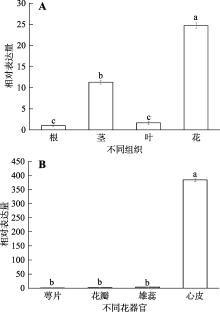
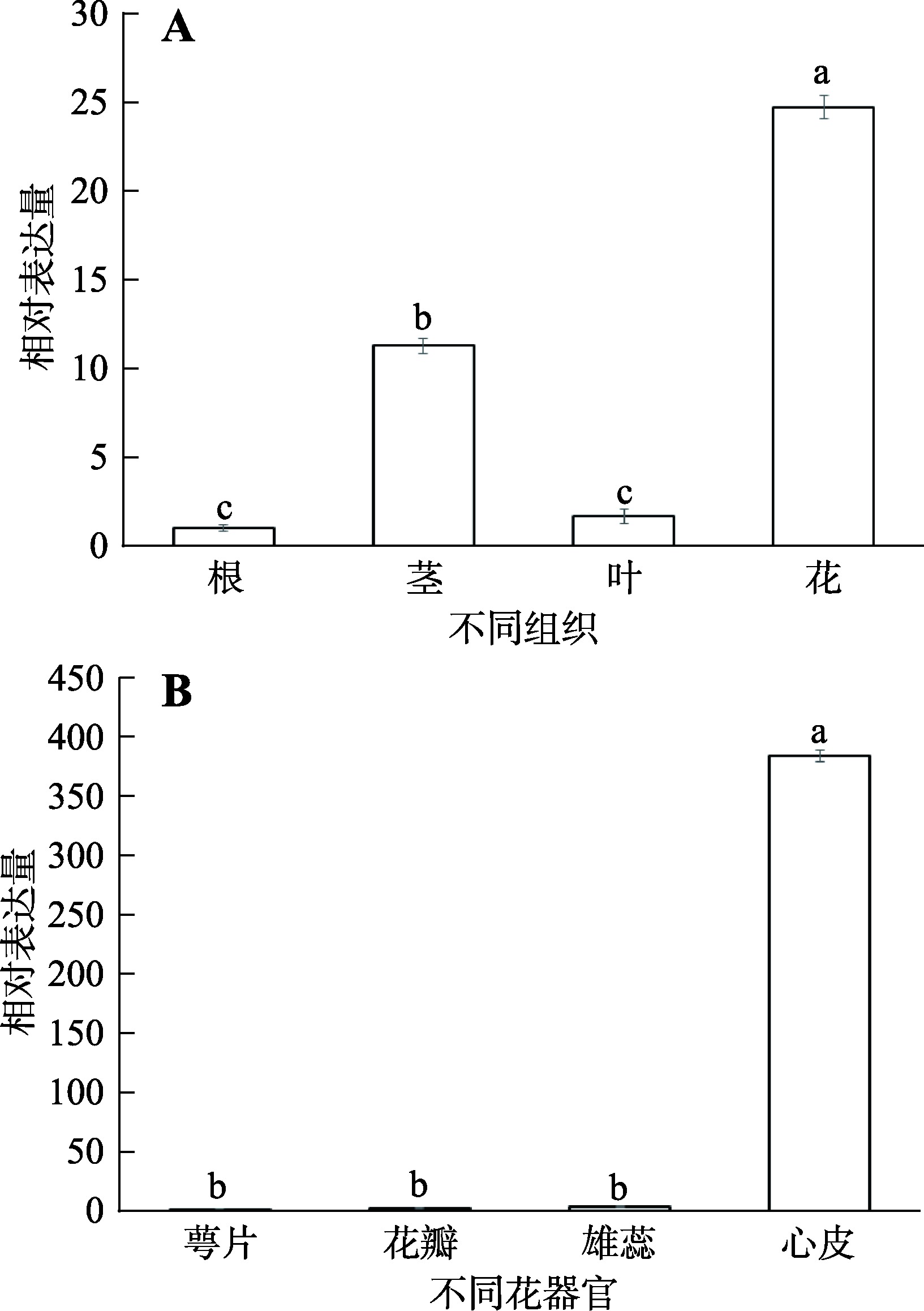
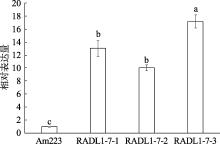
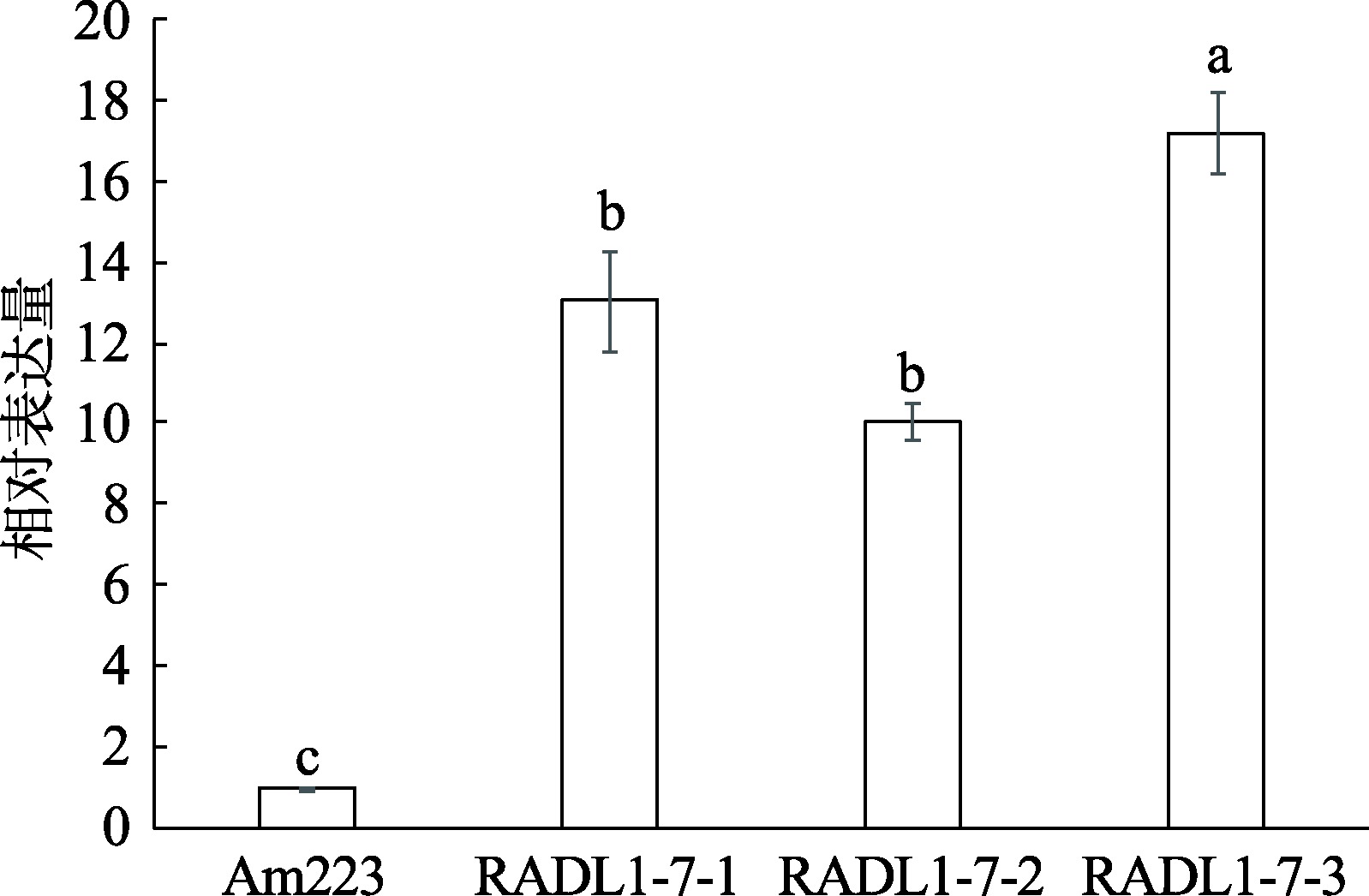
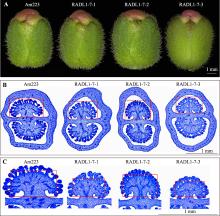
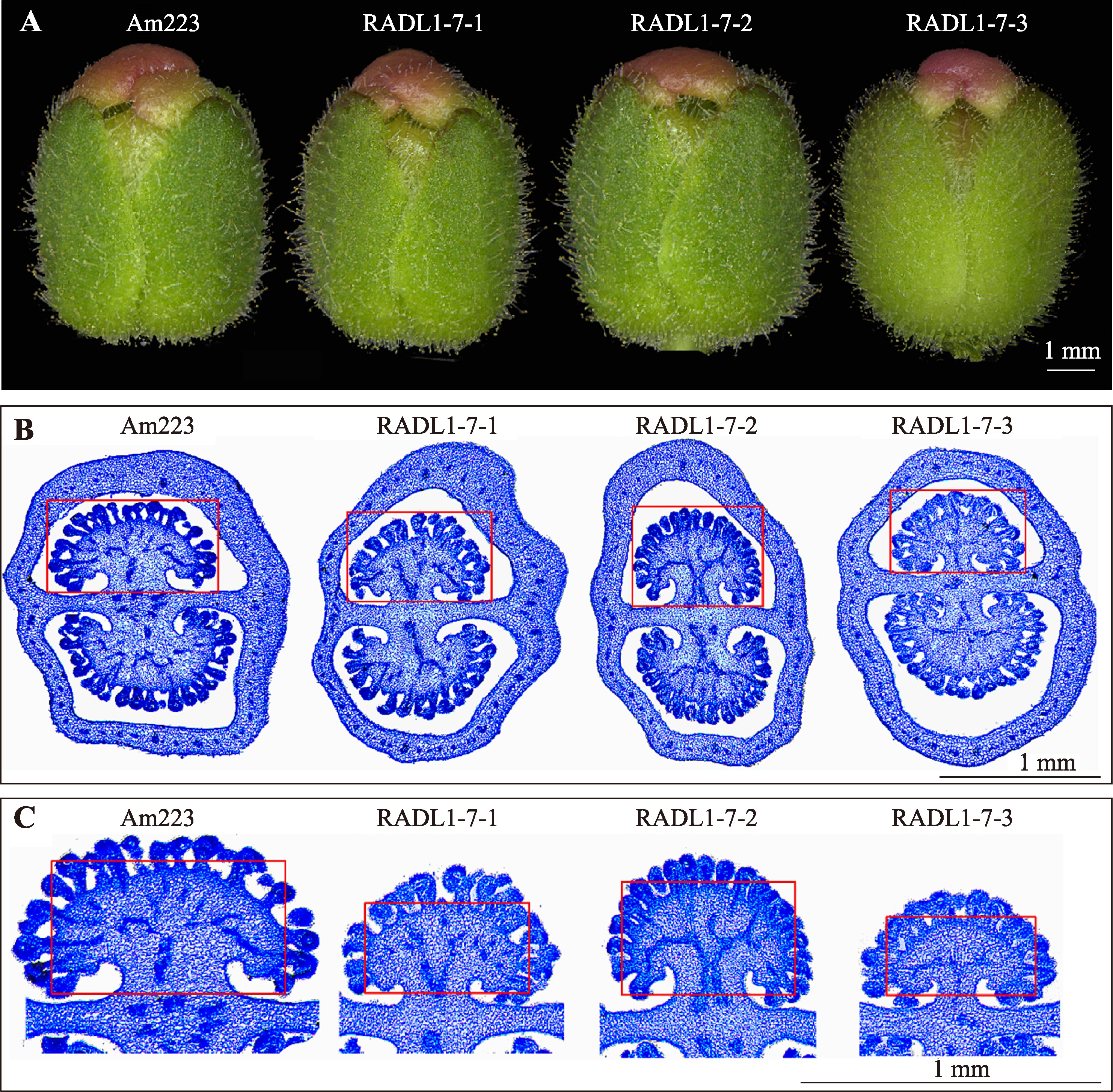
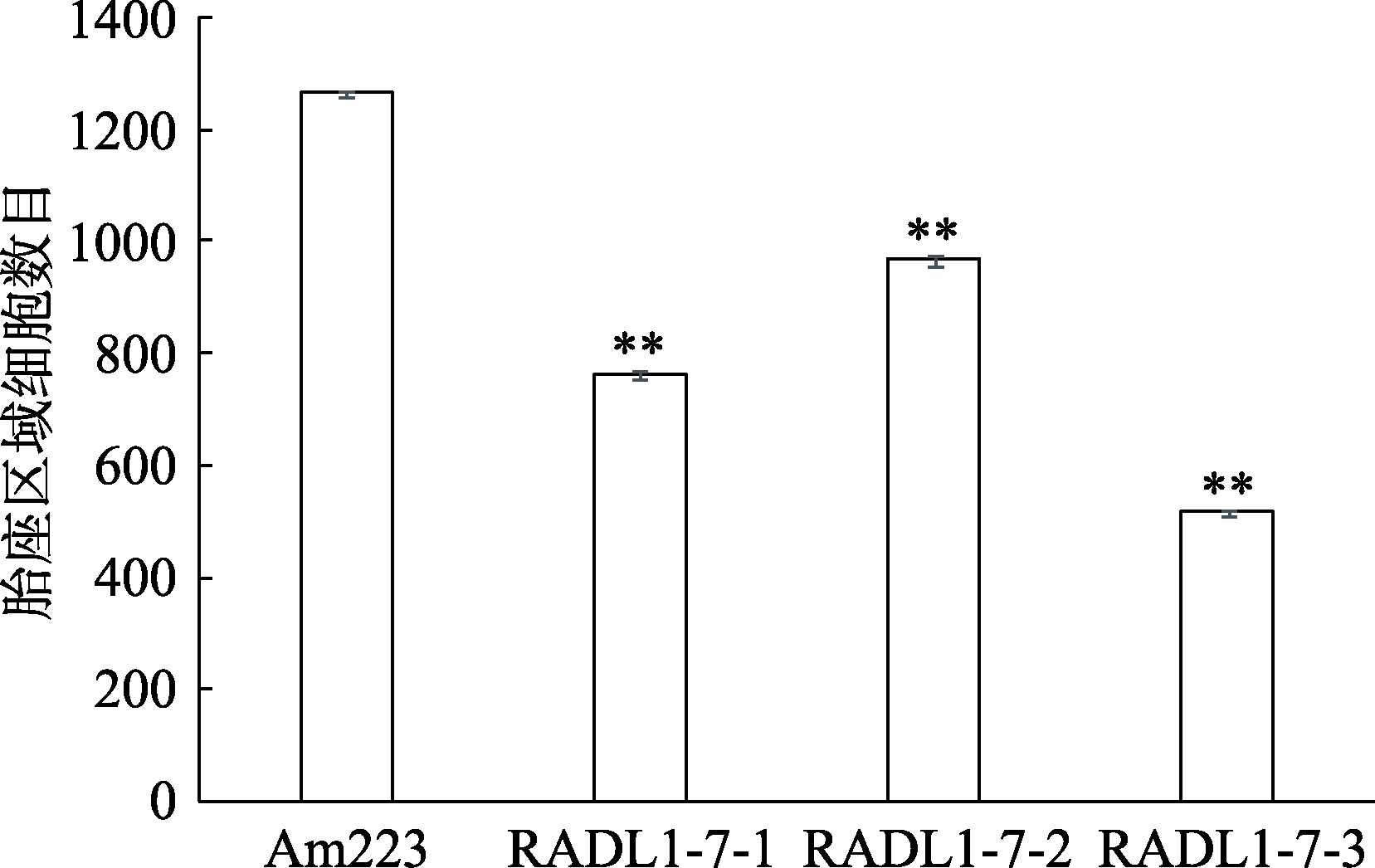
Hereditas(Beijing) ›› 2023, Vol. 45 ›› Issue (6): 526-535.doi: 10.16288/j.yczz.23-057
• Research Article • Previous Articles Next Articles
Cloning and functional analysis of RADIALIS-like 1 gene from Antirrhinum majus
Feifei Li( ), Yanmin Hao, Minlong Cui, Chunlan Piao(
), Yanmin Hao, Minlong Cui, Chunlan Piao( )
)
- Collaborative Innovation Center for Efficient and Green Production of Agriculture in Mountainous Areas of Zhejiang Province, College of Horticulture Science, Zhejiang A&F University, Hangzhou 311300, China
-
Received:2023-03-14Revised:2023-04-11Online:2023-06-20Published:2023-05-29 -
Contact:Piao Chunlan E-mail:1399745127@qq.com;2488335116@qq.com -
Supported by:Cooperative R & D Project of Zhejiang Academy of Agricultural Sciences(H20220312)
Cite this article
Feifei Li, Yanmin Hao, Minlong Cui, Chunlan Piao. Cloning and functional analysis of RADIALIS-like 1 gene from Antirrhinum majus[J]. Hereditas(Beijing), 2023, 45(6): 526-535.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
Table 1
The primer sequences of gene cloning and quantitative real-time PCR used in this study"
| 引物名称 | 引物序列(5′→3′) | 用途 |
|---|---|---|
| AmUBI | F: ATTGGTGCTGAGGTTGAGA | 内参基因 |
| R: ACAACTGACTCCAGCAAACG | ||
| Kan | F: AGATGGATTGCACGCAGGTTC | Kan引物鉴定 |
| R: GTGGTCGAATGGGCAGGTAG | ||
| AmRADL1 | F: AAGGATCCATGGCGTCGAGTTCGTTGTCAC | 基因克隆及荧光定量PCR引物 |
| R: AAGAGCTCTTAGCCACTAAAATAAATAAAAACAT |
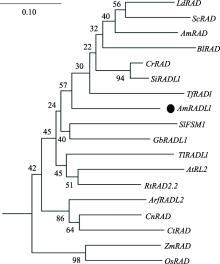
Fig. 2
Phylogenetic tree of AmRADL1 gene Ld:Lindernia dubia;Sc:Schaueria calytricha;Am:Antirrhinum majus;Bl:Bournea leiophylla;Cr:Conandron ramondioides;Si:Saintpaulia ionantha;Tf:Torenia fournieri;Sl:Solanum lycopersicum;Gb:Gossypium barbadense;Tl:Tropaeolum longifolium;At:Arabidopsis thaliana;Rt:Rhododendron taxifolium;Arf:Aristolochia fimbriata;Cn:Cocos nucifera;Ct:Cattleya trianae;Zm:Zea mays;Os:Oryza sativa。"
| [1] |
Thomson B, Wellmer F. Molecular regulation of flower development. Curr Top Dev Biol, 2019, 131: 185-210.
doi: S0070-2153(18)30086-3 pmid: 30612617 |
| [2] |
Ambawat S, Sharma P, Yadav NR, Yadav RC. MYB transcription factor genes as regulators for plant responses: an overview. Physiol Mol Biol Plants, 2013, 19(3): 307-321.
doi: 10.1007/s12298-013-0179-1 |
| [3] |
Qian JH, Li ZQ, Liao XF, Tang DF, Shi QQ, Zhou RY, Chen P. Advance on MYB transcription factors in regulating plant flower development. Letter Biotechnol, 2016, 27(2): 283-288.
doi: 10.1007/s10529-005-1811-0 |
|
钱景华, 李增强, 廖小芳, 汤丹峰, 史奇奇, 周瑞阳, 陈鹏. 调控植物花发育的MYB类转录因子研究进展. 生物技术通讯, 2016, 27(2): 283-288.
doi: 10.1007/s10529-005-1811-0 |
|
| [4] |
Coen ES, Meyerowitz EM. The war of the whorls: genetic interactions controlling flower development. Nature, 1991, 353(6339): 31-37.
doi: 10.1038/353031a0 |
| [5] |
Corley SB, Carpenter R, Copsey L, Coen E. Floral asymmetry involves an interplay between TCP and MYB transcription factors in Antirrhinum. Proc Natl Acad Sci USA, 2005, 102(14): 5068-5073.
pmid: 15790677 |
| [6] |
Wang SS, Koide Y, Kishima Y.How to establish a mutually beneficial relationship between a transposon and its host: lessons from Tam3 in Antirrhinum. Genes Genet Syst, 2022, 97(4): 177-184.
doi: 10.1266/ggs.22-00063 |
| [7] |
Schwinn K, Venail J, Shang YJ, Mackay S, Alm V, Butelli E, Oyama R, Bailey P, Davies K, Martin C. A small family of MYB-regulatory genes controls floral pigmentation intensity and patterning in the genus Antirrhinum. Plant Cell, 2006, 18(4): 831-851.
doi: 10.1105/tpc.105.039255 pmid: 16531495 |
| [8] |
Perez-Rodriguez M, Jaffe FW, Butelli E, Glover BJ, Martin C. Development of three different cell types is associated with the activity of a specific MYB transcription factor in the ventral petal of Antirrhinum majus flowers. Development, 2005, 132(2): 359-370.
doi: 10.1242/dev.01584 pmid: 15604096 |
| [9] |
Cui ML, Copsey L, Green AA, Bangham JA, Coen E. Quantitative control of organ shape by combinatorial gene activity. PLoS Biol, 2010, 8(11): e1000538.
doi: 10.1371/journal.pbio.1000538 |
| [10] |
Li MM, Zhang DF, Gao Q, Luo YF, Zhang H, Ma B, Chen CH, Whibley A, Zhang YE, Cao YH, Li Q, Guo H, Li JH, Song YZ, Zhang Y, Copsey L, Li Y, Li XX, Qi M, Wang JW, Chen Y, Wang D, Zhao JY, Liu GC, Wu B, Yu LL, Xu CY, Li J, Zhao SC, Zhang YJ, Hu SN, Liang CZ, Yin Y, Coen E, Xue YB. Genome structure and evolution of Antirrhinum majus L. Nat Plants, 2019, 5(2): 174-183.
doi: 10.1038/s41477-018-0349-9 |
| [11] |
Vincent CA, Coen ES. A temporal and morphological framework for flower development in Antirrhinum majus. Can J Bot, 2004, 82(5): 681-690.
doi: 10.1139/b04-042 |
| [12] |
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG.Clustal W and Clustal X version 2.0. Bioinformatics, 2007, 23(21): 2947-2948.
doi: 10.1093/bioinformatics/btm404 pmid: 17846036 |
| [13] |
Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol, 2018, 35(6): 1547-1549.
doi: 10.1093/molbev/msy096 pmid: 29722887 |
| [14] |
Cui ML, Handa T, Ezura H. An improved protocol for Agrobacterium-mediated transformation of Antirrhinum majus L. Mol Genet Genomics, 2003, 270(4): 296-302.
pmid: 14513365 |
| [15] | Hao YM, Chen KL, Feng LJ, Li FF, Cui ML, Piao CL.Cloning and functional analysis of SvAPETALA1 in Senecio vulgaris. J Zhejiang A F Univ, 2022, 39(4): 821-829. |
| 郝燕敏, 陈柯俐, 冯丽君, 李菲菲, 崔敏龙, 朴春兰. 欧洲千里光SvAPETALA1基因的克隆及功能分析. 浙江农林大学学报, 2022, 39(4): 821-829. | |
| [16] | Yang BC, Song ZH, Li CN, Jiang JH, Zhou YY, Wang RP, Wang Q, Ni C, Liang Q, Chen HD, Fan LM. RSM1, an Arabidopsis MYB protein, interacts with HY5/HYH to modulate seed germination and seedling development in response to abscisic acid and salinity. PLoS Genet, 2018, 14(12): e1007839. |
| [17] |
Barg R, Sobolev I, Eilon T, Gur A, Chmelnitsky I, Shabtai S, Grotewold E, Salts Y. The tomato early fruit specific gene Lefsm1 defines a novel class of plant-specific SANT/MYB domain proteins. Planta, 2005, 221(2): 197-211.
doi: 10.1007/s00425-004-1433-0 |
| [18] |
Su SH, Xiao W, Guo WX, Yao XR, Xiao JQ, Ye ZQ, Wang N, Jiao KY, Lei MQ, Peng QC, Hu XH, Huang X, Luo D. The CYCLOIDEA-RADIALIS module regulates petal shape and pigmentation, leading to bilateral corolla symmetry in Torenia fournieri (Linderniaceae). New Phytol, 2017, 215(4): 1582-1593.
doi: 10.1111/nph.2017.215.issue-4 |
| [19] |
Boyer LA, Latek RR, Peterson CL. The SANT domain: a unique histone-tail-binding module? Nat Rev Mol Cell Biol, 2004, 5(2): 158-163.
doi: 10.1038/nrm1314 |
| [20] |
Zhong JS, Preston JC, Hileman LC, Kellogg EA. Repeated and diverse losses of corolla bilateral symmetry in the Lamiaceae. Ann Bot, 2017, 119(7): 1211-1223.
doi: 10.1093/aob/mcx012 |
| [21] |
Zhang F, Liu X, Zuo KJ, Zhang JQ, Sun XF, Tang KX. Molecular cloning and characterization of a novel Gossypium barbadense L.RAD-like gene. Plant Mol Biol Rep, 2011, 29(2): 324-333.
doi: 10.1007/s11105-010-0234-9 |
| [22] |
Madrigal Y, Alzate JF, González F, Pabón-Mora N. Evolution of RADIALIS and DIVARICATA gene lineages in flowering plants with an expanded sampling in non-core eudicots. Am J Bot, 2019, 106(3): 334-351.
doi: 10.1002/ajb2.1243 pmid: 30845367 |
| [23] |
Lu SX, Knowles SM, Andronis C, Ong MS, Tobin EM.CIRCADIAN CLOCK ASSOCIATED1 and LATE ELONGATED HYPOCOTYL function synergistically in the circadian clock of Arabidopsis. Plant Physiol, 2009, 150(2): 834-843.
doi: 10.1104/pp.108.133272 |
| [24] |
Pesch M, Schultheiß I, Digiuni S, Uhrig JF, Hülskamp M.Mutual control of intracellular localisation of the patterning proteins AtMYC1, GL1 and TRY/CPC in Arabidopsis. Development, 2013, 140(16): 3456-3467
doi: 10.1242/dev.094698 pmid: 23900543 |
| [25] |
Dubos C, Le Gourrierec J, Baudry A, Huep G, Lanet E, Debeaujon I, Routaboul JM, Alboresi A, Weisshaar B, Lepiniec L.MYBL2 is a new regulator of flavonoid biosynthesis in Arabidopsis thaliana. Plant J, 2008, 55(6): 940-953.
doi: 10.1111/tpj.2008.55.issue-6 |
| [26] |
Boyden GS, Donoghue MJ, Howarth DG. Duplications and expression of RADIALIS-like genes in Dipsacales. Int J Plant Sci, 2012, 173(9): 971-983.
doi: 10.1086/667626 |
| [27] |
Preston JC, Kost MA, Hileman LC. Conservation and diversification of the symmetry developmental program among close relatives of snapdragon with divergent floral morphologies. New Phytol, 2009, 182(3): 751-762.
doi: 10.1111/j.1469-8137.2009.02794.x pmid: 19291006 |
| [28] | Valoroso MC, Paolo SD, Iazzetti G, Aceto S. Transcriptome-wide identification and expression analysis of DIVARICATA- and RADIALIS-like genes of the mediterranean Orchid Orchis italica. Genome Biol Evol, 2017, 9(6): evx101. |
| [29] | Guo SK, Zeng CR, Wu Y, Li J, LI XG, Yang YD. Cloning and expression analysis of coconut CnRADIALIS-like transcription factor. Chinese Journal of Tropical Crops, 2021, 42(09): 2478-2486. |
| 郭树宽, 曾春茹, 吴翼, 李静, 李新国, 杨耀东. 椰子CnRADIALIS-like转录因子的克隆与表达分析. 热带作物学报, 2021, 42(09): 2478-2486. | |
| [30] |
Baxter CEL, Costa MMR, Coen ES. Diversification and co-option of RAD-like genes in the evolution of floral asymmetry. Plant J, 2007, 52(1): 105-113.
doi: 10.1111/j.1365-313X.2007.03222.x |
| [31] |
Stevenson CE, Burton N, Costa MMR, Nath U, Dixon RA, Coen ES, Lawson DM. Crystal structure of the MYB domain of the RAD transcription factor from Antirrhinum majus. Proteins, 2006, 65(4): 1041-1045.
doi: 10.1002/prot.21136 |
| [32] | Ge J, Zhang Y, Zheng ZZ, Huang YW, Song HS. Research progress on CREB and the signal transduction pathway of its phosphorylation. J Anhui Agric Sci, 2010, 38(30): 16769-16771,16774. |
| 葛军, 张玉, 郑增长, 黄延旺, 宋红生. CREB转录因子及其磷酸化信号通路的研究进展. 安徽农业科学, 2010, 38(30): 16769-16771,16774. | |
| [33] |
Sengupta A, Hileman LC. A CYC-RAD-DIV-DRIF interaction likely pre-dates the origin of floral monosymmetry in Lamiales. Evodevo, 2022, 13(1): 3.
doi: 10.1186/s13227-021-00187-w pmid: 35093179 |
| [34] |
Machemer K, Shaiman O, Salts Y, Shabtai S, Sobolev I, Belausov E, Grotewold E, Barg R. Interplay of MYB factors in differential cell expansion, and consequences for tomato fruit development. Plant J, 2011, 68(2): 337-350.
doi: 10.1111/j.1365-313X.2011.04690.x |
| [35] |
Masuda K, Ikeda Y, Matsuura T, Kawakatsu T, Tao R, Kubo Y, Ushijima K, Henry IM, Akagi T. Reinvention of hermaphroditism via activation of a RADIALIS-like gene in hexaploid persimmon. Nat Plants, 2022, 8(3): 217-224.
doi: 10.1038/s41477-022-01107-z pmid: 35301445 |
| [36] |
Müller BM, Saedler H, Zachgo S. The MADS-box gene DEFH28 from Antirrhinum is involved in the regulation of floral meristem identity and fruit development. Plant J, 2001, 28(2): 169-179.
pmid: 11722760 |
| [37] | Callens C, Tucker MR, Zhang DB, Wilson ZA. Dissecting the role of MADS-box genes in monocot floral development and diversity. J Exp Bot, 2018, 69(10): 2435-2459. |
| [38] |
Zúñiga-Mayo VM, Gómez-Felipe A, Herrera-Ubaldo H, de Folter S. Gynoecium development: networks in Arabidopsis and beyond. J Exp Bot, 2019, 70(5): 1447-1460.
doi: 10.1093/jxb/erz026 pmid: 30715461 |
| [1] | Sinan Luan, Lele Liu, Jiayuan Zhou, Nurasya Imam, Minlong Cui, Chunlan Piao. Functional analysis of flower development related gene SvGLOBOSA from Senecio vulgaris [J]. Hereditas(Beijing), 2022, 44(6): 521-530. |
| [2] | Qingqian Ding,Xiaoting Wang,Liqin Hu,Xin Qi,Linhao Ge,Weiya XU,Zhaoshi Xu,Yongbin Zhou,Guanqing Jia,Xianmin Diao,Donghong Min,Youzhi Ma,Ming Chen. MYB-like transcription factor SiMYB42 from foxtail millet (Setaria italica L.) enhances Arabidopsis tolerance to low-nitrogen stress [J]. Hereditas(Beijing), 2018, 40(4): 327-338. |
| [3] |
Lizhen Zhang, Yong Zhang, Jinghua Hu, Zilong Wang, Zhijiang Zeng.
Molecular cloning and expression analysis of the tyramine receptor genes in Apis cerana cerana [J]. Hereditas(Beijing), 2018, 40(2): 155-161. |
| [4] | Jianzhong Chang, Fengxia Yan, Linyi Qiao, Jun Zheng, Fuyao Zhang, Qingshan Liu. Genome-wide identification and expression analysis of SBP-box gene family in Sorghum bicolor L. [J]. HEREDITAS(Beijing), 2016, 38(6): 569-580. |
| [5] | Yanbing Gu, Zhirui Ji, Fumei Chi, Zhuang Qiao, Chengnan Xu, Junxiang Zhang, Zongshan Zhou, Qinglong Dong. Genome-wide identification and expression analysis of the WRKY gene family in peach [J]. HEREDITAS(Beijing), 2016, 38(3): 254-270. |
| [6] | Jun Wu, Junhong Zhang, Menghui Huang, Minhui Zhu, Zaikang Tong. Expression analysis of miR164 and its target gene NAC1 in response to low nitrate availability in Betula luminifera [J]. HEREDITAS(Beijing), 2016, 38(2): 155-162. |
| [7] | Jianjiang Ma, Nuohan Wang, Man Wu, Wenfeng Pei, Wenkui Wang, Xingli Li, Jinfa Zhang, Shuxun Yu, Jiwen Yu. Genome-wide analysis of the LPAAT gene family in Gossypium raimondii and G. arboreum, and expression analysis of its orthologs in G. hirsutum [J]. HEREDITAS(Beijing), 2015, 37(7): 692-701. |
| [8] | Kun Zhou, Jinjin Zhang. Nitric oxide in plants and its role in regulating flower development [J]. HEREDITAS(Beijing), 2014, 36(7): 661-668. |
| [9] | Yunfei Liu, Hongjian Wan, Yuejian Yang, Yanping Wei, Zhimiao Li, Qingjing Ye, Rongqing Wang, Meiying Ruan, Zhuping Yao, Guozhi Zhou. Genome-wide identification and analysis of heat shock protein 90 in tomato [J]. HEREDITAS(Beijing), 2014, 36(10): 1043-1052. |
| [10] | HUANG Guo-Wen HAN Yu-Zhen FU Yong-Fu. Expression and functional analysis of SUA41gene in Arabidopsis thaliana [J]. HEREDITAS, 2013, 35(1): 93-100. |
| [11] | YANG Li-Wei, SHI Ji-Sen. Cloning and expression analysis of differentially expressed genes in Chinese fir stems treated by different concentrations of exogenous IAA [J]. HEREDITAS, 2012, 34(4): 472-484. |
| [12] | JIAO Sha-Sha, LIU Ka, LI Gang, GAO Jian-Feng, MA Run-Lin. Confirmation and expression analysis of three predicted genes in sheep MHC region [J]. HEREDITAS, 2011, 33(12): 1353-1358. |
| [13] | LUO Mao, ZHANG Zhi-Ming, GAO Jian, ZENG Xing, PAN Guang-Tang. The role of miR319 in plant development regulation [J]. HEREDITAS, 2011, 33(11): 1203-1211. |
| [14] | BAO Yong-Mei, LIU Yong-Hui, HU Dong-Qing, HUANG Ji, WANG Zhou-Fei, WANG Jian-Fei, ZHANG Gong-Sheng. Preparation, characterization and application of rice Qb-SNARE protein OsNPSN11 polyclonal antibody [J]. HEREDITAS, 2010, 32(9): 961-965. |
| [15] | DAO Qian-Yi, LI Zheng, HE Huan-Le, BO Dun-Song, CA Run. Expression analysis of the unisex-determine gene M in cucumber [J]. HEREDITAS, 2010, 32(6): 632-638. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||