Hereditas(Beijing) ›› 2021, Vol. 43 ›› Issue (10): 938-948.doi: 10.16288/j.yczz.21-185
• Research Article • Previous Articles Next Articles
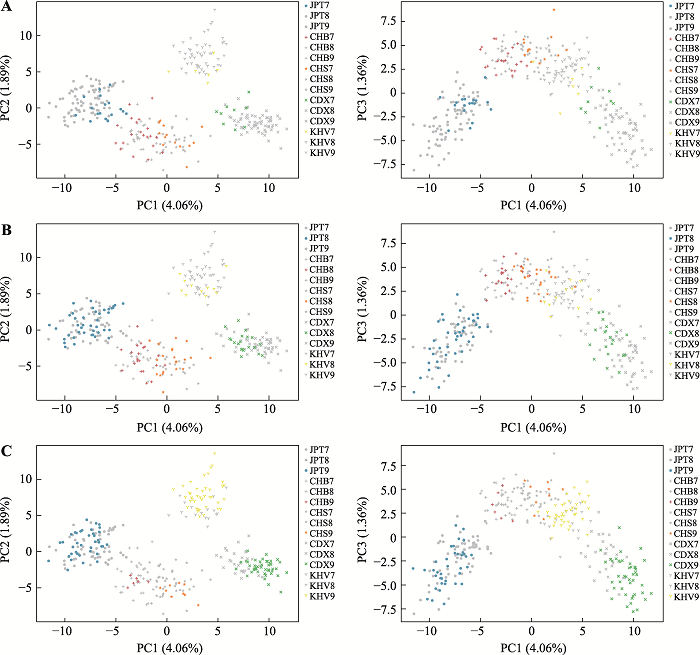
AI-SNPs screening based on the whole genome data and research on genetic structure differences of subcontinent populations
Haoyu Wang( ), Yuhan Hu(
), Yuhan Hu( ), Yueyan Cao, Qiang Zhu, Yuguo Huang, Xi Li, Ji Zhang
), Yueyan Cao, Qiang Zhu, Yuguo Huang, Xi Li, Ji Zhang
- West China School of Basic Medical Sciences and Forensic Medicine, Sichuan University, Chengdu 610041, China
-
Received:2021-05-26Revised:2021-07-23Online:2021-10-20Published:2021-08-04 -
Supported by:Supported by the National Natural Science Foundation of China Nos(81571861);Supported by the National Natural Science Foundation of China Nos(81630054)
Cite this article
Haoyu Wang, Yuhan Hu, Yueyan Cao, Qiang Zhu, Yuguo Huang, Xi Li, Ji Zhang. AI-SNPs screening based on the whole genome data and research on genetic structure differences of subcontinent populations[J]. Hereditas(Beijing), 2021, 43(10): 938-948.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
Table 1
The distribution of FST value of SNPs in dataset A"
| 数据集 | 最大FST值 | 最小FST值 | 总位点数 | FST≥0.25位点数 | 0.25>FST≥0.15位点数 | 0.15>FST≥0.05位点数 |
|---|---|---|---|---|---|---|
| A1(JPT-CHB) | 0.266925 | 0.095475 | 591 | 1 | 19 | 571 |
| A2(JPT-CHS) | 0.407479 | 0.111107 | 598 | 10 | 87 | 501 |
| A3(JPT-CDX) | 0.788409 | 0.16243 | 630 | 46 | 584 | 0 |
| A4(JPT-KHV) | 0.583637 | 0.141417 | 623 | 19 | 435 | 169 |
| A5(CHB-CHS) | 0.146048 | 0.05001 | 723 | 0 | 0 | 723 |
| A6(CHB-CDX) | 0.517659 | 0.109475 | 563 | 9 | 84 | 470 |
| A7(CHB-KHV) | 0.25611 | 0.087147 | 670 | 1 | 18 | 651 |
| A8(CHS-CDX) | 0.310909 | 0.081595 | 787 | 3 | 18 | 766 |
| A9(CHS-KHV) | 0.192399 | 0.069052 | 631 | 0 | 7 | 624 |
| A10(CDX-KHV) | 0.256551 | 0.067479 | 461 | 1 | 6 | 454 |
| [1] | Hellwege JN, Keaton JM, Giri A, Gao XY, Velez Edwards DR, Edwards TL. Population stratification in genetic association studies. Curr Protoc Hum Genet, 2017, 95: 1.22.1-1.22.23. |
| [2] | Schlebusch CM, Skoglund P, Sjödin P, Gattepaille LM, Hernandez D, Jay F, Li S, De Jongh M, Singleton A, Blum MG, Soodyall H, Jakobsson M . Genomic variation in seven Khoe-San groups reveals adaptation and complex African History. Science, 2012,338(6105):374-379. |
| [3] | Price AL, Zaitlen NA, Reich D, Patterson N . New approaches to population stratification in genome-wide association studies. Nat Rev Genet, 2010,11(7):459-463. |
| [4] | Gong X, Zhang C, Yiliyasi A, Shi Y, Yang XW, Nuersimanguli A, Guan YQ, Xu SH. A comparative analysis of genetic diversity of candidate genes associated with type 2 diabetes in worldwide populations. Hereditas (Beijing), 2016,38(6):544-565. |
| 弓弦, 张超, 伊利亚斯·艾萨, 时瑛, 杨雪唯, 努尔斯曼古丽·奥斯曼, 关亚群, 徐书华. 2型糖尿病易感候选基因在世界不同人群中的多样性比较分析. 遗传, 2016,38(6):544-565. | |
| [5] | Dai R, Zhang C, Cheng YJ, Chen WL, Li Q, Wang YM. Pharmacogenomics genetic differences between Wa and Blang ethnic groups in Yunnan. J Kunming Med Univ, 2020,41(5):33-40. |
| 代润, 张婵, 程瑜静, 陈婉璐, 李琦, 王玉明. 云南佤族和布朗族人群药物基因组学基因遗传差异. 昆明医科大学学报, 2020,41(5):33-40. | |
| [6] | Phillips C, Prieto L, Fondevila M, Salas A, Gómez-Tato A, Alvarez-Dios J, Alonso A, Blanco-Verea A, Brión M, Montesino M, Carracedo A, Lareu MV . Ancestry analysis in the 11-M Madrid bomb attack investigation. PLoS One, 2009,4(8):e6583. |
| [7] | Tian C, Plenge RM, Ransom M, Lee A, Villoslada P, Selmi C, Klareskog L, Pulver AE, Qi LH, Gregersen PK, Seldin MF . Analysis and application of European genetic substructure using 300 K SNP information. PLoS Genet, 2008,4(1):e4. |
| [8] | Enoch MA, Shen PH, Xu K, Hodgkinson C, Goldman D . Using ancestry-informative markers to define populations and detect population stratification. J Psychopharmacol, 2006,20(4):19-26. |
| [9] | Pritchard JK, Stephens M, Rosenberg NA, Donnelly P . Association mapping in structured populations. Am J Hum Genet, 2000,67(1):170-181. |
| [10] | Clarke L, Fairley S, Zheng-Bradley X, Streeter I, Perry E, Lowy E, Tassé AM, Flicek P . The international genome sample resource (IGSR): a worldwide collection of genome variation incorporating the 1000 genomes project data. Nucleic Acids Res, 2017,45(D1):D854-D859. |
| [11] | 1000 Genomes Project Consortium, Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, Korbel JO, Marchini JL, McCarthy S, McVean GA, Abecasis GR . A global reference for human genetic variation. Nature, 2015,526(7571):68-74. |
| [12] | Qin PF, Li ZQ, Jin WF, Lu DS, Lou HY, Shen JW, Jin L, Shi YY, Xu SH . A panel of ancestry informative markers to estimate and correct potential effects of population stratification in Han Chinese. Eur J Hum Genet, 2014,22(2):248-253 |
| [13] | Severe Covid-19 GWAS Group, Ellinghaus D, Degenhardt F, Bujanda L, Buti M, Albillos A, Invernizzi P, Fernández J, Prati D, Baselli G, Asselta R, Grimsrud MM, Milani C, Aziz F, Kässens J, May S, Wendorff M, Wienbrandt L, Uellendahl-Werth F, Zheng TH, Yi XL, de Pablo R, Chercoles AG, Palom A, Garcia-Fernandez AE, Rodriguez- Frias F, Zanella A, Bandera A, Protti A, Aghemo A, Lleo A, Biondi A, Caballero-Garralda A, Gori A, Tanck A, Carreras Nolla A, Latiano A, Fracanzani AL, Peschuck A, Julià A, Pesenti A, Voza A, Jiménez D, Mateos B, Nafria Jimenez B, Quereda C, Paccapelo C, Gassner C, Angelini C, Cea C, Solier A, Pestaña D, Muñiz-Diaz E, Sandoval E, Paraboschi EM, Navas E, García Sánchez F, Ceriotti F, Martinelli-Boneschi F, Peyvandi F, Blasi F, Téllez L, Blanco-Grau A, Hemmrich-Stanisak G, Grasselli G, Costantino G, Cardamone G, Foti G, Aneli S, Kurihara H, ElAbd H, My I, Galván-Femenia I, Martín J, Erdmann J, Ferrusquía-Acosta J, Garcia-Etxebarria K, Izquierdo- Sanchez L, Bettini LR, Sumoy L, Terranova L, Moreira L, Santoro L, Scudeller L, Mesonero F, Roade L, Rühlemann MC, Schaefer M, Carrabba M, Riveiro-Barciela M, Figuera Basso ME, Valsecchi MG, Hernandez-Tejero M, Acosta-Herrera M, D'Angiò M, Baldini M, Cazzaniga M, Schulzky M, Cecconi M, Wittig M, Ciccarelli M, Rodríguez-Gandía M, Bocciolone M, Miozzo M, Montano N, Braun N, Sacchi N, Martínez N, Özer O, Palmieri O, Faverio P, Preatoni P, Bonfanti P, Omodei P, Tentorio P, Castro P, Rodrigues PM, Blandino Ortiz A, de Cid R, Ferrer R, Gualtierotti R, Nieto R, Goerg S, Badalamenti S, Marsal S, Matullo G, Pelusi S, Juzenas S, Aliberti S, Monzani V, Moreno V, Wesse T, Lenz TL, Pumarola T, Rimoldi V, Bosari S, Albrecht W, Peter W, Romero-Gómez M, D'Amato M, Duga S, Banales JM, Hov JR, Folseraas T, Valenti L, Franke A, Karlsen TH . Genomewide association study of Severe Covid-19 with respiratory failure. N Engl J Med, 2020,383(16):1522-1534. |
| [14] | Foo JN, Tan LC, Irwan ID, Au WL, Low HQ, Prakash KM, Ahmad-Annuar A, Bei JX, Chan AY, Chen CM, Chen YC, Chung SJ, Deng H, Lim SY, Mok V, Pang H, Pei Z, Peng R, Shang HF, Song K, Tan AH, Wu YR, Aung T, Cheng CY, Chew FT, Chew SH, Chong SA, Ebstein RP, Lee J, Saw SM, Seow A, Subramaniam M, Tai ES, Vithana EN, Wong TY, Heng KK, Meah WY, Khor CC, Liu H, Zhang F, Liu J, Tan EK . Genome-wide association study of Parkinson's disease in East Asians. Hum Mol Genet, 2017,26(1):226-232. |
| [15] | Setakis E, Stirnadel H, Balding DJ . Logistic regression protects against population structure in genetic association studies. Genome Res, 2006,16(2):290-296. |
| [16] | Gaspar HA, Breen G . Probabilistic ancestry maps: a method to assess and visualize population substructures in genetics. BMC Bioinformatics, 2019,20(1):116. |
| [17] | Pritchard JK, Stephens M, Donnelly P . Inference of population structure using multilocus genotype data. Genetics, 2000,155(2):945-959. |
| [18] | Alexander DH, Novembre J, Lange K . Fast model-based estimation of ancestry in unrelated individuals. Genome Res, 2009,19(9):1655-1664. |
| [19] | Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D . Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet, 2006,38(8):904-909. |
| [20] | Phillips C, Salas A, Sánchez JJ, Fondevila M, Gómez-Tato A, Alvarez-Dios J, Calaza M, de Cal MC, Ballard D, Lareu MV, Carracedo A; SNPforID Consortium. Inferring ancestral origin using a single multiplex assay of ancestry-informative marker SNPs. Forensic Sci Int Genet, 2007,1(3-4):273-80. |
| [21] | Li CX, Pakstis AJ, Jiang L, Wei YL, Sun QF, Wu H, Bulbul O, Wang P, Kang LL, Kidd JR, Kidd KK . A panel of 74 AISNPs: improved ancestry inference within Eastern Asia. Forensic Sci Int Genet, 2016,23:101-110. |
| [22] | Liu j, Liu CC, Ma M, Wang L, Zhao WT, Ma Q, Ji AQ, Liu J, Li CX. The ancestry inference of Chinese populations using 74-plex SNPs system. Hereditas (Beijing), 2020,42(3):296-308. |
| 刘杨, 孙昌春, 马咪, 王玲, 赵雯婷, 马泉, 季安全, 刘京, 李彩霞. 74-plex SNPs复合检测体系在中国人群中的族群推断研究. 遗传, 2020,42(3):296-308. | |
| [23] | Qu SQ, Zhu J, Wang YJ, Yin L, Lv ML, Wang L, Jian H, Tan Y, Zhang RR, Liu YQ, Li F, Huang SC, Liang WB, Zhang L . Establishing a second-tier panel of 18 ancestry informative markers to improve ancestry distinctions among Asian populations. Forensic Sci Int Genet, 2019,41:159-167. |
| [24] | Bulbul O, Speed WC, Gurkan C, Soundararajan U, Rajeevan H, Pakstis AJ, Kidd KK . Improving ancestry distinctions among Southwest Asian populations. Forensic Sci Int Genet, 2018,35:14-20. |
| [25] | Shi CM, Liu Q, Zhao SL, Chen H . Ancestry informative SNP panels for discriminating the major East Asian populations: Han Chinese, Japanese and Korean. Ann Hum Genet, 2019,83(5):348-354 |
| [26] | Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, Handsaker RE, Lunter G, Marth GT, Sherry ST, McVean G, Durbin R; 1000 Genomes Project Analysis Group. The variant call format and VCFtools. Bioinformatics, 2011,27(15):2156-2158. |
| [27] | Weir BS, Cockerham CC . Estimating F-statistics for the analysis of population structure. Evolution, 1984,38(6):1358-1370. |
| [28] | Falush D, Stephens M, Pritchard JK . Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics. 2003,164(4):1567-87. |
| [29] | Earl DA, vonHoldt BM . Structure Harvester: a website and program for visualizing structure output and implementing the Evanno method. Conserv Genet Resour, 2012,4(2):359-361. |
| [30] | Jakobsson M, Rosenberg NA . Clumpp: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics, 2007,23(14):1801-1806. |
| [31] | Rosenberg NA . Distructd: a program for the graphical display of population structure. Mol Ecol Notes, 2004,4(1):137-138. |
| [32] | Zhou CX, Li M, Huai C, He L, Qin SY. Study on hereditary susceptibility genetic markers to anti-tuberculosis drug induced liver injury in Chinese population. Hereditas (Beijing), 2020,42(4):374-379. |
| 周晨希, 李沫, 怀聪, 贺林, 秦胜营. 中国人群中抗结核药物引发肝损伤的易感基因标记研究. 遗传, 2020,42(4):374-379. | |
| [33] | Sun YD, Tian ZZ, Zhou W, Li M, Huai C, He L, Qin SY. Genome-wide association study on liver function tests in Chinese. Hereditas(Beijing), 2021,43(3):249-260. |
| 孙一丹, 田子钊, 周伟, 李沫, 怀聪, 贺林, 秦胜营. 中国人群肝功能检测指标全基因组关联分析研究. 遗传, 2021,43(3):249-260. | |
| [34] | Wright S . The genetical structure of populations. Nature, 1951,15(4):323-354. |
| [35] | Holsinger KE, Weir BS . Genetics in geographically structured populations: defining, estimating and interpreting F(ST). Nat Rev Genet, 2009,10(9):639-650. |
| [36] | Santos C, Phillips C, Gomez-Tato A, Alvarez-Dios J, Carracedo Á, Lareu MV . Inference of ancestry in forensic analysis II: analysis of genetic data. Methods Mol Biol. 2016,1420:255-285. |
| [1] | HUAN Pin, ZHANG Xiao-Jun, LI Fu-Hua, ZHANG Xiang, DIAO Cui, LIU Bao-Zhong, XIANG Jian-Hai. Chromosomal localization and development of SNP markers of a serine protease gene in Farrer’s scallop (Chlamys farreri) [J]. HEREDITAS, 2009, 31(12): 1241-1247. |
| [2] | LAN Xian-Yong, CHEN Hong, TIAN Zhi-Quan, LIU Shao-Qing, ZHANG Yong-Bin, WANG Xin, FANG Xing-Tang . Correlations between SNP of LALBA gene and economic traits in Inner Mongolian white cashmere goat [J]. HEREDITAS, 2008, 30(2): 169-174. |
| [3] | ZHANG Lai-Jun, ZHENG Zi-Jian, ZHANG Ke-Jin, GAO Xiao-Cai, CHEN Chao, HUANG Shao-Ping, ZHANG Fu-Chang. Anassociation study between OPHN1 gene rs492933 polymorphism and mental retardation in children of the Qinba Mountain region [J]. HEREDITAS, 2008, 30(10): 1307-1311. |
| [4] | ZHAI Guang-Hua, WEN Ping, GUO Lan-Fang, CHEN Lu. Association of apolipoprotein A5 gene -1131T/C polymorphism with lipid metabolism and insulin resistance in patients with type Ⅱ dia-betes mellitus [J]. HEREDITAS, 2007, 29(5): 541-541―546. |
| [5] | LIU Min, LING Si-Hai, LI Wen-Biao, WANG Chuan-Yue, CHEN Da-Fang, WANG Gang. An association study between grin1, bdnf genes and bipolar disorder [J]. HEREDITAS, 2007, 29(1): 41-41―46. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||