Hereditas(Beijing) ›› 2023, Vol. 45 ›› Issue (7): 602-616.doi: 10.16288/j.yczz.23-115
• Research Article • Previous Articles Next Articles
Analysis of genetic diversity and genetic structure of Qinchuan cattle conservation population using whole-genome resequencing
Jun Ma1( ), Anping Fan2, Wusheng Wang2, Jinchuan Zhang2, Xiaojun Jiang2, Ruijun Ma2, Sheqiang Jia2, Fei Liu2, Chuchao Lei1, Yongzhen Huang1(
), Anping Fan2, Wusheng Wang2, Jinchuan Zhang2, Xiaojun Jiang2, Ruijun Ma2, Sheqiang Jia2, Fei Liu2, Chuchao Lei1, Yongzhen Huang1( )
)
- 1. College of Animal Science and Technology, Northwest A&F University, Yangling 712100, China
2. Shaanxi Provincial Livestock Breeding Farm, Baoji 722203, China
-
Received:2023-04-25Revised:2023-06-11Online:2023-07-20Published:2023-06-30 -
Contact:Yongzhen Huang E-mail:Junma96@163.com;hyzsci@nwafu.edu.cn -
Supported by:Supported by the Key R&D project of Shaanxi Province(2023-YBNY-141);Ministry of Finance and Ministry of Agriculture and Rural Affairs-National Modern Agricultural Industry Technology System(CARS-37)
Cite this article
Jun Ma, Anping Fan, Wusheng Wang, Jinchuan Zhang, Xiaojun Jiang, Ruijun Ma, Sheqiang Jia, Fei Liu, Chuchao Lei, Yongzhen Huang. Analysis of genetic diversity and genetic structure of Qinchuan cattle conservation population using whole-genome resequencing[J]. Hereditas(Beijing), 2023, 45(7): 602-616.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
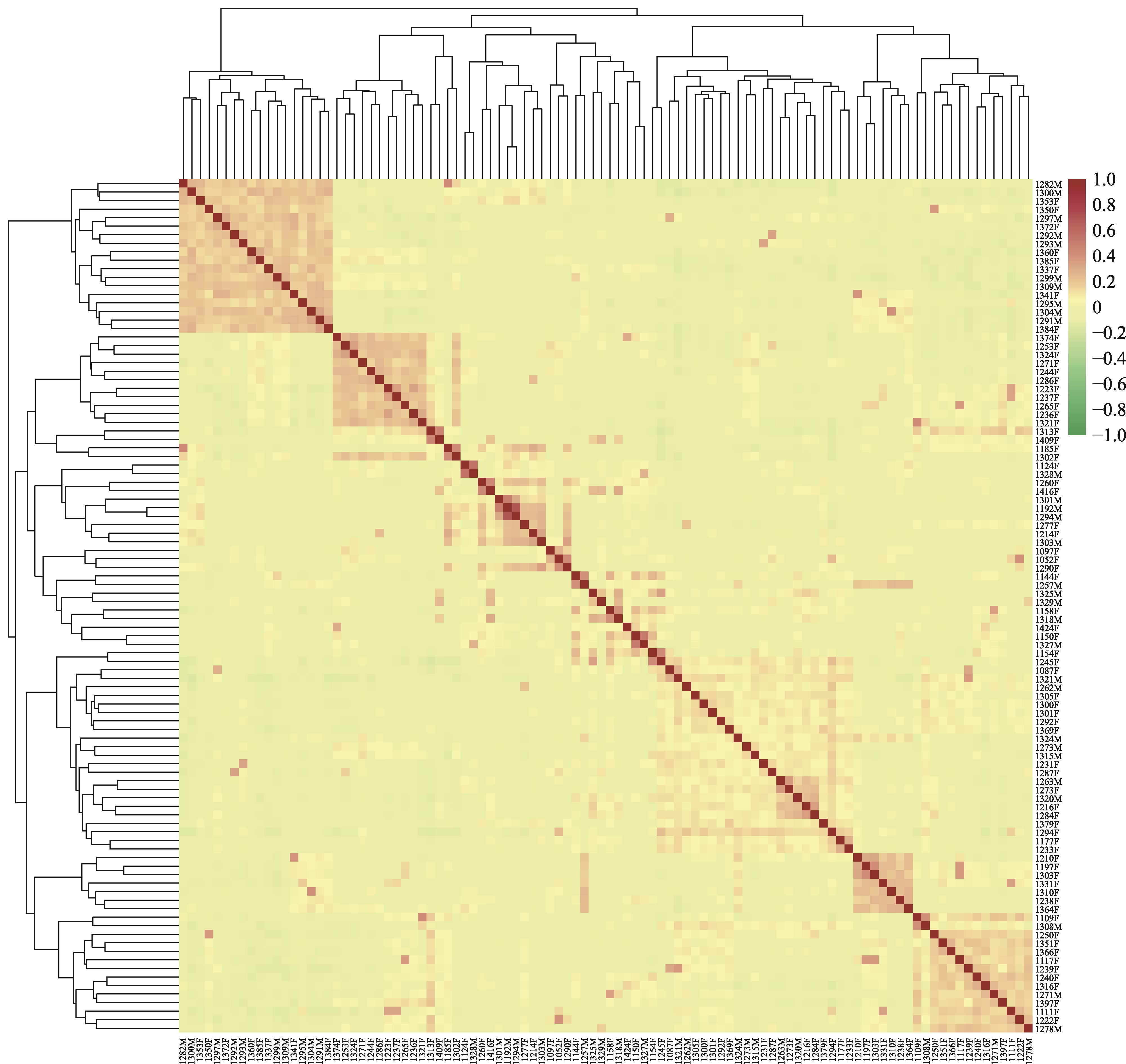
Table 1
Sample collection information of Qinchuan cattle"
| 性别 | 样本量 | 个体号 |
|---|---|---|
| 公 | 30 | 1192M、1257M、1262M、1263M、1271M、1273M、1278M、1282M、1291M、1292M、1293M、1294M、1295M、1297M、1299M、1300M、1301M、1303M、1304M、1308M、1309M、1315M、 1318M、1320M、1321M、1324M、1325M、1327M、1328M、1329M |
| 母 | 70 | 1052F、1087F、1097F、1109F、1111F、1117F、1124F、1144F、1150F、1154F、1158F、1177F、1185F、1197F、1210F、1214F、1216F、1222F、1223F、1231F、1233F、1236F、1237F、1238F、 1239F、1240F、1244F、1245F、1250F、1253F、1260F、1265F、1271F、1273F、1277F、1284F、1286F、1287F、1290F、1292F、1294F、1300F、1301F、1302F、1303F、1305F、1310F、1313F、1316F、1321F、1324F、1331F、1337F、1341F、1350F、1351F、1353F、1360F、1364F、1366F、1369F、1372F、1374F、1379F、1384F、1385F、1397F、1409F、1416F、1424F |
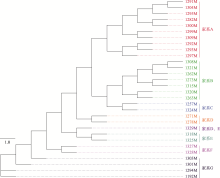
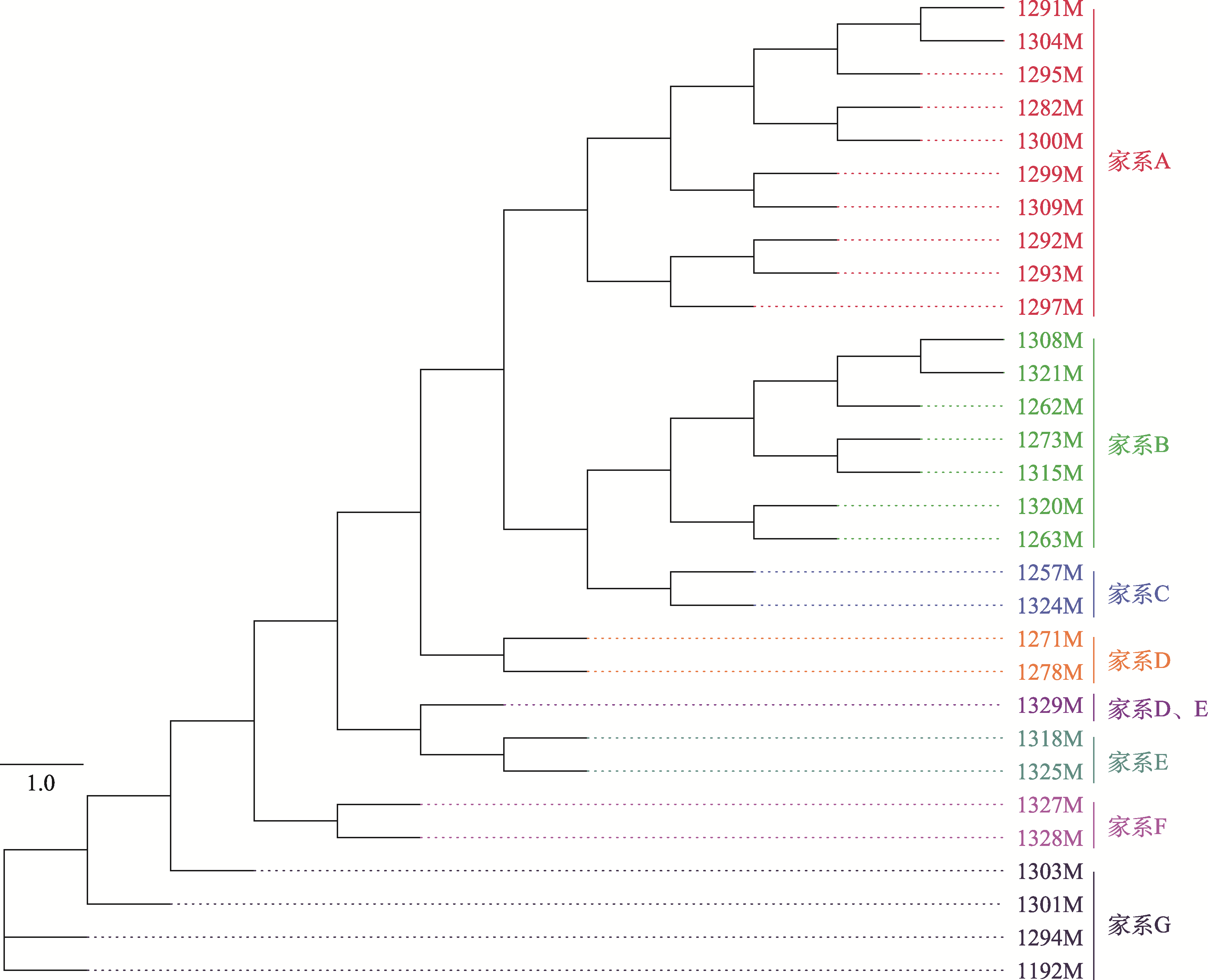
Table 3
Family conservation results of Qinchuan cattle"
| 家系名称 | 性别 | 数量 | 个体号 |
|---|---|---|---|
| 家系A | 公 | 10 | 1282M、1291M、1292M、1293M、1295M、1297M、1299M、1300M、1304M、1309M |
| 母 | 10 | 1144F、1185F、1337F、1341F、1350F、1353F、1360F、1372F、1384F、1385F | |
| 家系B | 公 | 7 | 1262M、1263M、1273M、1308M、1315M、1320M、1321M |
| 母 | 17 | 1087F、1109F、1144F、1177F、1216F、1231F、1233F、1273F、1284F、1287F、1292F、1294F、1300F、1301F、1305F、1369F、1379F | |
| 家系C | 公 | 2 | 1257M、1324M |
| 母 | 8 | 1144F、1197F、1210F、1238F、1303F、1310F、1331F、1364F | |
| 家系D | 公 | 3 | 1329M、1271M、1278M |
| 母 | 14 | 1109F、1111F、1117F、1158F、1222F、1239F、1240F、1250F、1313F、1316F、1351F、1366F、1397F、1409F | |
| 家系E | 公 | 3 | 1318M、1325M、1329M |
| 母 | 5 | 1154F、1158F、1245F、1409F、1424F | |
| 家系F | 公 | 2 | 1327M、1328M |
| 母 | 3 | 1124F、1144F、1150F | |
| 家系G | 公 | 4 | 1192M、1294M、1301M、1303M |
| 母 | 8 | 1052F、1097F、1185F、1214F、1260F、1277F、1290F、1416F | |
| 家系H | 公 | 0 | |
| 母 | 12 | 1223F、1236F、1237F、1244F、1253F、1265F、1271F、1286F、1302F、1321F、1324F、1374F |
| [1] | Wang B, Ma GJ, Zhang Y, Zhou FL, Cheng ZG, Hao P. External factors and improvement model of Qinchuan cattle hybrid improvement. Animal Husbandry and Veterinary Science (Electronic Edition), 2020, (21): 39-40. |
| 王勃, 马国际, 张岩, 周福来, 程助国, 郝鹏. 秦川牛杂交改良外部因素及改良模式. 畜牧兽医科学(电子版), 2020, (21): 39-40. | |
| [2] | Yan BY.Population genomics research on Chaidamu yellow cattle[Dissertation]. Lanzhou University, 2019. |
| 闫碧瑶.柴达木黄牛的群体基因组学研究[学位论文]. 兰州大学, 2019. | |
| [3] |
Bravo S, Larama G, Quiñones J, Paz E, Rodero E, Sepúlveda N. Genetic diversity and phylogenetic relationship among araucana creole sheep and Spanish sheep breeds. Small Ruminant Res, 2019, 172: 23-30.
doi: 10.1016/j.smallrumres.2019.01.007 |
| [4] | Jia XB, Ding P, Chen SY, Zhao SK, Wang J, Lai SJ. Analysis of MC1R, MITF, TYR, TYRP1, and MLPH genes polymorphism in four rabbit breeds with different coat colors. Animals (Basel), 2021, 11(1): 81. |
| [5] | Liu JX, Wei X, Deng TY, Xie R, Han JL, Du LX, Zhao FP, Wang LX. Genome-wide scan for run of homozygosity and identification of corresponding candidate genes in sheep populations. Acta Vet Zootech Sin, 2019, 50(8): 1554-1566. |
| 刘家鑫, 魏霞, 邓天宇, 谢锐, 韩建林, 杜立新, 赵福平, 王立贤. 绵羊全基因组ROH检测及候选基因鉴定. 畜牧兽医学报, 2019, 50(8): 1554-1566. | |
| [6] |
Broman KW, Weber JL. Long homozygous chromosomal segments in reference families from the centre d'Etude du polymorphisme humain. Am J Hum Genet, 1999, 65(6): 1493-1500.
doi: 10.1086/302661 pmid: 10577902 |
| [7] |
Al-Mamun HA, Clark SA, Kwan P, Gondro C. Genome- wide linkage disequilibrium and genetic diversity in five populations of Australian domestic sheep. Genet Sel Evol, 2015, 47: 90.
doi: 10.1186/s12711-015-0169-6 |
| [8] |
Purfield DC, Berry DP, McParland S, Bradley DG. Runs of homozygosity and population history in cattle. BMC Genet, 2012, 13: 70.
doi: 10.1186/1471-2156-13-70 pmid: 22888858 |
| [9] |
Browning SR, Browning BL. Identity by descent between distant relatives: detection and applications. Annu Rev Genet, 2012, 46: 617-633.
doi: 10.1146/annurev-genet-110711-155534 pmid: 22994355 |
| [10] | Liu G, Sun FZ, Zhu FX, Feng HY, Han X. Runs of homozygosity and its application on livestock genome study. Heredity(Beijing), 2019, 41(4): 304-317. |
| 刘刚, 孙飞舟, 朱芳贤, 冯海永, 韩旭. 连续性纯合片段在畜禽基因组研究中的应用. 遗传, 2019, 41(4): 304-317. | |
| [11] |
Marras G, Gaspa G, Sorbolini S, Dimauro C, Ajmone- Marsan P, Valentini A, Williams JL, Macciotta NPP. Analysis of runs of homozygosity and their relationship with inbreeding in five cattle breeds farmed in Italy. Anim Genet, 2015, 46(2): 110-121.
doi: 10.1111/age.12259 pmid: 25530322 |
| [12] | Ma J, Gao X, Li JY, Gao HJ, Wang ZZ, Zhang LP, Xu LY, Gao H, Li HW, Wang YH, Zhu B, Cai WT, Wang CY, Chen Y. Assessing the genetic background and selection signatures of Huaxi cattle using high-density SNP array. Animals (Basel), 2021, 11(12): 3469. |
| [13] |
Van der Nest MA, Hlongwane N, Hadebe K, Chan WY, van der Merwe NA, De Vos L, Greyling B, Kooverjee BB, Soma P, Dzomba EF, Bradfield M, Muchadeyi FC. Breed ancestry, divergence, admixture, and selection patterns of the Simbra crossbreed. Front Genet, 2020, 11: 608650.
doi: 10.3389/fgene.2020.608650 |
| [14] |
Zhang SJ, Yao Z, Li XM, Zhang ZJ, Liu X, Yang P, Chen NB, Xia XT, Lyu SJ, Shi QT, Wang EY, Ru BR, Jiang Y, Lei CZ, Chen H, Huang YZ. Assessing genomic diversity and signatures of selection in Pinan cattle using whole-genome sequencing data. BMC Genomics, 2022, 23(1): 460.
doi: 10.1186/s12864-022-08645-y pmid: 35729510 |
| [15] | Cai CB, Zhang XL, Zhang WF, Yang Y, Gao PF, Guo XH, Li BG, Cao GQ. Evaluation of genetic structure in Mashen pigs conserved population based on SNP chip. Acta Vet Zootech Sin, 2021, 52(4): 920-931. |
| 蔡春波, 张雪莲, 张万峰, 杨阳, 高鹏飞, 郭晓红, 李步高, 曹果清. 运用SNP芯片评估马身猪保种群体的遗传结构. 畜牧兽医学报, 2021, 52(4): 920-931. | |
| [16] | Liu B, Shen LY, Chen Y, Li Q, Liao K, Guo ZY, Zhang SH, Zhu L. Analysis of genetic structure of conservation population in Qingyu pig based on SNP chip. Acta Vet Zootech Sin, 2020, 51(2): 260-269. |
| 刘彬, 沈林園, 陈映, 李强, 廖坤, 郭芝忺, 张顺华, 朱砺. 基于SNP芯片分析青峪猪保种群体的遗传结构. 畜牧兽医学报, 2020, 51(2): 260-269. | |
| [17] | Long X, Chen L, Wu PX, Zhang TH, Pan HM, Zhang L, Wang JY, Guo ZY, Chai J. Evaluation of the genetic structure and selection signatures in Hechuan Black pigs conserved population. Acta Vet Zootech Sin, 2023, 54(5): 1854-1867. |
| 龙熙, 陈力, 吴平先, 张廷焕, 潘红梅, 张亮, 王金勇, 郭宗义, 柴捷. 合川黑猪保种群遗传结构及选择信号分析. 畜牧兽医学报, 2023, 54(5): 1854-1867. | |
| [18] |
Ma SL, Li XW, Li X, Xie SQ, Liu YL, Tang J, Jiang MF. Assessment of genetic structure of 3 Maiwa yak preserved populations based on genotyping-by-sequencing technology. Acta Prataculturae Sinica, 2022, 31(9): 183-194.
doi: 10.11686/cyxb2021464 |
|
马士龙, 李小伟, 李响, 谢书琼, 刘益丽, 唐娇, 江明锋. 基于GBS简化基因组测序评估3个麦洼牦牛保种群的遗传结构研究. 草业学报, 2022, 31(9): 183-194.
doi: 10.11686/cyxb2021464 |
|
| [19] |
Shi R, Zhang Y, Wang YC, Huang T, Lu GC, Yue T, Lu ZX, Huang XX, Wei XP, Feng ST, Chen J, Kagedeer Wulan, Abulizi Ruxianguli, Abulizi Ruxianguli. The evaluation of genomic homozygosity for Xinjiang inbred population by SNP panels. Hereditas(Beijing), 2020, 42(5): 493-506.
doi: 10.16288/j.yczz.20-071 pmid: 32431300 |
|
师睿, 张毅, 王雅春, 黄涛, 卢国昌, 岳涛, 卢振西, 黄锡霞, 卫新璞, 冯书堂, 陈军, 乌兰·卡格德尔, 茹先古丽·阿不力孜, 努尔胡马尔·木合塔尔. 利用SNP芯片信息评估新疆近交牛基因组纯合度. 遗传, 2020, 42(5): 493-506.
doi: 10.16288/j.yczz.20-071 pmid: 32431300 |
|
| [20] | Li YX, Yashengjiang Nasier, Sai Like Duman, Qian Y, Cao SX, Wang WL, Meng CH, Zhang J, Zhang JL. Evaluation of genetic diversity and genetic structure in Kirgiz sheep population cased on SNPs chip. Acta Vet Zootech Sin, 2023, 54(2): 572-583. |
| 李隐侠, 牙生江·那斯尔, 赛里克·都曼, 钱勇, 曹少先, 王伟列, 孟春花, 张俊, 张建丽. SNP芯片评估柯尔克孜羊群体遗传多样性和遗传结构. 畜牧兽医学报, 2023, 54(2): 572-583. | |
| [21] | Ma KY, Han JT, Bai YQ, Li TT, Ma YJ. Genetic diversity analysis of Yongdeng seven goats based on specific-locus amplified fragment sequencing. Acta Vet Zootech Sin, 2023, 54(5): 1939-1950. |
| 马克岩, 韩金涛, 白雅琴, 李讨讨, 马友记. 基于简化基因组测序的永登七山羊遗传多样性分析. 畜牧兽医学报, 2023, 54(5): 1939-1950. | |
| [22] |
Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics, 2014, 30(15): 2114-2120.
doi: 10.1093/bioinformatics/btu170 pmid: 24695404 |
| [23] | Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv: Genomics, 2013. |
| [24] |
Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup. The Sequence Alignment/Map format and SAMtools. Bioinformatics, 2009, 25(16): 2078-2079.
doi: 10.1093/bioinformatics/btp352 pmid: 19505943 |
| [25] |
McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res, 2010, 20(9): 1297- 1303.
doi: 10.1101/gr.107524.110 pmid: 20644199 |
| [26] |
Okonechnikov K, Conesa A, García-Alcalde F. Qualimap 2: advanced multi-sample quality control for high- throughput sequencing data. Bioinformatics, 2016, 32(2): 292-294.
doi: 10.1093/bioinformatics/btv566 pmid: 26428292 |
| [27] |
Li H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics, 2011, 27(21): 2987-2993.
doi: 10.1093/bioinformatics/btr509 pmid: 21903627 |
| [28] |
Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, Maller J, Sklar P, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet, 2007, 81(3): 559-575.
doi: 10.1086/519795 pmid: 17701901 |
| [29] | Zhao GY.Association and prediction of traits based on genomic homozygous segments in beef cattle[Dissertation]. Chinese Academy of Agricultural Sciences, 2021. |
| 赵国耀.基于肉牛基因组纯合片段的性状关联与预测[学位论文]. 中国农业科学院, 2021. | |
| [30] | Liu CL, Lu D, Zhou QY, Wan MC, Hao XD, Zhang XH, Zheng RQ, Ji HY. Analysis of population genetic structure of Hang pigs by high density SNP chips. Acta Vet Zootech Sin, 2022, 53(8): 2502-2513. |
| 刘晨龙, 卢丹, 周泉勇, 万明春, 郝晓东, 张献贺, 郑瑞强, 季华员. 利用高密度SNP芯片分析杭猪的群体遗传结构. 畜牧兽医学报, 2022, 53(8): 2502-2513. | |
| [31] |
Baum BR. PHYLIP: phylogeny inference package. Version 3.2. Joel felsenstein. The Quarterly Review of Biology, 1989, 64(4): 539-541.
doi: 10.1086/416571 |
| [32] |
Xu LY, Bickhart DM, Cole JB, Schroeder SG, Song JZ, Tassell CPV, Sonstegard TS, Liu GE. Genomic signatures reveal new evidences for selection of important traits in domestic cattle. Mol Biol Evol, 2015, 32(3): 711-725.
doi: 10.1093/molbev/msu333 pmid: 25431480 |
| [33] |
Edwards CE, Tessier BC, Swift JF, Bassüner B, Linan AG, Albrecht MA, Yatskievych GA. Conservation genetics of the threatened plant species Physaria filiformis (Missouri bladderpod) reveals strong genetic structure and a possible cryptic species. PLoS One, 2021, 16(3): e0247586.
doi: 10.1371/journal.pone.0247586 |
| [34] |
Liu F, Qu YK, Geng C, Wang AM, Zhang JH, Li JF, Chen KJ, Liu B, Tian HY, Yang WP, Yu YB. Analysis of the population structure and genetic diversity of the red swamp crayfish (Procambarus clarkii) in China using SSR markers. Electron J Biotechnol, 2020, 47: 59-71.
doi: 10.1016/j.ejbt.2020.06.007 |
| [35] |
Reed DH, Frankham R. Correlation between fitness and genetic diversity. Conserv Biol, 2003, 17(1): 230-237.
doi: 10.1046/j.1523-1739.2003.01236.x |
| [36] | Eldridge MDB, King JM, Loupis AK, Spencer PBS, Taylor AC, Pope LC, Hall GP. Unprecedented low levels of genetic variation and inbreeding depression in an island population of the Black-Footed Rock-Wallaby. Conserv Biol, 1999, 13(3): 531-541. |
| [37] |
Gao YH, Gautier M, Ding XD, Zhang H, Wang YC, Wang X, Faruque MO, Li JY, Ye SH, Gou X, Han JL, Lenstra JA, Zhang Y. Species composition and environmental adaptation of indigenous Chinese cattle. Sci Rep, 2017, 7(1): 16196.
doi: 10.1038/s41598-017-16438-7 pmid: 29170422 |
| [38] |
Zhang WG, Gao X, Zhang Y, Zhao YM, Zhang JB, Jia YT, Zhu B, Xu LY, Zhang LP, Gao HJ, Li JY, Chen Y. Genome-wide assessment of genetic diversity and population structure insights into admixture and introgression in Chinese indigenous cattle. BMC Genet, 2018, 19(1): 114.
doi: 10.1186/s12863-018-0705-9 pmid: 30572824 |
| [39] | Yuan J, Xu GQ, Zhou X, Xu SP, Li S, Li WM, Liu B. SNP chip-based monitoring of population conservation effect of Tongcheng pigs. Acta Vet Zootech Sin, 2022, 53(08): 2514-2523. |
| 袁娇, 徐国强, 周翔, 徐三平, 李胜, 黎望明, 刘榜. 基于SNP芯片监测通城猪的保种效果. 畜牧兽医学报, 2022, 53(08): 2514-2523. | |
| [40] |
Botstein D, White RL, Skolnick M, Davis RW. Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am J Hum Genet, 1980, 32(3): 314-331.
pmid: 6247908 |
| [41] |
Forutan M, Ansari Mahyari S, Baes C, Melzer N, Schenkel FS, Sargolzaei M. Inbreeding and runs of homozygosity before and after genomic selection in North American Holstein cattle. BMC Genomics, 2018, 19(1): 98.
doi: 10.1186/s12864-018-4453-z pmid: 29374456 |
| [42] |
Keller MC, Visscher PM, Goddard M. Quantification of inbreeding due to distant ancestors and its detection using dense single nucleotide polymorphism data. Genetics, 2011, 189: 237-249.
doi: 10.1534/genetics.111.130922 pmid: 21705750 |
| [43] | Wang Y, Dong RL, Li X, Cui C, Yu GH. Analysis of the genetic diversity and family structure of the Licha Black pig population on Jiaodong Peninsula, Shandong Province, China. Animals (Basel), 2022, 12(8): 1045. |
| [1] | Jiming Xu, Jianshu Zhu, Mengzhen Li, Han Hu, Chuanzao Mao. Progress on methods for acquiring flanking genomic sequence [J]. Hereditas(Beijing), 2022, 44(4): 313-321. |
| [2] | Jie Kou, Yan Li, Peng Wang, Hong Liu, Jiawen Liu, Juan Wang, Ye Wang, Liang Zhang, Fujun Shen. Optimization of microsatellite genotyping system used for genetic diversity evaluation of Ailuropoda melanoleuca [J]. Hereditas(Beijing), 2022, 44(3): 253-266. |
| [3] | Haoyu Wang, Yuhan Hu, Yueyan Cao, Qiang Zhu, Yuguo Huang, Xi Li, Ji Zhang. AI-SNPs screening based on the whole genome data and research on genetic structure differences of subcontinent populations [J]. Hereditas(Beijing), 2021, 43(10): 938-948. |
| [4] | Zhiwei Xu, Yunlin Wei, Xiuling Ji. Progress on phage genomics of Pseudomonas spp. [J]. Hereditas(Beijing), 2020, 42(8): 752-759. |
| [5] | Jianmin Zheng,Jiangtao Luo,Hongshen Wan,Shizhao Li,Manyu Yang,Jun Li,Ennian Yang,Yun Jiang,Yubin Liu,Xiangquan Wang,Zongjun Pu. Pedigree and development of wheat varieties in Sichuan Province [J]. Hereditas(Beijing), 2019, 41(7): 599-610. |
| [6] | Jiming Xu,Han Hu,Wenxuan Mao,Chuanzao Mao. Identifying T-DNA insertion site(s) of transgenic plants by whole-genome resequencing [J]. Hereditas(Beijing), 2018, 40(8): 676-682. |
| [7] | Jinhong Yuan, Junhua Li, Jiaojiao Yuan, Keli Jia, Shufen Li, Chuanliang Deng, Wujun Gao. The application of MutMap in forward genetic studies based on whole-genome sequencing [J]. Hereditas(Beijing), 2017, 39(12): 1168-1177. |
| [8] | Zhao Yongxin, Li Menghua. Research advances on the origin, evolution and genetic diversity of Chinese native sheep breeds [J]. Hereditas(Beijing), 2017, 39(11): 958-973. |
| [9] | Xian Gong, Chao Zhang, Aisa Yiliyasi, Ying Shi, Xuewei Yang, Aosiman Nuersimanguli, Yaqun Guan, Shuhua Xu. A comparative analysis of genetic diversity of candidate genes associated with type 2 diabetes in worldwide populations [J]. Hereditas(Beijing), 2016, 38(6): 543-559. |
| [10] | HUANG Yi-Min XIA Meng-Ying HUANG Shi. Evolutionary process unveiled by the maximum genetic diversity hypothesis [J]. HEREDITAS, 2013, 35(5): 599-606. |
| [11] | WEN Ying LU Xiao-Ping REN Rui MI Fu-Gui HAN Ping-An XUE Chun-Lei. Development of EST-SSR marker and genetic diversity analysis in Sorghum bicolor×Sorghum sudanenes [J]. HEREDITAS, 2013, 35(2): 225-232. |
| [12] | LI Duo CHAI Zhi-Xin JI Qiu-Mei ZHANG Cheng-Fu XIN Jin-Wei ZHONG Jin-Cheng. Genetic diversity of DNA microsatellite for Tibetan Yak [J]. HEREDITAS, 2013, 35(2): 175-184. |
| [13] | MA Zhi-Jie ZHONG Jin-Cheng HAN Jian-Lin XU Jing-Tao LIU Zhong-Na BAI Wen-Lin. Research progress on molecular genetic diversity of the yak (Bos grunniens) [J]. HEREDITAS, 2013, 35(2): 151-160. |
| [14] | FU Jian-Jun LI Jia-Le SHEN Yu-Bang WANG Rong-Quan XUAN Yun-Feng XU Xiao-Yan CHEN Yong. Genetic variation analysis of wild populations of grass carp (Ctenopharyngodon idella) using microsatellite markers [J]. HEREDITAS, 2013, 35(2): 192-201. |
| [15] | WANG Xiao-Qing WANG Chuan-Chao DENG Qiong-Ying LI Hui. Genetic analysis of Y chromosome and mitochondrial DNA poly-morphism of Mulam ethnic group in Guangxi, China [J]. HEREDITAS, 2013, 35(2): 168-174. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||