Hereditas(Beijing) ›› 2022, Vol. 44 ›› Issue (4): 313-321.doi: 10.16288/j.yczz.21-415
• Review • Previous Articles Next Articles
Progress on methods for acquiring flanking genomic sequence
Jiming Xu( ), Jianshu Zhu, Mengzhen Li, Han Hu, Chuanzao Mao(
), Jianshu Zhu, Mengzhen Li, Han Hu, Chuanzao Mao( )
)
- Institute of Plant Biology, College of Life Science, Zhejiang University, Hangzhou 310058, China
-
Received:2021-12-02Revised:2022-03-08Online:2022-04-20Published:2022-03-25 -
Contact:Mao Chuanzao E-mail:xujiming@zju.edu.cn;mcz@zju.edu.cn -
Supported by:Supported by the National Natural Science Foundation of China No(32002121)
Cite this article
Jiming Xu, Jianshu Zhu, Mengzhen Li, Han Hu, Chuanzao Mao. Progress on methods for acquiring flanking genomic sequence[J]. Hereditas(Beijing), 2022, 44(4): 313-321.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
Table 1
The comparison of methods for cloning flanking sequence"
| 方法 | DNA酶切 | DNA用量 | 限制因素 | 非特异性产物 | 灵敏度 | 操作复杂性 |
|---|---|---|---|---|---|---|
| 质粒拯救 | 需要 | 2~5 μg | 酶切位点选择;连接或环化效率 | 菌落假阳性 | 低 | 复杂 |
| 反向PCR | 需要 | 2~5 μg | 酶切位点选择;连接或环化效率 | 非特异性连接 | 低 | 复杂 |
| 接头介导PCR | 需要 | 2~5 μg | 酶切位点选择;连接或环化效率 | 单引物扩增 | 中等 | 复杂 |
| 半随机引物PCR | 不需要 | 20~200 ng | 简并引物设计;插入载体完整性 | 单引物扩增 | 高 | 中等 |
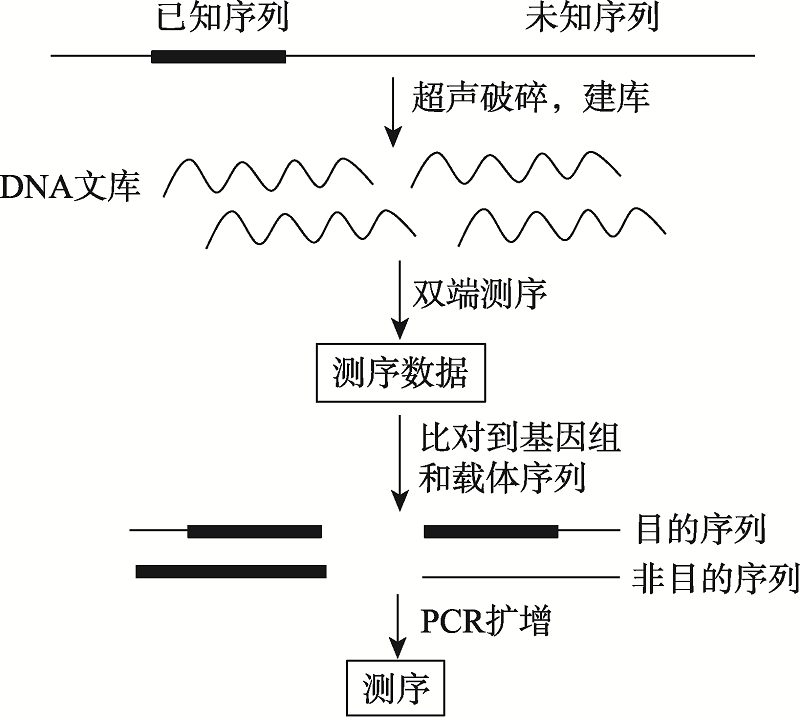
| 基因组重测序 | 不需要 | 2~5 μg | 测序深度;插入位置 | 无 | 非常高 | 简单 |
| [1] | Wei Q, Ao Y, Yang MM, Chen T, Han H, Zhang XJ, Wang R, Xia QJ, Jiang FF, Li Y. Identification of genomic insertion of dominant-negative GHR mutation transgenes in Wuzhishan pig using whole genome sequencing method. Hereditas(Beijing), 2021, 43(12):1149-1158. |
| 魏强, 奥岩, 杨漫漫, 陈涛, 韩虎, 张兴举, 王然, 夏秋菊, 姜芳芳, 李勇. 利用全基因组重测序技术鉴定五指山猪GHR突变体转基因插入位点. 遗传, 2021, 43(12):1149-1158. | |
| [2] |
Liu B, Su Q, Tang MQ, Yuan XD, An LJ. Progress of the PCR amplification techniques for chromosome walking. Hereditas(Beijing), 2006, 28(5):587-595.
pmid: 16735240 |
|
刘博, 苏乔, 汤敏谦, 袁晓东, 安利佳. 应用于染色体步移的PCR扩增技术的研究进展. 遗传, 2006, 28(5):587-595.
pmid: 16735240 |
|
| [3] |
Perucho M, Hanahan D, Lipsich L, Wigler M. Isolation of the chicken thymidine kinase gene by plasmid rescue. Nature, 1980, 285(5762):207-210.
doi: 10.1038/285207a0 |
| [4] | Zhang JC, Zhang XW, Dai XZ. Construction of rescue plasmid with T-DNA tag. J Anhui Agri Sci, 2007, 35(34):11016-11018. |
| 张金谌, 张学文, 戴雄泽. 质粒拯救型T-DNA标签质粒的构建. 安徽农业科学, 2007, 35(34):11016-11018. | |
| [5] | Li ZM, Zhang HK, Cao JS, He ZH. Construction of an activation tagging library of Arabidopsis and cloning for mutant genes. J Plant Physiol Mol Biol, 2005, 31(5):499-506. |
| 李志邈, 张海扩, 曹家树, 何祖华. 拟南芥激活标记突变体库的构建及突变体基因的克隆. 植物生理与分子生物学学报, 2005, 31(5):499-506. | |
| [6] |
Li AH, Zhang YF, Wu CY, Tang W, Wu R, Dai ZY, Liu GQ, Zhang HX, Pan XB. Screening for and genetic analysis on T-DNA-inserted mutant pool in rice. Acta Genet Sin, 2006, 33(4):319-329.
doi: 10.1016/S0379-4172(06)60057-7 |
| [7] |
Mizobuchi M, Frohman LA. Rapid amplification of genomic DNA ends. Biotechniques, 1993, 15(2):214-216.
pmid: 8373580 |
| [8] |
Ochman H, Gerber AS, Hartl DL. Genetic applications of an inverse polymerase chain reaction. Genetics, 1988, 120(3):621-623.
doi: 10.1093/genetics/120.3.621 pmid: 2852134 |
| [9] |
Kohda T, Taira K. A simple and efficient method to determine the terminal sequences of restriction fragments containing known sequences. DNA Res, 2000, 7(2):151-155.
pmid: 10819332 |
| [10] |
Chen L, Tu ZM, Hussain J, Cong L, Yan YJ, Jin L, Yang GX, He GY. Isolation and heterologous transformation analysis of a pollen-specific promoter from wheat (Triticum aestivum L.). Mol Biol Rep, 2010, 37(2):737-744.
doi: 10.1007/s11033-009-9582-7 |
| [11] |
Forster C, Arthur E, Crespi S, Hobbs SL, Mullineaux P, Casey R. Isolation of a pea (Pisum sativum) seed lipoxygenase promoter by inverse polymerase chain reaction and characterization of its expression in transgenic tobacco. Plant Mol Biol, 1994, 26(1):235-248.
pmid: 7948873 |
| [12] | Han ZY, Wang XQ, Shen GZ. Cloning of foreign gene's flanking sequences in transgenic rice by inverse PCR. Acta Agric Shanghai, 2001, 17(2):27-32. |
| 韩志勇, 王新其, 沈革志. 反向PCR克隆转基因水稻的外源基因旁侧序列. 上海农业学报, 2001, 17(2):27-32. | |
| [13] |
Ohshima K, Mukai Y, Shiraki H, Suzumiya J, Tashiro K, Kikuchi M. Clonal integration and expression of human T-cell lymphotropic virus type I in carriers detected by polymerase chain reaction and inverse PCR. Am J Hematol, 1997, 54(4):306-312.
pmid: 9092686 |
| [14] |
Jones DH. Panhandle PCR. PCR Methods Appl, 1995, 4(5):S195-S201.
doi: 10.1101/gr.4.5.s195 pmid: 7580908 |
| [15] |
Myrick KV, Gelbart WM. Universal fast walking for direct and versatile determination of flanking sequence. Gene, 2002, 284(1-2):125-131.
pmid: 11891053 |
| [16] |
Wang SM, He J, Cui ZL, Li SP. Self-formed adaptor PCR: A simple and efficient method for chromosome walking. Appl Environ Microbiol, 2007, 73(15):5048-5051.
doi: 10.1128/AEM.02973-06 |
| [17] |
Shyamala V, Ames GF. Genome walking by single- specific-primer polymerase chain reaction: SSP-PCR. Gene, 1989, 84(1):1-8.
pmid: 2691331 |
| [18] |
Lagerström M, Parik J, Malmgren H, Stewart J, Pettersson U, Landegren U. Capture PCR: efficient amplification of DNA fragments adjacent to a known sequence in human and YAC DNA. PCR Methods Appl, 1991, 1(2):111-119.
pmid: 1842928 |
| [19] |
Arnold C, Hodgson IJ. Vectorette PCR: a novel approach to genomic walking. PCR Methods Appl, 1991, 1(1):39-42.
pmid: 1842919 |
| [20] |
Kilstrup M, Kristiansen KN. Rapid genome walking: a simplified oligo-cassette mediated polymerase chain reaction using a single genome-specific primer. Nucleic Acids Res, 2000, 28(11):E55.
doi: 10.1093/nar/28.11.e55 pmid: 10871354 |
| [21] |
Siebert PD, Chenchik A, Kellogg DE, Lukyanov KA, Lukyanov SA. An improved PCR method for walking in uncloned genomic DNA. Nucleic Acids Res, 1995, 23(6):1087-1088.
pmid: 7731798 |
| [22] |
Tan GH, Gao Y, Shi M, Zhang XY, He SP, Chen ZL, An CC. SiteFinding-PCR: a simple and efficient PCR method for chromosome walking. Nucleic Acids Res, 2005, 33(13):e122.
doi: 10.1093/nar/gni124 |
| [23] |
Wang Z, Ye SF, Li JJ, Zheng B, Bao MZ, Ning GG. Fusion primer and nested integrated PCR (FPNI-PCR): a new high-efficiency strategy for rapid chromosome walking or flanking sequence cloning. BMC Biotechnol, 2011, 11:109.
doi: 10.1186/1472-6750-11-109 pmid: 22093809 |
| [24] |
García-Cerdán JG, Schmid EM, Takeuchi T, McRae I, McDonald KL, Yordduangjun N, Hassan AM, Grob P, Xu CS, Hess HF, Fletcher DA, Nogales E, Niyogi KK. Chloroplast Sec14-like 1 (CPSFL1) is essential for normal chloroplast development and affects carotenoid accumulation in Chlamydomonas. Proc Natl Acad Sci USA, 2020, 117(22):12452-12463.
doi: 10.1073/pnas.1916948117 |
| [25] |
Hsu PJ, Tan MC, Shen HL, Chen YH, Wang YY, Hwang SG, Chiang MH, Le QV, Kuo WS, Chou YC, Lin SY, Jauh GY, Cheng WH. The nucleolar protein SAHY1 is involved in pre-rRNA processing and normal plant growth. Plant Physiol, 2021, 185(3):1039-1058.
doi: 10.1093/plphys/kiaa085 |
| [26] |
Currall BB, Chen M, Sallari RC, Cotter M, Wong KE, Robertson NG, Penney KL, Lunardi A, Reschke M, Hickox AE, Yin YB, Wong GT, Fung J, Brown KK, Williamson RE, Sinnott-Armstrong NA, Kammin T, Ivanov A, Zepeda-Mendoza CJ, Shen J, Quade BJ, Signoretti S, Arnos KS, Banks AS, Patsopoulos N, Liberman MC, Kellis M, Pandolfi PP, Morton CC. Loss of LDAH associated with prostate cancer and hearing loss. Hum Mol Genet, 2018, 27(24):4194-4203.
doi: 10.1093/hmg/ddy310 |
| [27] |
Liu Q, Li L, Cheng HT, Yao LX, Wu J, Huang H, Ning W, Kai GY. The basic helix-loop-helix transcription factor TabHLH1 increases chlorogenic acid and luteolin biosynthesis in Taraxacum antungense Kitag. Hortic Res, 2021, 8(1):195.
doi: 10.1038/s41438-021-00630-y |
| [28] |
Parker JD, Rabinovitch PS, Burmer GC. Targeted gene walking polymerase chain reaction. Nucleic Acids Res, 1991, 19(11):3055-3060.
pmid: 2057362 |
| [29] |
Liu YG, Whittier RF. Thermal asymmetric interlaced PCR: automatable amplification and sequencing of insert end fragments from P1 and YAC clones for chromosome walking. Genomics, 1995, 25(3):674-681.
pmid: 7759102 |
| [30] |
Gong WK, Zhou Y, Wang R, Wei XL, Zhang L, Dai Y, Zhu Z. Analysis of T-DNA integration events in transgenic rice. J Plant Physiol, 2021, 266:153527.
doi: 10.1016/j.jplph.2021.153527 |
| [31] |
Jaser SKK, Perazza CA, Fávaro LCL, Goto MA, de Oliveira AM, Hallerman E, Hilsdorf AWS. Identification and analysis of a novel microsatellite marker within the growth hormone gene promoter of Colossoma macropomum(Characiformes: Characidae) detected by TAIL-PCR. J Appl Ichthyol, 2021, 37(3):439-448.
doi: 10.1111/jai.14170 |
| [32] |
Feng XB, Zheng ZW, Zhang X, Gu J, Feng QL, Huang LH. Discovering genes responsible for silk synthesis in Bombyx mori by piggyBac-based random insertional mutagenesis. Insect Sci, 2019, 26(5):821-830.
doi: 10.1111/1744-7917.12595 |
| [33] | Guan LY, Lu RY, Wu ZJ, Zhong GW, Zhang SZ. Precise expression of afmed15 is crucial for asexual development, virulence, and survival of aspergillus fumigatus. mSphere, 2020, 5(5):e00771-20. |
| [34] |
Liu YG, Chen YL. High-efficiency thermal asymmetric interlaced PCR for amplification of unknown flanking sequences. Biotechniques, 2007, 43(5): 649-650,652,654 passim.
doi: 10.2144/000112601 |
| [35] | Hou N, He HQ, Dong M, Xu RQ, Wan YS, Jin WJ, Liu HB. The exogenous gene integrated structure and event-specific detection of insect resistant transgenic cotton. Mol Plant Breed, 2012, 10(3):317-323. |
| 侯娜, 贺辉群, 董美, 徐荣旗, 宛煜嵩, 金芜军, 刘好宝. 转基因抗虫棉外源DNA插入整合结构分析和转化事件特异性检测方法的建立. 分子植物育种, 2012, 10(3):317-323. | |
| [36] |
Chen M, Xu ZS, Xia LQ, Li LC, Cheng XG, Dong JH, Wang QY, Ma YZ. Cold-induced modulation and functional analyses of the DRE-binding transcription factor gene, GmDREB3, in soybean(Glycine max L.). J Exp Bot, 2009, 60(1):121-135.
doi: 10.1093/jxb/ern269 |
| [37] |
Schouten HJ, Vande Geest H, Papadimitriou S, Bemer M, Schaart JG, Smulders MJM, Perez GS, Schijlen E. Re-sequencing transgenic plants revealed rearrangements at T-DNA inserts, and integration of a short T-DNA fragment, but no increase of small mutations elsewhere. Plant Cell Rep, 2017, 36(3):493-504.
doi: 10.1007/s00299-017-2098-z pmid: 28155116 |
| [38] |
Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics, 2009, 25(14):1754-1760.
doi: 10.1093/bioinformatics/btp324 |
| [39] | Guo BF, Guo Y, Hong HL, Qiu LJ. Identification of genomic insertion and flanking sequence of G2-EPSPS and GAT transgenes in soybean using whole genome sequencing method. Front Plant Sci, 2016, 7:1009. |
| [40] |
Polko JK, Temanni MR, van Zanten M, van Workum W, Iburg S, Pierik R, Voesenek LACJ, Peeters AJM. Illumina sequencing technology as a method of identifying T-DNA insertion loci in activation-tagged Arabidopsis thaliana plants. Mol Plant, 2012, 5(4):948-950.
doi: 10.1093/mp/sss022 |
| [41] | Xu JM, Hu H, Mao WX, Mao CZ. Identifying T-DNA insertion site(s) of transgenic plants by whole-genome resequencing. Hereditas(Beijing), 2018, 40(8):676-682. |
| 徐纪明, 胡晗, 毛文轩, 毛传澡. 利用重测序技术获取转基因植物T-DNA插入位点. 遗传, 2018, 40(8):676-682. | |
| [42] |
Sun L, Ge YB, Sparks JA, Robinson ZT, Cheng XF, Wen JQ. TDNAscan: a software to identify complete and truncated T-DNA insertions. Front Genet, 2019, 10:685.
doi: 10.3389/fgene.2019.00685 |
| [43] |
Peng C, Mei YT, Ding L, Wang XF, Chen XY, Wang JM, Xu JF. Using combined methods of genetic mapping and nanopore-based sequencing technology to analyze the insertion positions of G10evo-EPSPS and Cry1Ab/Cry2Aj transgenes in maize. Front Plant Sci, 2021, 12:690951.
doi: 10.3389/fpls.2021.690951 |
| [44] |
Li SJ, Jia SG, Hou LL, Nguyen H, Sato S, Holding D, Cahoon E, Zhang C, Clemente T, Yu B. Mapping of transgenic alleles in soybean using a nanopore-based sequencing strategy. J Exp Bot, 2019, 70(15):3825-3833.
doi: 10.1093/jxb/erz202 |
| [45] |
Nicholls PK, Bellott DW, Cho TJ, Pyntikova T, Page DC. Locating and characterizing a transgene integration site by Nanopore sequencing. G3 (Bethesda), 2019, 9(5):1481-1486.
doi: 10.1534/g3.119.300582 |
| [1] | Jun Ma, Anping Fan, Wusheng Wang, Jinchuan Zhang, Xiaojun Jiang, Ruijun Ma, Sheqiang Jia, Fei Liu, Chuchao Lei, Yongzhen Huang. Analysis of genetic diversity and genetic structure of Qinchuan cattle conservation population using whole-genome resequencing [J]. Hereditas(Beijing), 2023, 45(7): 602-616. |
| [2] | Jiming Xu,Han Hu,Wenxuan Mao,Chuanzao Mao. Identifying T-DNA insertion site(s) of transgenic plants by whole-genome resequencing [J]. Hereditas(Beijing), 2018, 40(8): 676-682. |
| [3] | Jinhong Yuan, Junhua Li, Jiaojiao Yuan, Keli Jia, Shufen Li, Chuanliang Deng, Wujun Gao. The application of MutMap in forward genetic studies based on whole-genome sequencing [J]. Hereditas(Beijing), 2017, 39(12): 1168-1177. |
| [4] | Dingding Zhao, Zhongying Qiao, Xiao Cheng, Jianping Wang, Cuicui Jiao, Bingyao Sun. Molecular identification of one event of Ds excision and re-insertion at two loci in rice genome [J]. HEREDITAS(Beijing), 2014, 36(12): 1249-1255. |
| [5] | YANG Lin, FU Feng-Ling, LI Wan-Chen. T-DNA integration patterns in transgenic plants mediated by Agrobacterium tumefaciens [J]. HEREDITAS, 2011, 33(12): 1327-1334. |
| [6] | SUN Bing-Yao, TAN Jian-Zhong, LU Xiao-Ping, QU Chun-Xiang, WAN Zhi-Gang, GU Fu-Gen. Amplification of Ds Flanking Sequences from Rice Genomic DNA of Hybrids of Ac × Ds Lines and the Analysis of Ds Insertions [J]. HEREDITAS, 2006, 28(12): 1555-1555~1561. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||