Hereditas(Beijing) ›› 2021, Vol. 43 ›› Issue (2): 160-168.doi: 10.16288/j.yczz.20-319
• Research Article • Previous Articles Next Articles
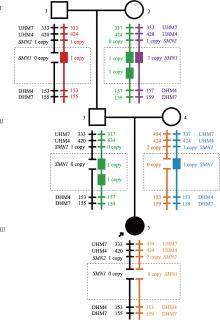
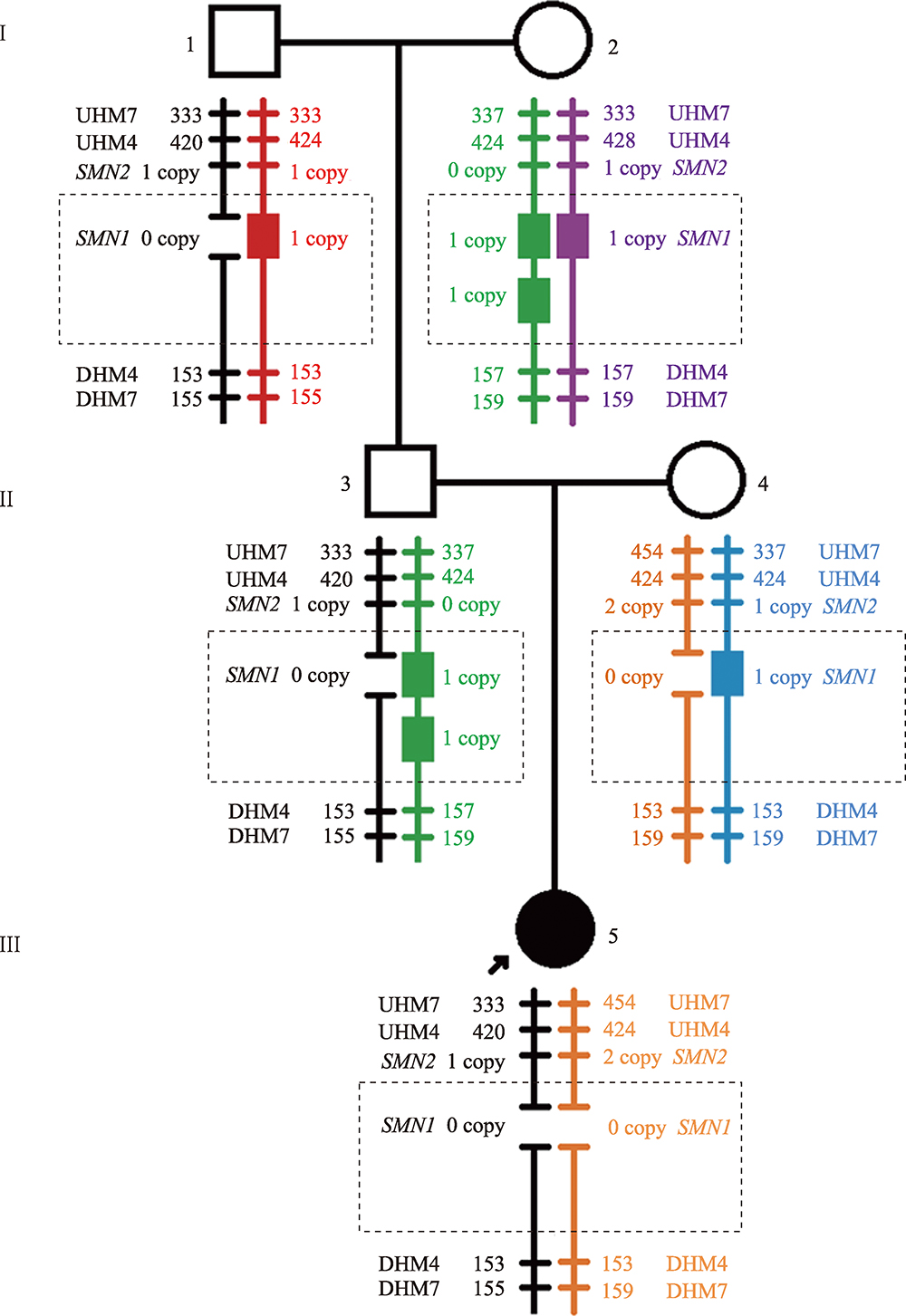
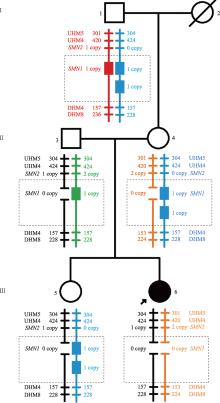
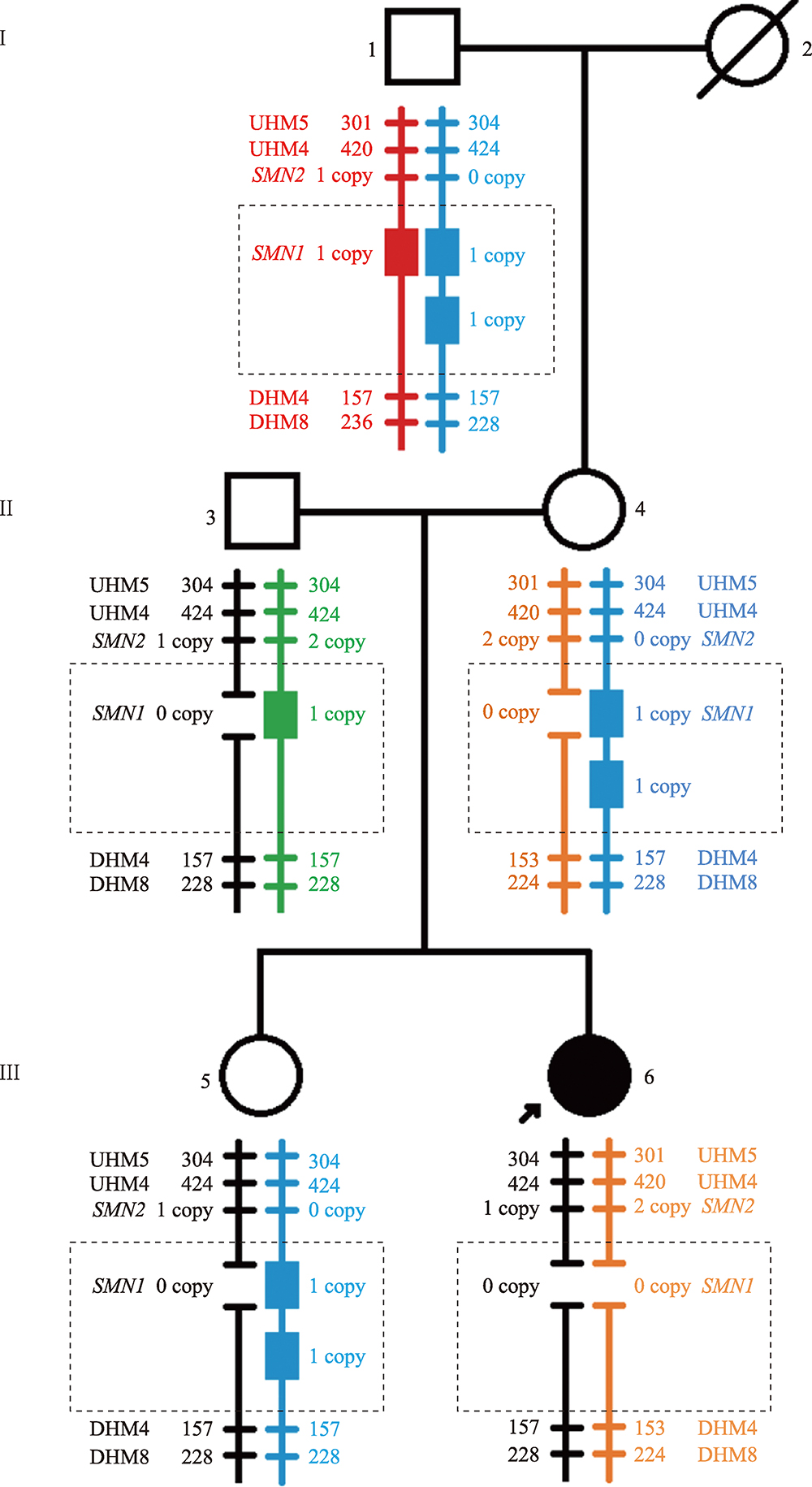
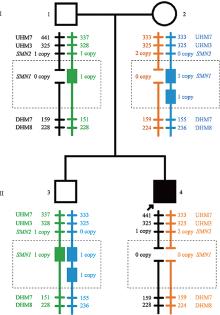
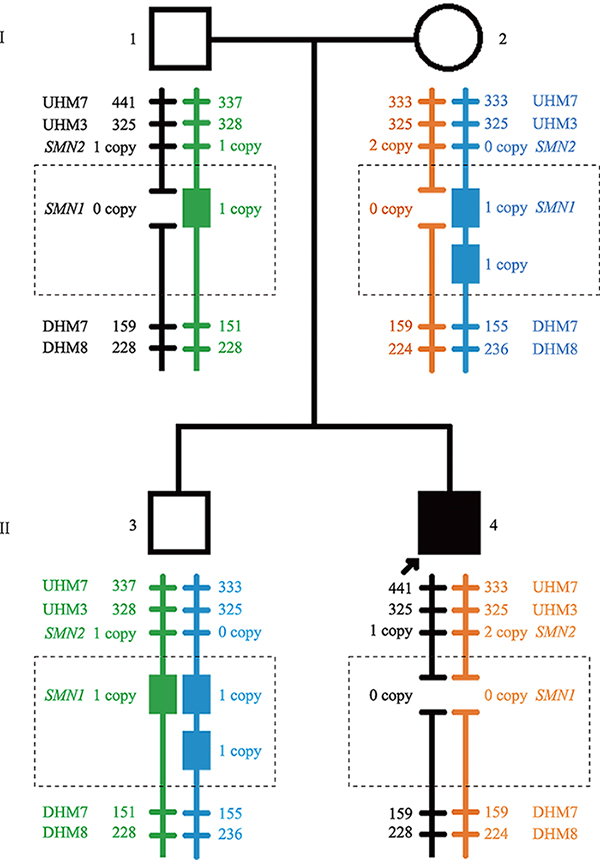
Familial study of spinal muscular atrophy carriers with SMN1 (2+0) genotype
Yanyan Cao1( ), Miaomiao Cheng1, Fang Song1(
), Miaomiao Cheng1, Fang Song1( ), Yujin Qu1, Jinli Bai1, Hong Wang1
), Yujin Qu1, Jinli Bai1, Hong Wang1
- 1 Department of Medical Genetics, Capital Institute of Pediatrics, Beijing 100020, China
-
Received:2020-10-17Online:2021-02-16Published:2020-12-24 -
Supported by:the National Key Research and Development Program of China(2016YFC0901505);CAMS Initiative for Innovative Medicine(2016-I2M-1-008);the China Postdoctoral Science Foundation(2018M630108);National Natural Science Foundation of China(81500979);Beijing Natural Science Foundation(5163028)
Cite this article
Yanyan Cao, Miaomiao Cheng, Fang Song, Yujin Qu, Jinli Bai, Hong Wang.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
| [1] |
Zhu SY, Xiong F, Chen YJ, Yan TZ, Zeng J, Li L, Zhang YN, Chen WQ, Bao XH, Zhang C, Xu XM. Molecularcharacterization of SMN copy number derived fromcarrier screening and from core families with SMA in a Chinesepopulation. Eur J Hum Genet , 2010, 18: 978-984.
doi: 10.1038/ejhg.2010.54 pmid: 20442745 |
| [2] | Luo MJ, Liu L, Peter I, Zhu J, Scott SA, Zhao GP, Eversley C, Kornreich R, Desnick RJ, Edelmann L. An Ashkenazi Jewish SMN1 haplotype specific to duplication alleles improves pan-ethnic carrier screening for spinal muscular atrophy. Genet Med , 2014, 16(2): 149-156. |
| [3] | Verhaart IEC, Robertson A, Wilson IJ, Aartsma-Rus A, Cameron S, Jones CC, Cook SF, Lochmüller H. Prevalence, incidence and carrier frequency of 5q-linked spinal muscular atrophy - a literature review. Orphanet J Rare Dis , 2017, 12(1): 124. |
| [4] | McAndrew PE, Parsons DW, Simard LR, Rochette C, Ray PN, Mendell JR, Prior TW, Burghes AH. Identification ofproximal spinal muscular atrophy carriers and patients byanalysis of SMNT and SMNC gene copy number. Am J HumGenet , 1997, 60(6): 1411-1422. |
| [5] |
Chen KL, Wang YL, Rennert H, Joshi I, Mills JK, Leonard DG, Wilson RB. Duplications and de novo deletions of the SMNt gene demonstrated by fluorescence-based carrier testingfor spinal muscular atrophy. Am J Med Genet , 1999, 85(5): 463-469.
pmid: 10405443 |
| [6] | Yan H, Papadopoulos N, Marra G, Perrera C, Jiricny J, Boland CR, Lynch HT, Chadwick RB, de la Chapelle A, Berg K, Eshleman JR, Yuan W, Markowitz S, LakenSJ, Lengauer C, Kinzler KW, Vogelstein B. Conversion of diploidy to haploidy. Nature , 2000, 403(6771): 723-724. |
| [7] |
Mailman MD, Hemingway T, Darsey RL, Glasure CE, Huang Y, Chadwick RB, Heinz JW, Papp AC, Snyder PJ, Sedra MS, Schafer RW, Abuelo DN, Reich EW, Theil KS, Burghes AH, de la Chapelle A, Prior TW. Hybrids monosomal for human chromosome 5 reveal the presence of a spinal muscular atrophy (SMA) carrier with two SMN1 copies on one chromosome. Hum Genet , 2001, 108(2): 109-115.
pmid: 11281448 |
| [8] |
Burlet P, Gigarel N, Magen M, Drunat S, Benachi A, Hesters L, Munnich A, Bonnefont JP, Steffann J. Single- sperm analysis for recurrence risk assessment of spinal muscular atrophy. Eur J Hum Genet , 2010, 18(4): 505-508.
doi: 10.1038/ejhg.2009.198 pmid: 19904299 |
| [9] |
Cao YY, Qu YJ, Bai JL, Cheng MM, Jin YW, Wang H, Song F. Transmission characteristics of SMN from 227 spinal muscular atrophy core families in China. J Hum Genet , 2020, 65(5): 469-473. To define the relationship between the survival motor neuron 1 gene (SMN1) and SMN2, and explore the variability of these two genes within the generations, SMN1 and SMN2 copy numbers were determined for 227 SMA families. The association analysis indicated that there was a negative correlation between the copy number of SMN1 and SMN2 (Spearman = -0.472, P < 0.001) in 227 SMA children and 454 of their parents. The average SMN copies from father and mother in each SMA family were used to represent the copy number in the parent's generation. Subsequently, SMN transmission analysis showed that the similar distribution trend of SMN1 and SMN2 copy number was not only in the SMA children and their parents' generation but also in the non-SMA families. Moreover, when the SMN2 copy number was one in the parent's generation, 75% of their SMA children had type I and 25% of them had type II/III. However, when the SMN2 copies were three in the parent's generation, all of their SMA children were type II/III. Therefore, the diversity of SMN copies was mostly inherited and the SMN2 copy number in the parent's generation could predict the disease severity of SMA children to some extent.
doi: 10.1038/s10038-020-0730-1 pmid: 32051521 |
| [10] |
Chen X, Sanchis-Juan A, French CE, Connell AJ, Delon I, Kingsbury Z, Chawla A, Halpern AL, Taft RJ, BioResource N, Bentley DR, Butchbach MER, Raymond FL, Eberle MA. Spinal muscular atrophy diagnosis and carrier screening from genome sequencing data. Genet Med , 2020, 22(5): 945-953.
pmid: 32066871 |
| [11] |
Zhao SM, Wang WY, Wang YS, Han R, Fan CN, Ni PX, Guo FY, Zeng FW, Yang QN, Yang Y, Sun Y, Zhang XH, Chen Y, Zhu BS, Cai WW, Chen S, Cai R, Guo XL, Zhang CL, Zhou YQ, Huang SD, Liu YH, Chen BY, Yan SH, Chen YJ, Ding HM, Shang X, Xu XM, Sun J, Peng ZY. NGS-based spinal muscular atrophy carrier screening of 10,585diverse couples in China: a pan-ethnic study. Eur J Hum Genet , 2020, doi: 10.1038/s41431-020-00714-8.
pmid: 33446828 |
| [12] | Alías L, Bernal S, Calucho M, Martínez E, March F, Gallano P, Fuentes-Prior P, Abuli A, Serra-Juhe C, Tizzano EF. Utility of two SMN1 variants to improve spinal muscular atrophy carrier diagnosis and genetic counselling. Eur J Hum Genet , 2018, 26(10): 1554-1557. |
| [1] | Yerong Li, Juan Lv, Yuguo Wang, Jianxin Tan, Binbin Shao, Jingjing Zhang. Application of multiplex competitive PCR combined with capillary electrophoresis in carrier screening of spinal muscular atrophy [J]. Hereditas(Beijing), 2022, 44(7): 618-628. |
| [2] | Xueqian Wang, Qingzhen Zhang, Peng Cheng, Tingting Dong, Weiguo Li, Zhe Zhou, Shengqi Wang. Genetic polymorphisms of 66 InDel loci in the Chinese Han population [J]. Hereditas(Beijing), 2022, 44(4): 335-345. |
| [3] | Haoyu Wang, Yuhan Hu, Yueyan Cao, Qiang Zhu, Yuguo Huang, Xi Li, Ji Zhang. AI-SNPs screening based on the whole genome data and research on genetic structure differences of subcontinent populations [J]. Hereditas(Beijing), 2021, 43(10): 938-948. |
| [4] | Yuzhuo Wang, Yiming Zhang, Xiaolian Dong, Xuecai Wang, Jianfu Zhu, Na Wang, Feng Jiang, Yue Chen, Qingwu Jiang, Chaowei Fu. Modification effects of T2DM-susceptible SNPs on the reduction of blood glucose in response to lifestyle interventions [J]. Hereditas(Beijing), 2020, 42(5): 483-492. |
| [5] | Qing Zhang,Jie Ping,Haoxiang Zhang,Bo Kang, ,Gangqiao Zhou. Genetic association of MKL1 gene polymorphisms with the high-altitude adaptation [J]. Hereditas(Beijing), 2019, 41(7): 634-643. |
| [6] | Jieyu Zhang,Yanan Chu,Bingjie Zou,Yanjie Zhang,Liying Feng. Detection of UGT1A1*6 single nucleotide polymorphism in oral swap samples using the cascade invader assay-based real-time PCR [J]. Hereditas(Beijing), 2018, 40(8): 668-675. |
| [7] | Zhe-ye Peng,Zi-jun Tang,Min-zhu Xie. Research progress in machine learning methods for gene-gene interaction detection [J]. Hereditas(Beijing), 2018, 40(3): 218-226. |
| [8] | Lili Liu, Aiwei Guo, Peifu Wu, Fenfen Chen, Yajin Yang, Qin Zhang. Regulation of VPS28 gene knockdown on the milk fat synthesis in Chinese Holstein dairy [J]. Hereditas(Beijing), 2018, 40(12): 1092-1100. |
| [9] | Wang Shiming, Song Xiao, Zhao Xueying, Chen Hongyan, Wang Jiucun, Wu Junjie, Gao Zhiqiang, Qian Ji, Bai Chunxue, Li Qiang, Han Baohui, Lu Daru. Association between polymorphisms of autophagy pathway and responses in non-small cell lung cancer patients treated with platinum-based chemotherapy [J]. Hereditas(Beijing), 2017, 39(3): 250-262. |
| [10] | Bengang Wang, Zhi Lv, Qian Xu, Yongfeng Liu. Interactions among polymorphisms of NER genes prompt the risk of transplantation rejection [J]. Hereditas(Beijing), 2017, 39(1): 22-31. |
| [11] | Mei Fu, Kehui Xu, Wenming Xu. Research advances of Dicer in regulating reproductive function [J]. HEREDITAS(Beijing), 2016, 38(7): 612-622. |
| [12] | Xian Gong, Chao Zhang, Aisa Yiliyasi, Ying Shi, Xuewei Yang, Aosiman Nuersimanguli, Yaqun Guan, Shuhua Xu. A comparative analysis of genetic diversity of candidate genes associated with type 2 diabetes in worldwide populations [J]. Hereditas(Beijing), 2016, 38(6): 543-559. |
| [13] | Zhen Liu, Jianhong Xu. The application of the high throughput sequencing technology in the transposable elements [J]. HEREDITAS(Beijing), 2015, 37(9): 885-898. |
| [14] | Dayan Niu, Weili Yan. The application of genetic risk score in genetic studies of complex human diseases [J]. HEREDITAS(Beijing), 2015, 37(12): 1204-1210. |
| [15] | Qin Chu, Dong Li, Shiyu Hou, Wanhai Shi, Lin Liu, Yachun Wang. Direct sequencing of DNA pooling for screening highly informative SNPs in dairy cattle [J]. HEREDITAS(Beijing), 2014, 36(7): 691-696. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||