遗传 ›› 2021, Vol. 43 ›› Issue (8): 775-791.doi: 10.16288/j.yczz.21-132
收稿日期:2021-04-11
修回日期:2021-05-12
出版日期:2021-08-20
发布日期:2021-06-30
作者简介:王玲,硕士研究生,专业方向:生物学。E-mail: 基金资助:
Ling Wang( ), Jinhuan Li, Haiyan Huang(
), Jinhuan Li, Haiyan Huang( ), Qiang Wu(
), Qiang Wu( )
)
Received:2021-04-11
Revised:2021-05-12
Published:2021-08-20
Online:2021-06-30
Supported by:摘要:
三维基因组染色质架构蛋白CTCF (CCCTC-binding factor)能够介导增强子与基因启动子的远距离染色质相互作用,也可以结合调控区域的绝缘子发挥增强子绝缘功能,对发育中的基因表达调控具有重要作用。同源框基因家族(Homeobox gene family, Hox)编码一类控制动物发育的关键转录因子,在发育中主要沿胚胎首尾轴(head-to-tail axis)呈时空线性表达。在哺乳动物中,Hox基因分为HoxA、HoxB、HoxC和HoxD四个基因簇,在中枢神经系统、骨骼和四肢发育中发挥重要功能。HoxD基因簇主要调控四肢发育,受位于其两侧调控域内的增强子调节,沿肢体近远轴(proximal-distal axis)呈时空线性表达。在人类基因组中,HOXD基因簇及其两侧的调控区域分布有串联排列的CTCF结合位点(简称CTCF位点),参与9个HOXD基因的表达调控。本研究以HOXD基因簇为模式基因,探究CTCF对发育基因(developmental genes)转录调控的影响。利用CRISPR DNA片段编辑技术在人HEK293T细胞中获得一系列的串联反向CTCF位点删除的单细胞克隆株。RNA-seq实验揭示CTCF位点删除后HOXD基因表达下降。定量高分辨率染色体构象捕获实验显示,HOXD与上游增强子簇的远距离染色质相互作用增强,与下游增强子簇的远距离染色质相互作用减弱。综上所述,串联反向的CTCF位点通过其绝缘子功能维持上下游增强子簇对HOXD基因簇表达调控的平衡,为探究动物发育过程中Hox基因表达的精准调控机制提供参考。
王玲, 李金环, 黄海燕, 吴强. 串联反向CTCF位点的系列删除揭示增强子调控HOXD基因簇表达的平衡[J]. 遗传, 2021, 43(8): 775-791.
Ling Wang, Jinhuan Li, Haiyan Huang, Qiang Wu. Serial deletions of tandem reverse CTCF sites reveal balanced HOXD regulatory landscape of enhancers[J]. Hereditas(Beijing), 2021, 43(8): 775-791.
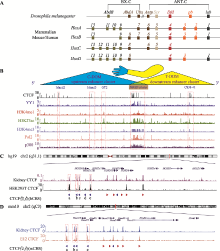
图1
CTCF位点在Hox基因簇及其调控区域内的分布 A:果蝇及哺乳动物Hox基因家族的基因组结构。果蝇Hox基因簇分为触足复合群(ANT-C)和双胸复合群(BX-C),其中ANT-C包含Antp、Scr、Dfd、pb和lab基因,BX-C包含AbdB、AbdA和Ubx基因。哺乳动物具有4个Hox基因簇:HoxA、HoxB、HoxC和HoxD,共包含39个基因,其中HoxD基因簇包括HoxD1、HoxD3、HoxD4、HoxD8~HoxD13。果蝇及哺乳动物相同颜色的Hox基因为直系同源基因(orthologues),它们起源于同一祖先基因。B:人胚胎肾细胞系HEK293T中的CTCF蛋白、架构蛋白YY1、增强子标记H3K4me1和H3K27ac、启动子标记H3K4me3、转录活性标记PolⅡ和p300在人HOXD基因簇及其调控区域内的分布。红色虚线框指示增强子Island2、Island5、GT2、CS38-41和HOXD基因簇所在区域。哺乳动物的HoxD基因簇位于3′端粒侧TAD (T-DOM)和5′中心粒侧TAD (C-DOM)交界处,并受到上下游增强子簇(upstream enhancer cluster和downstream enhancer cluster)的调控。C:人类HOXD基因簇区域CTCF位点的分布。人肾脏和胚胎肾细胞系HEK293T的CTCF ChIP-seq结合峰分布图:HOXD基因簇C-DOM和T-DOM交界区域均具有串联排列的CTCF位点。红色虚线框依次指示CBS a、b、c和e所在区域。D:小鼠HoxD基因簇区域CTCF位点的分布。小鼠肾脏和第12.5天胚胎肢芽CTCF ChIP-seq数据显示小鼠HoxD基因簇中心粒侧对应的位置具有串联反向排列的4个CTCF位点。图C和D中箭头代表CTCF位点,其中红色箭头代表正向CTCF位点,蓝色箭头代表反向CTCF位点。"
表1
引物序列"
| 类型 | 引物名称 | 序列(5′→3′) |
|---|---|---|
| PCR | a1F | TTCCAGCACCTCGGCTTTGTC |
| a1R | CCCACTTTCCACCTCTGTCCTG | |
| b1F | GTCCGCCCGTGAGCTTCTGAA | |
| b1R1 | CTCACAGCAGCCGAAACCG | |
| c1F | TGATGCAGCCTCTGTGACCG | |
| c1R | AGTTTTCCCGTGGCGTCTGA | |
| e1F | TTCCCTGTCCCAGCTTGATTTC | |
| e1R | TCAACAGTGAAGGGCGGTGC | |
| e2F | CAAGCCACTCTCCCGCCACTA | |
| e2R | TCGCTCTCGTCCTCTCTTGGG | |
| e2R1 | GGCTCCTGCACTGAGACCACA | |
| sgRNA | sgRNA a1F | ACCGCGAAGAGTGCGGGAGAACGG |
| sgRNA a1R | AAACCCGTTCTCCCGCACTCTTCG | |
| sgRNA b1F | ACCGGGCGCATCAGGAATGTAAG | |
| sgRNA b1R | AAACCTTACATTCCTGATGCGCC | |
| sgRNA c1F | ACCGCAGGCGAAGTGCGGTTTCCA | |
| sgRNA c1R | AAACTGGAAACCGCACTTCGCCTG | |
| sgRNA e1F | ACCGAACTGTGCTCAAACGCTCTC | |
| sgRNA e1R | AAACGAGAGCGTTTGAGCACAGTT | |
| sgRNA e2F | ACCGGAGGCGCAAACAGCTGTTGT | |
| sgRNA e2R | AAACACAACAGCTGTTTGCGCCTC | |
| Index | Island5-bioprimer | 5′biotin-AAACACAAATGCATCAACCTG |
| GT2-bioprimer | 5′biotin-GAGCCAAACTGTACCCCTAGC | |
| HOXD9-bioprimer | 5′biotin-ACCGACTAGTTCGCAGGCT | |
| Island5-P5 | AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCTATGCATCTCATGAAGCTGGCATCT | |
| GT2-P5 | AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCTGTAACTTTAGCTAAACCAAGGCCT | |
| HOXD9-P5 | AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCTCTGCAGCCTCCACCATTG | |
| P7-index-1 | CAAGCAGAAGACGGCATACGAGATCGAGTAATGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT | |
| P7-index-2 | CAAGCAGAAGACGGCATACGAGATTCTCCGGAGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT | |
| P7-index-3 | CAAGCAGAAGACGGCATACGAGATAATGAGCGGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT | |
| P7-index-4 | CAAGCAGAAGACGGCATACGAGATGGAATCTCGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT | |
| P7-index-5 | CAAGCAGAAGACGGCATACGAGATTTCTGAATGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT | |
| P7-index-6 | CAAGCAGAAGACGGCATACGAGATACGAATTCGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT | |
| P7-index-7 | CAAGCAGAAGACGGCATACGAGATAGCTTCAGGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT | |
| P7-index-8 | CAAGCAGAAGACGGCATACGAGATGCGCATTAGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT | |
| P7-index-9 | CAAGCAGAAGACGGCATACGAGATCATAGCCGGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT | |
| P7-index-10 | CAAGCAGAAGACGGCATACGAGATTTCGCGGAGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT | |
| P7-index-11 | CAAGCAGAAGACGGCATACGAGATGCGCGAGAGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT | |
| P7-index-12 | CAAGCAGAAGACGGCATACGAGATCTATCGCTGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT | |
| P7-index-13 | CAAGCAGAAGACGGCATACGAGATAGAGTACTGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT | |
| P7-index-14 | CAAGCAGAAGACGGCATACGAGATGCTCCGTAGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT | |
| P7-index-15 | CAAGCAGAAGACGGCATACGAGATCATGAGAGGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT | |
| P7-index-16 | CAAGCAGAAGACGGCATACGAGATTGAATCGCGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT | |
| P7-index-17 | CAAGCAGAAGACGGCATACGAGATGTCTGACTGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT | |
| P7-index-18 | CAAGCAGAAGACGGCATACGAGATCTGAATGCGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT | |
| P7-index-19 | CAAGCAGAAGACGGCATACGAGATCGCTTCTGGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT | |
| P7-index-20 | CAAGCAGAAGACGGCATACGAGATTCGCATGAGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT | |
| P7-index-21 | CAAGCAGAAGACGGCATACGAGATAATAGCAGGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT | |
| P7-index-22 | CAAGCAGAAGACGGCATACGAGATGTCGCGTAGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT | |
| P7-index-23 | CAAGCAGAAGACGGCATACGAGATACGCGATAGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT | |
| P7-index-24 | CAAGCAGAAGACGGCATACGAGATTGATCGATGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT | |
| P7-index-25 | CAAGCAGAAGACGGCATACGAGATCCGCATGAGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT | |
| P7-index-26 | CAAGCAGAAGACGGCATACGAGATCCACAATCGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT | |
| P7-index-27 | CAAGCAGAAGACGGCATACGAGATGATGTTCGGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT |
图2
CBS e删除改变增强子与启动子间的远程互作从而影响HOXD基因簇的基因表达 A:利用CRISPR DNA片段编辑技术获得CBS e删除的单细胞克隆株(e#113和e#126)的示意图。针对CBS e设计一对sgRNA(sgRNA e1和sgRNA e2),Cas9核酸酶在sgRNA e1和sgRNA e2的介导下特异性识别CBS e两侧靶向序列后进行切割,形成的两个切口被修复后连接在一起获得CBS e删除的编辑细胞。B:采用特异性引物PCR后进行凝胶电泳实验鉴定单细胞克隆株。引物对e1F/e2R在野生型(WT)细胞中扩增出1560 bp的片段,在e#113和e#126细胞株中扩增出CBS e删除后的729 bp片段(红色虚线框指示目的条带)。C:TA克隆并进行桑格测序确定单细胞克隆株的基因型。引物对e1F/e2R在e#113和e#126细胞株中扩增的产物经TA克隆后的测序结果图。D:RNA-seq数据分析比较WT细胞、e#113和e#126细胞株中HOXD基因簇表达水平。*:P<0.05;**:P<0.01;FPKM:fragments per kilobase of transcript per million mapped reads。E:在染色质构象捕获(QHR-4C)实验中,以增强子Island5为观测点(viewpoint,VP),分析e#113和e#126细胞株中Island5与HOXD基因簇启动子及调控元件Island2、GT2和CS38-41的远程互作。将e#113和e#126细胞株的数据分别与WT进行log2处理,红色实线框内为Island5与HOXD基因簇启动子之间的染色质相互作用,黑色虚线框依次指示调控元件Island2、GT2和CS38-41。F:以增强子GT2为VP,分析CBS e删除后GT2与HOXD基因簇启动子的远程互作。红色实线框内为GT2与HOXD基因簇启动子间的染色质相互作用放大图,黑色虚线框依次指示调控元件Island2、Island5和CS38-41。G:以HOXD9启动子为VP,分析e#113和e#126细胞株中调控元件Island2、Island5、GT2、CS38-41与HOXD9启动子之间的染色质相互作用。黑色虚线框指示Island2,红色实线框内分别为Island5、GT2和CS38-41与HOXD9启动子之间的染色质相互作用放大图。"
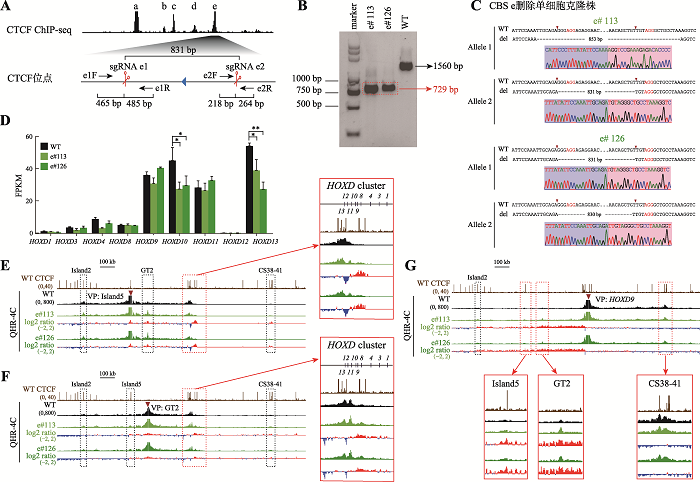
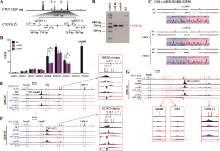
图3
CBS c-e删除引起上游增强子与近端HOXD基因相互作用的增强 A:设计一对sgRNA(sgRNA c1和sgRNA e2)对HEK293T野生型细胞进行编辑获得CBS c-e删除的单细胞克隆株(c-e#49和c-e#54)的示意图。B:用引物c1F和e2R PCR鉴定c-e#49和c-e#54单细胞克隆株的凝胶电泳图。C:TA克隆鉴定c-e#49和c-e#54单细胞克隆株基因型的测序结果图。D:RNA-seq数据分析比较WT细胞、c-e#49和c-e#54细胞中HOXD基因簇表达水平。E:QHR-4C实验中,以Island5为VP,分析c-e#49和c-e#54细胞株中Island5与HOXD基因簇启动子的远程互作。红色实线框内为Island5与HOXD基因簇启动子之间的染色质相互作用放大图,黑色虚线框依次指示调控元件Island2、GT2和CS38-41。F:以GT2为VP,分析CBS c- e删除后GT2与HOXD基因簇启动子的远程互作。红色实线框内为GT2与HOXD基因簇启动子间的染色质相互作用放大图,黑色虚线框依次指示调控元件Island2、Island5和CS38-41。G:以HOXD9启动子为VP,c-e#49和c-e#54细胞株中Island2、Island5、GT2、CS38-41与HOXD9启动子之间的染色质相互作用。黑色虚线框指示Island2,红色实线框内分别为Island5、GT2和CS38-41与HOXD9启动子之间的染色质相互作用放大图。"
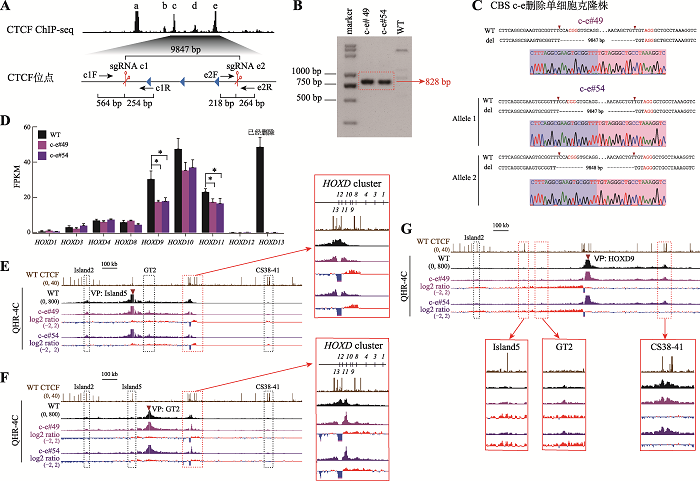
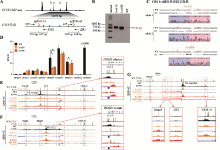
图4
串联排列CTCF位点对增强子远程互作和HOXD基因表达具有叠加效应 A:利用CRISPR/Cas9编辑系统获得CBS b-e删除的单细胞克隆株(b-e#59和b-e#80)的示意图。B:用引物b1F和e2R1进行PCR鉴定b-e#59和b-e#80单细胞克隆株的凝胶电泳图。C:TA克隆鉴定b-e#59和b-e#80单细胞克隆株基因型的测序结果图。D:RNA-seq数据分析比较WT细胞、b-e#59和b-e#80细胞株细胞中HOXD基因簇表达水平。E:QHR-4C实验中,以Island5为VP,分析CBS b-e删除对Island5与HOXD基因簇启动子远程互作的影响。红色实线框内为Island5与HOXD基因簇启动子之间的染色质相互作用放大图,黑色虚线框依次指示调控元件Island2、GT2和CS38-41。F:以GT2为VP,分析b-e#59和b-e#80细胞株中GT2与HOXD基因簇启动子的远程互作。红色实线框内为GT2与HOXD基因簇启动子间的染色质相互作用放大图,黑色虚线框依次指示调控元件Island2、Island5和CS38-41。G:以HOXD9启动子为VP,分析b-e#59和b-e#80细胞株中Island5、GT2、CS38-41与HOXD9启动子间的染色质相互作用。黑色虚线框指示Island2,红色实线框内分别为Island5、GT2和CS38-41与HOXD9启动子之间的染色质相互作用放大图。"
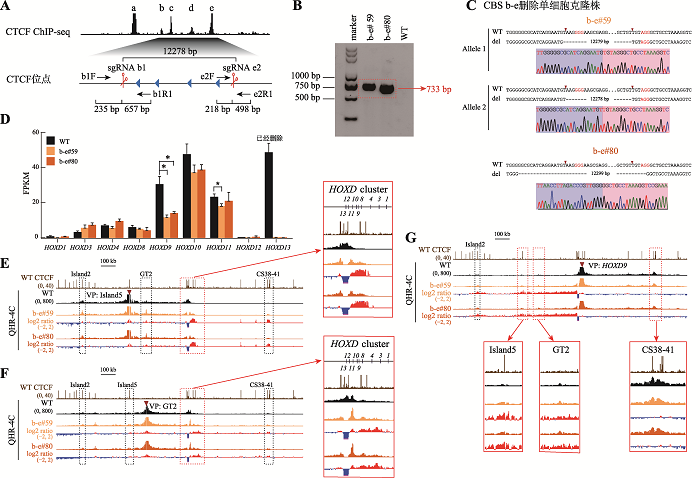
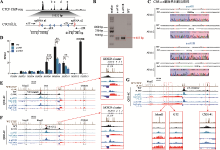
图5
删除全部反向CTCT位点破坏增强子调控HOXD基因表达的平衡 A:获得CBS a-e删除的单细胞克隆株(a-e#93和a-e#108)的示意图。B:用引物a1F和e2R PCR鉴定a-e#93和a-e#108细胞株的凝胶电泳图。C:TA克隆鉴定a-e#93和a-e#108单细胞克隆株基因型的测序结果图。D:RNA-seq数据分析比较WT细胞、a-e#93和a-e#108细胞株细胞中HOXD基因簇转录水平。E:QHR-4C实验中,以Island5为VP,分析CBS a-e删除对Island5与HOXD基因簇启动子及两侧调控区域染色质相互作用的影响。红色实线框内为Island5与HOXD基因簇启动子之间的染色质相互作用放大图,黑色虚线框依次指示调控元件Island2、GT2和CS38-41。F:以GT2为VP,分析a-e#93和a-e#108细胞株中GT2与HOXD基因簇启动子的染色质相互作用。红色实线框内为GT2与HOXD基因簇启动子间的染色质相互作用放大图,黑色虚线框依次指示调控元件Island2、Island5和CS38-41。G:以HOXD9启动子为VP,分析CBS a-e删除后HOXD9的表达调控模式。黑色虚线框指示Island2,红色实线框内为Island5、GT2、CS38-41与HOXD9基因簇启动子间的染色质相互作用放大图。"
| [1] |
Wu Q, Liu PF, Wang LY. Many facades of CTCF unified by its coding for three-dimensional genome architecture. J Genet Genomics, 2020, 47(8):407-424.
doi: 10.1016/j.jgg.2020.06.008 |
| [2] |
Guo Y, Monahan K, Wu HY, Gertz J, Varley KE, Li W, Myers RM, Maniatis T, Wu Q. CTCF/cohesin-mediated DNA looping is required for protocadherin alpha promoter choice. Proc Natl Acad Sci USA, 2012, 109(51):21081-21086.
doi: 10.1073/pnas.1219280110 |
| [3] |
Guo Y, Xu Q, Canzio D, Shou J, Li JH, Gorkin DU, Jung I, Wu HY, Zhai YN, Tang YX, Lu YC, Wu YH, Jia ZL, Li W, Zhang MQ, Ren B, Krainer AR, Maniatis T, Wu Q. CRISPR inversion of CTCF sites alters genome topology and enhancer/promoter function. Cell, 2015, 162(4):900-910.
doi: 10.1016/j.cell.2015.07.038 pmid: 26276636 |
| [4] | Zhai YN, Xu Q, Guo Y, Wu Q. Characterization of a cluster of CTCF-binding sites in a protocadherin regulatory region. Hereditas(Beijing), 2016, 38(4):323-336. |
| 翟亚男, 许泉, 郭亚, 吴强. 原钙粘蛋白基因簇调控区域中成簇的CTCF结合位点分析. 遗传, 2016, 38(4):323-336. | |
| [5] |
Yin M, Wang J, Wang M, Li X, Zhang M, Wu Q, Wang Y. Molecular mechanism of directional CTCF recognition of a diverse range of genomic sites. Cell Res, 2017, 27(11):1365-1377.
doi: 10.1038/cr.2017.131 |
| [6] | Guo Y, Wu Q. Inversion of CTCF binding sites by DNA fragment editing alters genome topology and enhancer/ promoter functions. Hereditas(Beijing), 2015, 37(10):1073-1074. |
| 郭亚, 吴强. 采用DNA片段编辑技术反转CTCF结合位点改变基因组拓扑结构和增强子与启动子功能. 遗传, 2015, 37(10):1073-1074. | |
| [7] |
Filippova GN, Fagerlie S, Klenova EM, Myers C, Dehner Y, Goodwin G, Neiman PE, Collins SJ, Lobanenkov VV. An exceptionally conserved transcriptional repressor, CTCF, employs different combinations of zinc fingers to bind diverged promoter sequences of avian and mammalian c-myc oncogenes. Mol Cell Biol, 1996, 16(6):2802-2813.
pmid: 8649389 |
| [8] |
Chen HB, Tian Y, Shu WJ, Bo XC, Wang SQ. Comprehensive identification and annotation of cell type-specific and ubiquitous CTCF-binding sites in the human genome. PLoS One, 2012, 7(7):e41374.
doi: 10.1371/journal.pone.0041374 |
| [9] |
Nasmyth K. Disseminating the genome: Joining, resolving, and separating sister chromatids during mitosis and meiosis. Annu Rev Genet, 2001, 35:673-745.
pmid: 11700297 |
| [10] |
Kim Y, Shi ZB, Zhang HS, Finkelstein IJ, Yu HT. Human cohesin compacts DNA by loop extrusion. Science, 2019, 366(6471):1345-1349.
doi: 10.1126/science.aaz4475 |
| [11] |
Lu YJ, Shou J, Jia ZL, Wu YH, Li JH, Guo Y, Wu Q. Genetic evidence for asymmetric blocking of higher-order chromatin structure by CTCF/cohesin. Protein Cell, 2019, 10(12):914-920.
doi: 10.1007/s13238-019-00656-y |
| [12] | Zheng XF, Huang HY, Wu Q. Chromatin architectural protein CTCF regulates gene expression of the UGT1 cluster. Hereditas(Beijing), 2019, 41(6):509-523. |
| 郑晓飞, 黄海燕, 吴强. 染色质架构蛋白CTCF调控UGT1基因簇的表达. 遗传, 2019, 41(6):509-523. | |
| [13] |
Jia ZL, Li JW, Ge X, Wu YH, Guo Y, Wu Q. Tandem CTCF sites function as insulators to balance spatial chromatin contacts and topological enhancer-promoter selection. Genome Biol, 2020, 21(1):75.
doi: 10.1186/s13059-020-01984-7 |
| [14] | Wu YH, Jia ZL, Ge X, Wu Q. Three-dimensional genome architectural CCCTC-binding factor makes choice in duplicated enhancers at Pcdhα locus. Sci China Life Sci, 2020, 63(6):835-844. |
| [15] |
Dekker J, Mirny L. The 3D genome as moderator of chromosomal communication. Cell, 2016, 164(6):1110-1121.
doi: S0092-8674(16)30073-3 pmid: 26967279 |
| [16] |
Nichols MH, Corces VG. A tethered-inchworm model of SMC DNA translocation. Nat Struct Mol Biol, 2018, 25(10):906-910.
doi: 10.1038/s41594-018-0135-4 |
| [17] |
Wu Q, Jia ZL. Wiring the brain by clustered protocadherin neural codes. Neurosci Bull, 2021, 37(1):117-131.
doi: 10.1007/s12264-020-00578-4 |
| [18] | Lin SG, Ba ZQ, Alt FW, Zhang Y. RAG chromatin scanning during V(D)J recombination and chromatin loop extrusion are related processes. Adv Immunol, 2018, 139:93-135. |
| [19] |
Chen L, Carico Z, Shih HY, Krangel MS. A discrete chromatin loop in the mouse Tcra-Tcrd locus shapes the TCRdelta and TCRalpha repertoires. Nat Immunol, 2015, 16(10):1085-1093.
doi: 10.1038/ni.3232 |
| [20] | Majumder K, Koues OI, Chan EAW, Kyle KE, Horowitz JE, Yang-Iott K, Bassing CH, Taniuchi I, Krangel MS, Oltz EM. Lineage-specific compaction of Tcrb requires a chromatin barrier to protect the function of a long-range tethering element. J Exp Med, 2015, 212(1):107-120. |
| [21] | Rodríguez-Carballo E, Lopez-Delisle L, Zhan Y, Fabre PJ, Beccari L, El-Idrissi I, Huynh THN, Ozadam H, Dekker J, Duboule D. The HoxD cluster is a dynamic and resilient TAD boundary controlling the segregation of antagonistic regulatory landscapes. Genes Dev, 2017, 31(22):2264-2281. |
| [22] | Rodríguez-Carballo E, Lopez-Delisle L, Yakushiji- Kaminatsui N, Ullate-Agote A, Duboule D. Impact of genome architecture on the functional activation and repression of Hox regulatory landscapes. BMC Biol, 2019, 17(1):55. |
| [23] | Rodríguez-Carballo E, Lopez-Delisle L, Willemin A, Beccari L, Gitto S, Mascrez B, Duboule D. Chromatin topology and the timing of enhancer function at the HoxD locus. Proc Natl Acad Sci USA, 2020, 117(49):31231-31241. |
| [24] |
Jia ZL, Wu Q. Clustered protocadherins emerge as novel susceptibility loci for mental disorders. Front Neurosci, 2020, 14:587819.
doi: 10.3389/fnins.2020.587819 |
| [25] |
Heger P, Marin B, Bartkuhn M, Schierenberg E, Wiehe T. The chromatin insulator CTCF and the emergence of metazoan diversity. Proc Natl Acad Sci USA, 2012, 109(43):17507-17512.
doi: 10.1073/pnas.1111941109 |
| [26] |
Lewis EB. A gene complex controlling segmentation in Drosophila. Nature, 1978, 276(5688):565-570.
doi: 10.1038/276565a0 |
| [27] | Mallo M. Reassessing the role of Hox genes during vertebrate development and evolution. Trends Genet, 2018, 34(3):209-217. |
| [28] |
Kmita M, Duboule D. Organizing axes in time and space; 25 years of colinear tinkering. Science, 2003, 301(5631):331-333.
doi: 10.1126/science.1085753 |
| [29] | Andrey G, Montavon T, Mascrez B, Gonzalez F, Noordermeer D, Leleu M, Trono D, Spitz F, Duboule D. A switch between topological domains underlies HoxD genes collinearity in mouse limbs. Science, 2013, 340(6137):1234167. |
| [30] | Beccari L, Yakushiji-Kaminatsui N, Woltering JM, Necsulea A, Lonfat N, Rodríguez-Carballo E, Mascrez B, Yamamoto S, Kuroiwa A, Duboule D. A role for Hox13 proteins in the regulatory switch between TADs at the HoxD locus. Genes Dev, 2016, 30(10):1172-1186. |
| [31] | Montavon T, Soshnikova N, Mascrez B, Joye E, Thevenet L, Splinter E, de Laat W, Spitz F, Duboule D. A regulatory archipelago controls Hox genes transcription in digits. Cell, 2011, 147(5):1132-1145. |
| [32] | Lonfat N, Montavon T, Darbellay F, Gitto S, Duboule D. Convergent evolution of complex regulatory landscapes and pleiotropy at Hox loci. Science, 2014, 346(6212):1004-1006. |
| [33] |
Long HK, Prescott SL, Wysocka J. Ever-changing landscapes: Transcriptional enhancers in development and evolution. Cell, 2016, 167(5):1170-1187.
doi: 10.1016/j.cell.2016.09.018 |
| [34] |
Schoenfelder S, Fraser P. Long-range enhancer-promoter contacts in gene expression control. Nat Rev Genet, 2019, 20(8):437-455.
doi: 10.1038/s41576-019-0128-0 pmid: 31086298 |
| [35] |
Kim S, Shendure J. Mechanisms of interplay between transcription factors and the 3D genome. Mol Cell, 2019, 76(2):306-319.
doi: 10.1016/j.molcel.2019.08.010 |
| [36] | Noordermeer D, Leleu M, Splinter E, Rougemont J, De Laat W, Duboule D. The dynamic architecture of Hox gene clusters. Science, 2011, 334(6053):222-225. |
| [37] |
Li JH, Shou J, Guo Y, Tang YX, Wu YH, Jia ZL, Zhai YN, Chen ZF, Xu Q, Wu Q. Efficient inversions and duplications of mammalian regulatory DNA elements and gene clusters by CRISPR/Cas9. J Mol Cell Biol, 2015, 7(4):284-298.
doi: 10.1093/jmcb/mjv016 |
| [38] |
Chang N, Sun C, Gao L, Zhu D, Xu X, Zhu X, Xiong JW, Xi JJ. Genome editing with RNA-guided Cas9 nuclease in zebrafish embryos. Cell Res, 2013, 23(4):465-472.
doi: 10.1038/cr.2013.45 |
| [39] |
Doudna JA, Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science, 2014, 346(6213):1258096.
doi: 10.1126/science.1258096 |
| [40] | Liu PF, Wu Q. Probing 3D genome by CRISPR/Cas9. Hereditas(Beijing), 2020, 42(1):18-31. |
| 刘沛峰, 吴强. CRISPR/Cas9基因编辑在三维基因组研究中的应用. 遗传, 2020, 42(1):18-31. | |
| [41] | Li JH, Shou J, Wu Q. DNA fragment editing of genomes by CRISPR/Cas9. Hereditas(Beijing), 2015, 37(10):992-1002. |
| 李金环, 寿佳, 吴强. CRISPR/Cas9系统在基因组DNA片段编辑中的应用. 遗传, 2015, 37(10):992-1002. | |
| [42] |
Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with tophat and cufflinks. Nat Protoc, 2012, 7(3):562-578.
doi: 10.1038/nprot.2012.016 pmid: 22383036 |
| [43] | Guo XQ, Chen FZ, Gao F, Li L, Liu K, You LJ, Hua C, Yang F, Liu WL, Peng CH, Wang LN, Yang XX, Zhou FY, Tong JW, Cai J, Li ZY, Wan B, Zhang L, Yang T, Zhang MW, Yang LL, Yang YW, Zeng WJ, Wang B, Wei XF, Xu X. CNSA: A data repository for archiving omics data. Database (Oxford), 2020; 2020: baaa055. |
| [44] | Chen FZ, You LJ, Yang F, Wang LN, Guo XQ, Gao F, Hua C, Tan C, Fang L, Shan RQ, Zeng WJ, Wang B, Wang R, Xu X, Wei XF. CNGBdb: China National Genebank Database. Hereditas(Beijing), 2020, 42(08):799-809. |
| 陈凤珍, 游丽金, 杨帆, 王丽娜, 郭学芹, 高飞, 华聪, 谈聪, 方林, 单日强, 曾文君, 王博, 王韧, 徐讯, 魏晓锋. CNGBdb: 国家基因库生命大数据平台. 遗传, 2020, 42(8):799-809. | |
| [45] |
Pearson JC, Lemons D, McGinnis W. ModulatingHox gene functions during animal body patterning. Nat Rev Genet, 2005, 6(12):893-904.
pmid: 16341070 |
| [46] |
Lonfat N, Duboule D. Structure, function and evolution of topologically associating domains (TADs) atHox loci. FEBS Lett, 2015, 589(20):2869-2876.
doi: 10.1016/j.febslet.2015.04.024 |
| [47] | Holland PW, Garcia-Fernàndez J, Williams NA, Sidow A. Gene duplications and the origins of vertebrate development. Dev Suppl, 1994, 125-133. |
| [48] |
Shou J, Li J, Liu Y, Wu Q. Precise and predictable CRISPR chromosomal rearrangements reveal principles of Cas9-mediated nucleotide insertion. Mol Cell, 2018, 71(4):498-509 e4.
doi: S1097-2765(18)30466-0 pmid: 30033371 |
| [49] |
Fu YF, Foden JA, Khayter C, Maeder ML, Reyon D, Joung JK, Sander JD. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat Biotechnol, 2013, 31(9):822-826.
doi: 10.1038/nbt.2623 |
| [50] |
Perry MW, Boettiger AN, Levine M. Multiple enhancers ensure precision of gap gene-expression patterns in the Drosophila embryo. Proc Natl Acad Sci USA, 2011, 108(33):13570-13575.
doi: 10.1073/pnas.1109873108 |
| [51] |
Frankel N, Davis GK, Vargas D, Wang S, Payre F, Stern DL. Phenotypic robustness conferred by apparently redundant transcriptional enhancers. Nature, 2010, 466(7305):490-493.
doi: 10.1038/nature09158 |
| [52] |
Barolo S. Shadow enhancers: Frequently asked questions about distributed cis-regulatory information and enhancer redundancy. Bioessays, 2012, 34(2):135-141.
doi: 10.1002/bies.201100121 pmid: 22083793 |
| [53] |
Buecker C, Wysocka J. Enhancers as information integration hubs in development: Lessons from genomics. Trends Genet, 2012, 28(6):276-284.
doi: 10.1016/j.tig.2012.02.008 |
| [54] |
Jolma A, Yin YM, Nitta KR, Dave K, Popov A, Taipale M, Enge M, Kivioja T, Morgunova E, Taipale J. DNA- dependent formation of transcription factor pairs alters their binding specificity. Nature, 2015, 527(7578):384-388.
doi: 10.1038/nature15518 |
| [55] |
Wang N, Jia ZL, Wu Q. RFX5 regulates gene expression of the Pcdhα cluster.Hereditas(Beijing), 2020, 42(8):760-774.
doi: 10.16288/j.yczz.20-184 pmid: 32952112 |
|
王娜, 甲芝莲, 吴强. RFX5调控原钙粘蛋白α基因簇的表达. 遗传, 2020, 42(8):760-774.
doi: 10.16288/j.yczz.20-184 pmid: 32952112 |
|
| [56] | Malik S, Roeder RG. The metazoan mediator co-activator complex as an integrative hub for transcriptional regulation. Nat Rev Genet, 2010, 11(11):761-772. |
| [57] |
Bolt CC, Duboule D. The regulatory landscapes of developmental genes. Development, 2020, 147(3): dev171736.
doi: 10.1242/dev.171736 |
| [58] |
Neijts R, Deschamps J. At the base of colinearHox gene expression: Cis-features and trans-factors orchestrating the initial phase of Hox cluster activation. Dev Biol, 2017, 428(2):293-299.
doi: S0012-1606(16)30864-8 pmid: 28728680 |
| [59] |
Xu DF, Ma RS, Zhang JH, Liu ZJ, Wu B, Peng JH, Zhai YN, Gong QG, Shi YY, Wu JH, Wu Q, Zhang ZY, Ruan K. Dynamic nature of CTCF tandem 11 zinc fingers in multivalent recognition of DNA as revealed by NMR spectroscopy. J Phys Chem Lett, 2018, 9(14):4020-4028.
doi: 10.1021/acs.jpclett.8b01440 |
| [1] | 安梦婷, 郭冠麟, 吴杰, 孙文靖, 贾学渊. 基于生物信息学分析胃癌双微体中增强子的调控机制[J]. 遗传, 2025, 47(5): 558-572. |
| [2] | 李轲, 周晓蓉, 朱东丽, 陈晓峰, 郭燕. 系统性红斑狼疮易感区域FAM167A-BLK遗传变异的调控机制研究[J]. 遗传, 2025, 47(11): 1244-1255. |
| [3] | 杨敏, 林思远, 杨长淇, 陈瑶生, 何祖勇. SOX9及其增强子在哺乳动物性别决定中的研究进展[J]. 遗传, 2024, 46(9): 677-689. |
| [4] | 王纪龙, 李青, 战廷正. 自转录活性调节区测序技术在增强子发现研究中的应用[J]. 遗传, 2024, 46(8): 589-602. |
| [5] | 陈秀丽, 黄海燕, 吴强. 靶向敲除β-珠蛋白基因座控制区增强子HS2对K562细胞转录组的影响[J]. 遗传, 2022, 44(9): 783-797. |
| [6] | 徐思远, 寿佳, 吴强. HS5-1增强子eRNA PEARL对原钙粘蛋白α基因簇的表达调控[J]. 遗传, 2022, 44(8): 695-764. |
| [7] | 漆思晗, 王棨临, 张俊有, 刘倩, 李春燕. 增强子调控癌症发生发展的机制研究[J]. 遗传, 2022, 44(4): 275-288. |
| [8] | 万星琦, 魏婉珍, 郭胜良, 崔一笑, 景雪莹, 黄露杰, 马捷. BMP2基因远程调控元件的功能分析[J]. 遗传, 2022, 44(12): 1141-1147. |
| [9] | 周聪, 周强伟, 成盛, 李国亮. CTCF在介导三维基因组形成及调控基因表达中的研究进展[J]. 遗传, 2021, 43(9): 816-821. |
| [10] | 何象龙, 李金环, 吴强. HOXD基因簇内一系列CTCF位点反转揭示绝缘子功能[J]. 遗传, 2021, 43(8): 758-774. |
| [11] | 刘倩, 李春燕. 增强子的鉴定及其在肿瘤研究中的应用[J]. 遗传, 2020, 42(9): 817-831. |
| [12] | 秦中勇, 石晓, 曹平平, 褚鹰, 管蔚, 杨楠, 程禾, 孙玉洁. 细胞凋亡反应中NOXA基因启动子发挥增强子功能调节BCL2基因表达[J]. 遗传, 2020, 42(11): 1110-1121. |
| [13] | 吴志强, 米泽云. 超级增强子在肿瘤研究中的进展[J]. 遗传, 2019, 41(1): 41-51. |
| [14] | 李俊涛,赵薇,李丹丹,冯静,巴贵,宋天增,张红平. miR-101a靶向EZH2促进山羊骨骼肌卫星细胞的分化[J]. 遗传, 2017, 39(9): 828-836. |
| [15] | 程霄,杨琼,谭镇东,谭娅,蒲红州,赵雪,张顺华,朱砺. 增强子RNA研究现状[J]. 遗传, 2017, 39(9): 784-797. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||

www.chinagene.cn
备案号:京ICP备09063187号-4
总访问:,今日访问:,当前在线:
