Hereditas(Beijing) ›› 2024, Vol. 46 ›› Issue (2): 168-180.doi: 10.16288/j.yczz.23-226
• Research Article • Previous Articles
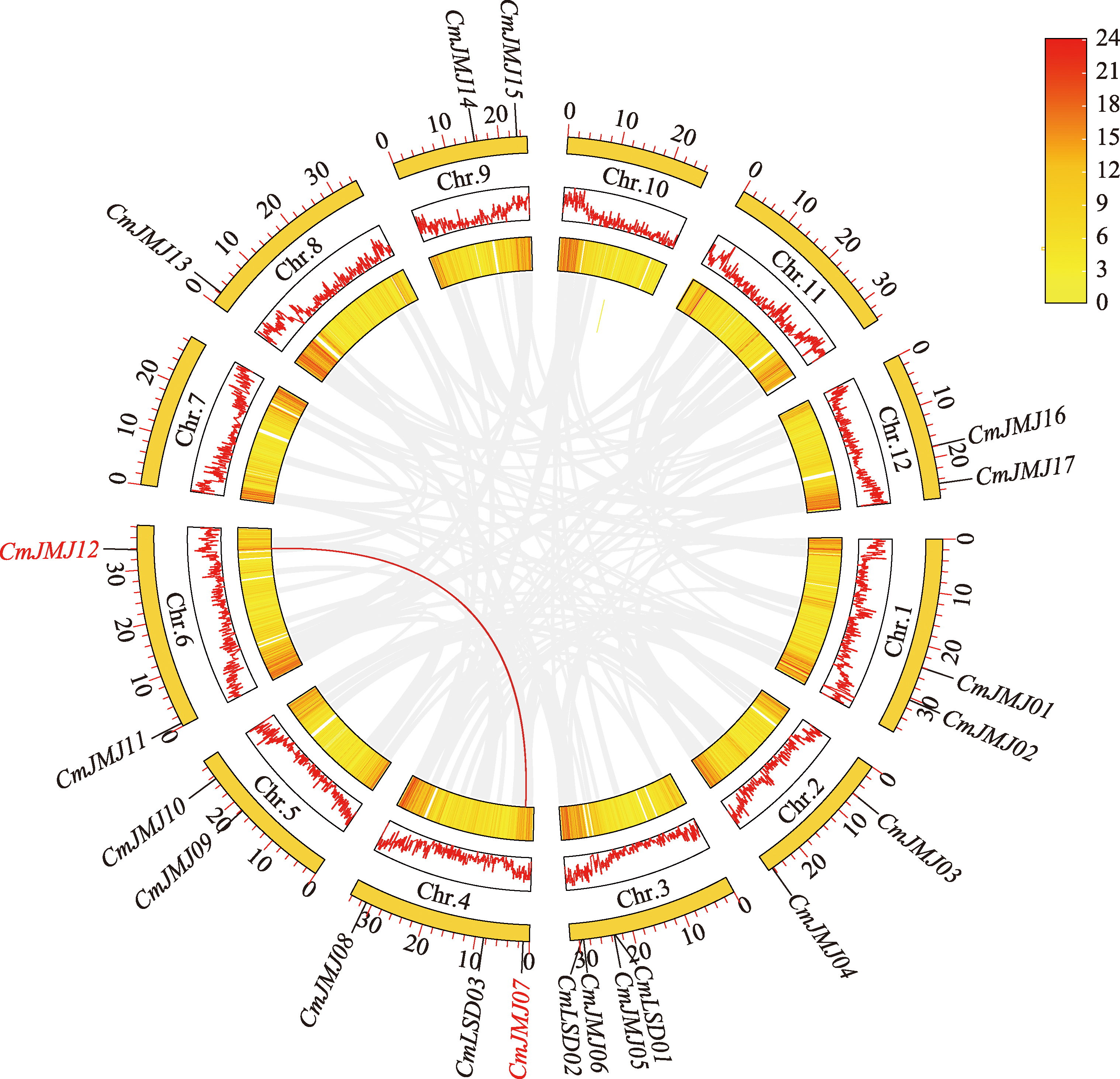
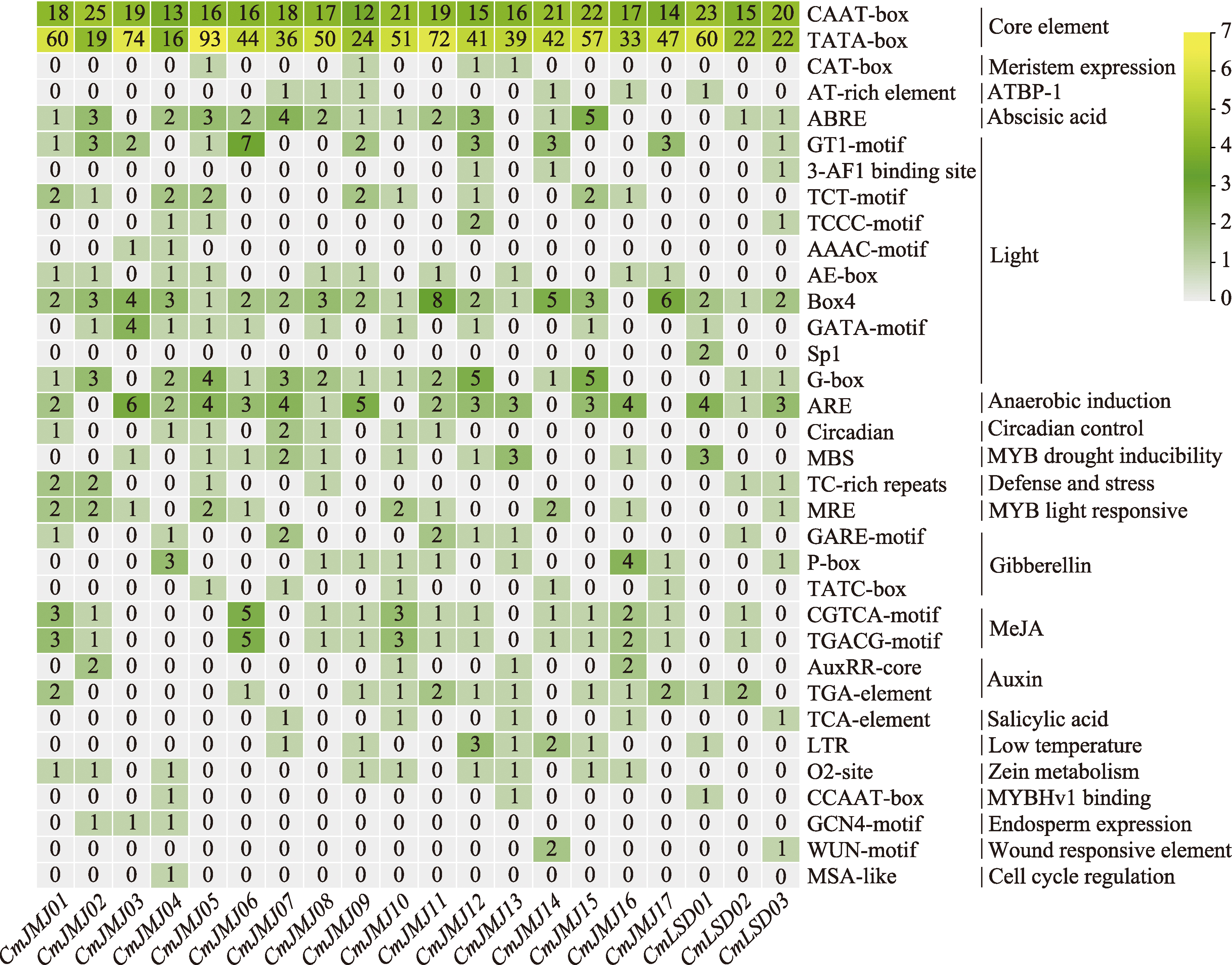
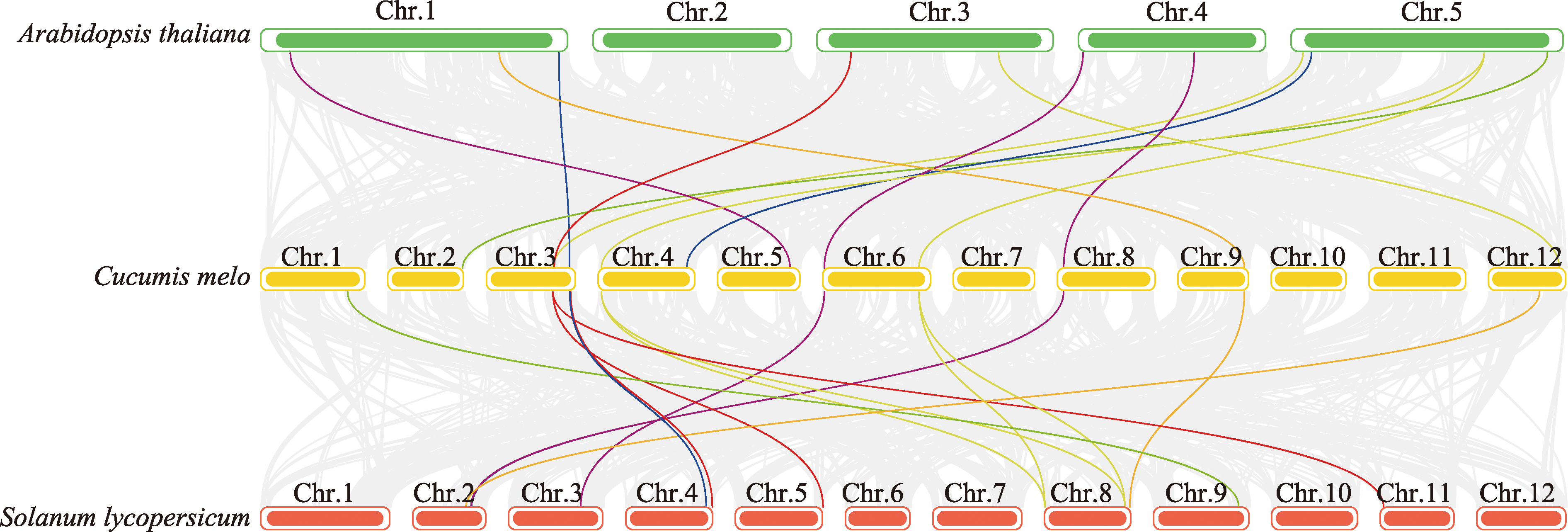
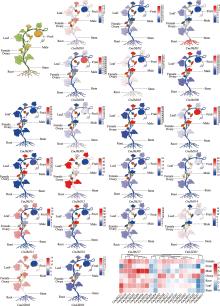
Identification and characterization analysis of the HDM gene family in melon (Cucumis melo L.)
Zhi Zhang( ), Jing Zhang, Jin Zhang, Agula Hasi, Jinfeng Hao(
), Jing Zhang, Jin Zhang, Agula Hasi, Jinfeng Hao( )
)
- Inner Mongolia Key Laboratory of Herbage & Endemic Crop Biotechnology, School of Life Sciences, Inner Mongolia University, Hohhot 010070, China
-
Received:2023-08-23Revised:2023-09-25Online:2024-02-20Published:2023-10-25 -
Contact:Jinfeng Hao E-mail:1819296047@qq.com;haojindd@163.com -
Supported by:Inner Mongolia Natural Science Foundation(2020MS03025)
Cite this article
Zhi Zhang, Jing Zhang, Jin Zhang, Agula Hasi, Jinfeng Hao. Identification and characterization analysis of the HDM gene family in melon (Cucumis melo L.)[J]. Hereditas(Beijing), 2024, 46(2): 168-180.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
| [1] |
Qian SZ, Wang YX, Ma H, Zhang LS. Expansion and functional divergence of Jumonji C-containing histone demethylases: significance of duplications in ancestral angiosperms and vertebrates. Plant Physiol, 2015, 168(4): 1321-1337.
doi: 10.1104/pp.15.00520 pmid: 26059336 |
| [2] |
Luger K, Richmond TJ. The histone tails of the nucleosome. Curr Opin Genet Dev, 1998, 8(2): 140-146.
pmid: 9610403 |
| [3] |
Chang YN, Zhu C, Jiang J, Zhang HM, Zhu JK, Duan CG. Epigenetic regulation in plant abiotic stress responses. J Integr Plant Biol, 2020, 62(5): 563-580.
doi: 10.1111/jipb.12901 |
| [4] |
Suganuma T, Workman JL. Crosstalk among histone modifications. Cell, 2008, 135(4): 604-607.
doi: 10.1016/j.cell.2008.10.036 pmid: 19013272 |
| [5] |
Qian YX, Chen CL, Jiang LY, Zhang J, Ren QY. Genome-wide identification, classification and expression analysis of the JmjC domain-containing histone demethylase gene family in maize. BMC Genomics, 2019, 20(1): 1-16.
doi: 10.1186/s12864-018-5379-1 |
| [6] |
Li ZW, Jiang GX, Liu XC, Ding XC, Zhang DD, Wang XW, Zhou YJ, Yan HL, Li TT, Wu KQ, Jiang YM, Duan XW. Histone demethylase SlJMJ6 promotes fruit ripening by removing H3K27 methylation of ripening-related genes in tomato. New Phytol, 2020, 227(4): 1138-1156.
doi: 10.1111/nph.16590 pmid: 32255501 |
| [7] |
Zhang Y, Reinberg D. Transcription regulation by histone methylation: interplay between different covalent modifications of the core histone tails. Genes Dev, 2001, 15(18): 2343-2360.
doi: 10.1101/gad.927301 |
| [8] |
Chang BS, Chen Y, Zhao YM, Bruick RK. JMJD6 is a histone arginine demethylase. Science, 2007, 318(5849): 444-447.
doi: 10.1126/science.1145801 pmid: 17947579 |
| [9] |
Liu CY, Lu FL, Cui X, Cao XF. Histone methylation in higher plants. Annu Rev Plant Biol, 2010, 61(1): 395-420.
doi: 10.1146/arplant.2010.61.issue-1 |
| [10] |
Allis CD, Berger SL, Cote J, Dent S, Jenuwien T, Kouzarides T, Pillus L, Reinberg D, Shi Y, Shiekhattar R, Shilatifard A, Workman J, Zhang Y. New nomenclature for chromatin-modifying enzymes. Cell, 2007, 131(4): 633-636.
doi: 10.1016/j.cell.2007.10.039 pmid: 18022353 |
| [11] |
Strahl BD, Ohba R, Cook RG, Allis CD. Methylation of histone H3 at lysine 4 is highly conserved and correlates with transcriptionally active nuclei in tetrahymena. Proc Natl Acad Sci USA, 1999, 96(26): 14967-14972.
doi: 10.1073/pnas.96.26.14967 pmid: 10611321 |
| [12] | Martin C, Zhang Y. The diverse functions of histone lysine methylation. Nat Rev Mol Cell Biol, 2005, 6(11): 838-849. |
| [13] |
Tsukada Y, Fang J, Erdjument-Bromage H, Warren ME, Borchers CH, Tempst P, Zhang Y. Histone demethylation by a family of JmjC domain-containing proteins. Nature, 2006, 439(7078): 811-816.
doi: 10.1038/nature04433 |
| [14] |
Shi YJ, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA, Shi Y.Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell, 2004, 119(7): 941-953.
doi: 10.1016/j.cell.2004.12.012 pmid: 15620353 |
| [15] |
Forneris F, Binda C, Vanoni MA, Mattevi A, Battaglioli E. Histone demethylation catalysed by LSD1 is a flavin- dependent oxidative process. FEBS Lett, 2005, 579(10): 2203-2207.
doi: 10.1016/j.febslet.2005.03.015 |
| [16] | Accari SL, Fisher PR. Emerging roles of JmjC domain-containing proteins. Int Rev Cell Mol Biol, 2015, 319 (2015): 165-220. |
| [17] | Klose RJ, Kallin EM, Zhang Y. JmjC domain-containing proteins and histone demethylation. Nat Rev Genet, 2006, 7(9): 715-727. |
| [18] |
Dong YW, Lu J, Liu JY, Jalal A, Wang CQ. Genome-wide identification and functional analysis of JmjC domain- containing genes in flower development of Rosa chinensis. Plant Mol Biol, 2020, 102(4-5): 417-430.
doi: 10.1007/s11103-019-00955-2 |
| [19] |
Lu FL, Li GL, Cui X, Liu CY, Wang XJ, Cao XF. Comparative analysis of JmjC domain-containing proteins reveals the potential histone demethylases in Arabidopsis and rice. J Integr Plant Biol, 2008, 50(7): 886-896.
doi: 10.1111/jipb.2008.50.issue-7 |
| [20] | Yang HC, Han ZF, Cao Y, Fan D, Li H, Mo HX, Feng Y, Liu L, Wang Z, Yue YL, Cui SJ, Chen S, Chai JJ, Ma LG. A companion cell-dominant and developmentally regulated H3K4 demethylase controls flowering time in Arabidopsis via the repression of FLC expression. PLoS Genet, 2012, 8(4): 1002664. |
| [21] |
Lu FL, Cui X, Zhang SB, Liu CY, Cao XF.JMJ 14 is an H3K4 demethylase regulating flowering time in Arabidopsis. Cell Res, 2010, 20(3): 387-390.
doi: 10.1038/cr.2010.27 |
| [22] | Wang XL, Gao J, Gao S, Song Y, Yang Z, Kuai BK.The H3K27me3 demethylase REF6 promotes leaf senescence through directly activating major senescence regulatory and functional genes in Arabidopsis. PLoS Genet, 2019, 15(4): 1008068. |
| [23] |
Liu P, Zhang SB, Zhou B, Luo X, Zhou XF, Cai B, Jin YH, Niu D, Lin JX, Cao XF, Jin JB.The histone H3K4 demethylase JMJ16 represses leaf senescence in Arabidopsis. Plant Cell, 2019, 31(2): 430-443.
doi: 10.1105/tpc.18.00693 |
| [24] |
Lu SX, Knowles SM, Webb CJ, Celaya RB, Cha C, Siu JP, Tobin EM. The Jumonji C domain-containing protein JMJ30 regulates period length in the Arabidopsis circadian clock. Plant Physiol, 2011, 155(2): 906-915.
doi: 10.1104/pp.110.167015 |
| [25] | Li W, Liu H, Cheng ZJ, Su YH, Han HN, Zhang Y, Zhang XS. DNA methylation and histone modifications regulate de novo shoot regeneration in Arabidopsis by modulating WUSCHEL expression and auxin signaling. PLoS Genet, 2011, 7(8): 1002243. |
| [26] | Chen QF, Chen XS, Wang Q, Zhang FB, Lou ZY, Zhang QF, Zhou DX. Structural basis of a histone H3 lysine 4 demethylase required for stem elongation in rice. PLoS Genet, 2013, 9(1): 1003239. |
| [27] |
Yamaguchi N, Matsubara S, Yoshimizu K, Seki M, Hamada k, Kamitani M, Kurita Y, Nomura Y, Nagashima K, Inagaki S, Suzuki T, Gan ES, To T, Kakutani T, Nagano AJ, Satake A, Ito T. H3K27me3 demethylases alter HSP22 and HSP17.6C expression in response to recurring heat in Arabidopsis. Nat Commun, 2021, 12(1): 3480.
doi: 10.1038/s41467-021-23766-w |
| [28] |
Song T, Zhang Q, Wang HQ, Han JB, Xu ZQ, Yan SN, Zhu ZG. OsJMJ703, a rice histone demethylase gene, plays key roles in plant development and responds to drought stress. Plant Physiol Biochem, 2018, 132: 183-188.
doi: 10.1016/j.plaphy.2018.09.007 |
| [29] | Shin AY, Kim YM, Koo N, Lee SM, Nahm S, Kwon SY. Transcriptome analysis of the oriental melon (Cucumis melo L. var. makuwa) during fruit development. Peer J, 2017, 5: 2834. |
| [30] |
Wu B, Wang L, Pan GY, Li T, Li X, Hao JH.Genome- wide characterization and expression analysis of the auxin response factor (ARF) gene family during melon (Cucumis melo L.) fruit development. Protoplasma, 2020, 257(3): 979-992.
doi: 10.1007/s00709-020-01484-2 |
| [31] |
Han YP, Li XY, Cheng L, Liu YC, Wang H, Ke DX, Yuan HY, Zhang LS, Wang L. Genome-wide analysis of soybean JmjC domain-containing proteins suggests evolutionary conservation following whole-genome duplication. Front Plant Sci, 2016, 7: 1800.
pmid: 27994610 |
| [32] |
Gu TT, Han YH, Huang RR, McAvoy RJ, Li Y. Identification and characterization of histone lysine methylation modifiers in Fragaria vesca. Sci Rep, 2016, 6: 23581.
doi: 10.1038/srep23581 pmid: 27049067 |
| [33] |
Jiang DH, Yang WN, He YH, Amasino RM. Arabidopsis relatives of the human lysine-specific demethylase1 repress the expression of FWA and FLOWERING LOCUS C and thus promote the floral transition. Plant Cell, 2007, 19(10): 2975-2987.
doi: 10.1105/tpc.107.052373 |
| [34] |
Sun QW, Zhou DX. Rice jmjC domain-containing gene JMJ706 encodes H3K9 demethylase required for floral organ development. Proc Natl Acad Sci USA, 2008, 105(36): 13679-13684.
doi: 10.1073/pnas.0805901105 pmid: 18765808 |
| [35] |
Cui XK, Jin P, Cui X, Gu LF, Lu ZK, Xue YM, Wei LY, Qi JF, Song XW, Luo M, An G, Cao XF. Control of transposon activity by a histone H3K4 demethylase in rice. Proc Natl Acad Sci USA, 2013, 110(5): 1953-1958.
doi: 10.1073/pnas.1217020110 pmid: 23319643 |
| [36] |
Ding XC, Liu XC, Jiang GX, Li ZW, Song YB, Zhang DD, Jiang YM, Duan XW. SlJMJ7 orchestrates tomato fruit ripening via crosstalk between H3K4me3 and DML2- mediated DNA demethylation. New Phytol, 2022, 233(3): 1202-1219.
doi: 10.1111/nph.v233.3 |
| [1] | Feifei Li, Yanmin Hao, Minlong Cui, Chunlan Piao. Cloning and functional analysis of RADIALIS-like 1 gene from Antirrhinum majus [J]. Hereditas(Beijing), 2023, 45(6): 526-535. |
| [2] | Qingqian Ding,Xiaoting Wang,Liqin Hu,Xin Qi,Linhao Ge,Weiya XU,Zhaoshi Xu,Yongbin Zhou,Guanqing Jia,Xianmin Diao,Donghong Min,Youzhi Ma,Ming Chen. MYB-like transcription factor SiMYB42 from foxtail millet (Setaria italica L.) enhances Arabidopsis tolerance to low-nitrogen stress [J]. Hereditas(Beijing), 2018, 40(4): 327-338. |
| [3] |
Lizhen Zhang, Yong Zhang, Jinghua Hu, Zilong Wang, Zhijiang Zeng.
Molecular cloning and expression analysis of the tyramine receptor genes in Apis cerana cerana [J]. Hereditas(Beijing), 2018, 40(2): 155-161. |
| [4] | Jianzhong Chang, Fengxia Yan, Linyi Qiao, Jun Zheng, Fuyao Zhang, Qingshan Liu. Genome-wide identification and expression analysis of SBP-box gene family in Sorghum bicolor L. [J]. HEREDITAS(Beijing), 2016, 38(6): 569-580. |
| [5] | Yanbing Gu, Zhirui Ji, Fumei Chi, Zhuang Qiao, Chengnan Xu, Junxiang Zhang, Zongshan Zhou, Qinglong Dong. Genome-wide identification and expression analysis of the WRKY gene family in peach [J]. HEREDITAS(Beijing), 2016, 38(3): 254-270. |
| [6] | Jun Wu, Junhong Zhang, Menghui Huang, Minhui Zhu, Zaikang Tong. Expression analysis of miR164 and its target gene NAC1 in response to low nitrate availability in Betula luminifera [J]. HEREDITAS(Beijing), 2016, 38(2): 155-162. |
| [7] | Jianjiang Ma, Nuohan Wang, Man Wu, Wenfeng Pei, Wenkui Wang, Xingli Li, Jinfa Zhang, Shuxun Yu, Jiwen Yu. Genome-wide analysis of the LPAAT gene family in Gossypium raimondii and G. arboreum, and expression analysis of its orthologs in G. hirsutum [J]. HEREDITAS(Beijing), 2015, 37(7): 692-701. |
| [8] | Yunfei Liu, Hongjian Wan, Yuejian Yang, Yanping Wei, Zhimiao Li, Qingjing Ye, Rongqing Wang, Meiying Ruan, Zhuping Yao, Guozhi Zhou. Genome-wide identification and analysis of heat shock protein 90 in tomato [J]. HEREDITAS(Beijing), 2014, 36(10): 1043-1052. |
| [9] | XIE Yu-Xiao GAO Shi-Zheng ZHAO Su-Mei. Function of comparative gene identification-58 (CGI-58) on lipid metabolism in animals [J]. HEREDITAS, 2013, 35(5): 595-598. |
| [10] | HUANG Guo-Wen HAN Yu-Zhen FU Yong-Fu. Expression and functional analysis of SUA41gene in Arabidopsis thaliana [J]. HEREDITAS, 2013, 35(1): 93-100. |
| [11] | YANG Li-Wei, SHI Ji-Sen. Cloning and expression analysis of differentially expressed genes in Chinese fir stems treated by different concentrations of exogenous IAA [J]. HEREDITAS, 2012, 34(4): 472-484. |
| [12] | ZHANG Lei, CHAO Jiang-Chao, CUI Meng-Meng, CHEN Ya-Qiong, ZONG Feng, SUN Yu-Ge. Bioinformatic prediction of conserved microRNAs and their target genes in eggplant (Solanum melongena L.) [J]. HEREDITAS, 2011, 33(7): 776-784. |
| [13] | JIAO Sha-Sha, LIU Ka, LI Gang, GAO Jian-Feng, MA Run-Lin. Confirmation and expression analysis of three predicted genes in sheep MHC region [J]. HEREDITAS, 2011, 33(12): 1353-1358. |
| [14] | . Construction of a melon genetic map with fruit and seed QTLs [J]. HEREDITAS, 2011, 33(12): 1398-1408. |
| [15] | BAO Yong-Mei, LIU Yong-Hui, HU Dong-Qing, HUANG Ji, WANG Zhou-Fei, WANG Jian-Fei, ZHANG Gong-Sheng. Preparation, characterization and application of rice Qb-SNARE protein OsNPSN11 polyclonal antibody [J]. HEREDITAS, 2010, 32(9): 961-965. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||