol, 2021, 105(2): 725-739. |
| [20] | Liu XY, Yu XJ, Xia J, Lv JS, Xu JX, Dai BL, Xu XQ, Xu JM. Erythritol production by Yarrowia lipolytica from okara pretreated with the in-house enzyme pools of fungi. Bioresour Technol, 2017, 244(Pt 1): 1089-1095. |
| [21] | Walker C, Dien B, Giannone RJ, Slininger P, Thompson SR, Trinh CT. Exploring proteomes of robust Yarrowia lipolytica isolates cultivated in biomass hydrolysate reveals key processes impacting mixed sugar utilization, lipid accumulation, and degradation. mSystems, 2021, 6(4): e0044321. |
| [22] | Liu LQ, Alper HS. Draft genome sequence of the oleaginous yeast Yarrowia lipolytica PO1f, a commonly used metabolic engineering host. Genome Announc, 2014, 2(4): e00652-14. |
| [23] | Park YK, Ledesma-Amaro R. What makes Yarrowia lipolytica well suited for industry? Trends Biotechnol, 2023, 41(2): 242-254. |
| [24] | Silva JP, Ticona ARP, Hamann PRV, Quirino BF, Noronha EF. Deconstruction of lignin: from enzymes to microorganisms. Molecules, 2021, 26(8): 2299. |
| [25] | Celińska E, Nicaud JM, Białas W. Hydrolytic secretome engineering in Yarrowia lipolytica for consolidated bioprocessing on polysaccharide resources: review on starch, cellulose, xylan, and inulin. Appl Microbiol Biotechnol, 2021, 105(3): 975-989. |
| [26] | Ryu S, Hipp J, Trinh CT. Activating and elucidating metabolism of complex sugars in Yarrowia lipolytica. Appl Environ Microbiol, 2015, 82(4): 1334-1345. |
| [27] | Moreno AD, Ibarra D, Fernández JL, Ballesteros M.Different laccase detoxification strategies for ethanol production from lignocellulosic biomass by the thermotolerant yeast Kluyveromyces marxianus CECT 10875. Bioresour Technol, 2012, 106: 101-109. |
| [28] | Sánchez C. Lignocellulosic residues: biodegradation and bioconversion by fungi. Biotechnol Adv, 2009, 27(2): 185-194. |
| [29] | Peng MF, Li PJ, Shan Y, Chen YQ, Yang DJ, Lei LC, Huang ZH, Yu KX. Comparative genome analysis of carbohydrate activity enzymes zymogram of Lactobacillus from Guangxi pickle. Food Ferment Ind, 2021, 47(4): 68-73. |
| 彭明芳, 李培骏, 单杨, 陈玉秋, 杨岱峻, 雷丽嫦, 黄芝辉, 余孔新. 比较基因组揭示广西酸菜乳杆菌碳水化合物活性酶谱. 食品与发酵工业, 2021, 47(4): 68-73. | |
| [30] | Lazar Z, Neuvéglise C, Rossignol T, Devillers H, Morin N, Robak M, Nicaud JM, Crutz-Le Coq AM. Characterization of hexose transporters in Yarrowia lipolytica reveals new groups of sugar porters involved in yeast growth. Fungal Genet Biol, 2017, 100: 1-12. |
| [31] | Hapeta P, Szczepańska P, Witkowski T, Nicaud JM, Crutz-Le Coq AM, Lazar Z. The Role of hexokinase and hexose transporters in preferential use of glucose over fructose and downstream metabolic pathways in the yeast Yarrowia lipolytica. Int J Mol Sci, 2021, 22(17): 9282. |
| [32] | Cheng HL, Wang SQ, Bilal M, Ge XM, Zhang C, Fickers P, Cheng HR. Identification, characterization of two NADPH-dependent erythrose reductases in the yeast Yarrowia lipolytica and improvement of erythritol productivity using metabolic engineering. Microb Cell Fact, 2018, 17(1): 133. |
| [33] | Qiu XL, Gu Y, Du GC, Zhang J, Xu P, Li JH. Conferring thermotolerant phenotype to wild-type Yarrowia lipolytica improves cell growth and erythritol production. Biotechnol Bioeng, 2021, 118(8): 3117-3127. |
| [34] | Yang LB, Zhan XB, Zheng ZY, Wu JR, Gao MJ, Lin CC. A novel osmotic pressure control fed-batch fermentation strategy for improvement of erythritol production by Yarrowia lipolytica from glycerol. Bioresour Technol, 2014, 151: 120-127. |
| [35] | Dickson RC, Sumanasekera C, Lester RL. Functions and metabolism of sphingolipids in Saccharomyces cerevisiae. Prog Lipid Res, 2006, 45(6): 447-465. |
| [36] | Caspeta L, Nielsen J. Thermotolerant yeast strains adapted by laboratory evolution show trade-off at ancestral temperatures and preadaptation to other stresses. mBio, 2015, 6(4): e00431. |
| [37] | Li B, Liu N, Zhao XB. Response mechanisms of Saccharomyces cerevisiae to the stress factors present in lignocellulose hydrolysate and strategies for constructing robust strains. Biotechnol Biofuels Bioprod, 2022, 15(1): 28. |
| [38] | Caspeta L, Chen Y, Ghiaci P, Feizi A, Buskov S, Hallström BM, Petranovic D, Nielsen J. Altered sterol composition renders yeast thermotolerant. Science, 2014, 346(6205): 75-78. |
| [1] | Xiaocui Li, Kaicheng Kang, Xianzhong Huang, Yongbin Fan, Miaomiao Song, Yunjie Huang, Jiajia Ding. Genome-wide identification, phylogenetic analysis and expression profiling of the MKK gene family in Arabidopsis pumila [J]. Hereditas(Beijing), 2020, 42(4): 403-421. |
| [2] | Yifan Wang,Zhen Li,Jiaowen Pan,Yingxiu Li,Qingguo Wang,Guan Yan'an,Wei Liu. Cloning and functional analysis of the SiRLK35 gene in Setaria italic L. [J]. Hereditas(Beijing), 2017, 39(5): 413-422. |
| [3] | LI Chao-Bei, HU Li-Li, WANG Zhen-Dong, ZHONG Chu-Qi, LEI Lei. Regulation of compaction initiation in mouse embryo [J]. HEREDITAS, 2009, 31(12): 1177-1184. |
| [4] | . Plant non-host resistance: current progress and future prospect [J]. HEREDITAS, 2008, 30(8): 977-982. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||