Hereditas(Beijing) ›› 2021, Vol. 43 ›› Issue (9): 880-889.doi: 10.16288/j.yczz.21-073
• Research Article • Previous Articles Next Articles
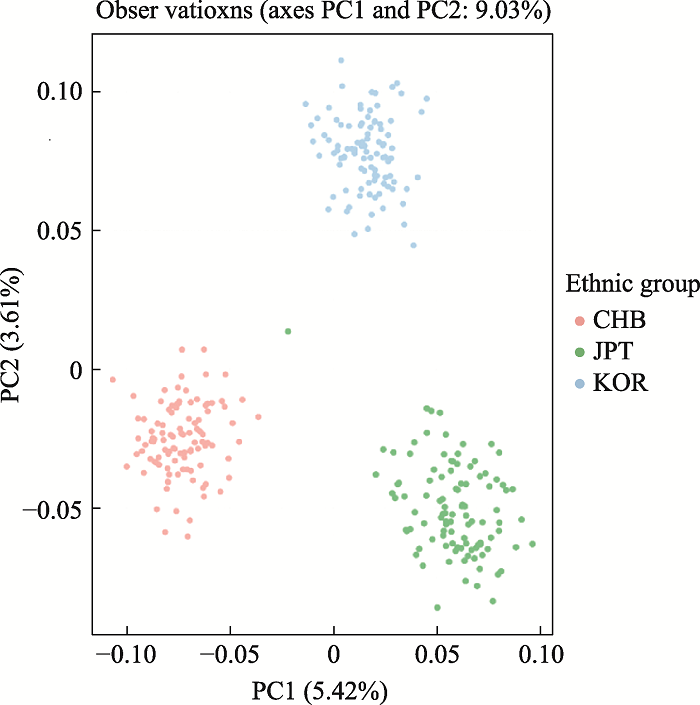
49AISNP: a study on the ancestry inference of the three ethnic groups in the north of East Asia
Xiaoyuan Guo1,2( ), Changchun Sun1,2, Siyao Xue1,2, Hui Zhao2, Li Jiang2(
), Changchun Sun1,2, Siyao Xue1,2, Hui Zhao2, Li Jiang2( ), Caixia Li1,2(
), Caixia Li1,2( )
)
- 1. Shanxi Medical University, Taiyuan 030001, China
2. Key Laboratory of Forensic Genetics, Beijing Engineering Research Center of Crime Scene Evidence Examination, National Engineering Laboratory for Forensic Science, Institute of Forensic Science, Beijing 100038, China
-
Received:2021-02-25Revised:2021-05-31Online:2021-09-20Published:2021-07-01 -
Contact:Jiang Li,Li Caixia E-mail:827633812@qq.com;jl@mail.bnu.edu.cn;licaixia@tsinghua.org.cn -
Supported by:Supported by the National Natural Science Foundation of China No(81772027)
Cite this article
Xiaoyuan Guo, Changchun Sun, Siyao Xue, Hui Zhao, Li Jiang, Caixia Li. 49AISNP: a study on the ancestry inference of the three ethnic groups in the north of East Asia[J]. Hereditas(Beijing), 2021, 43(9): 880-889.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
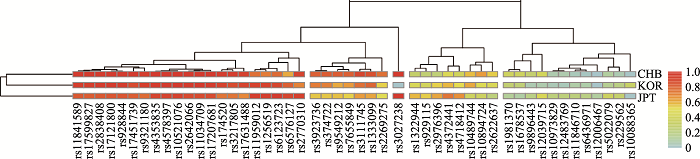
Table 2
Information of the 49AISNP"
| 序号 | rs号 | 染色体 | 第37版序列位置 | 等位基因 | Fst | 序号 | rs号 | 染色体 | 第37版序列位置 | 等位基因 | Fst |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | rs10088365 | 8 | 10097398 | G/A | 0.2709 | 26 | rs2622637 | 8 | 106505971 | G/A/T | 0.0951 |
| 2 | rs10489744 | 1 | 165380623 | G/A | 0.0690 | 27 | rs2642066 | 17 | 65639014 | G/T | 0.0910 |
| 3 | rs10521076 | 9 | 108878562 | C/T | 0.0775 | 28 | rs2838408 | 21 | 45246422 | A/G | 0.0750 |
| 4 | rs10894724 | 11 | 133547942 | T/A/G | 0.0812 | 29 | rs2976396 | 8 | 143764001 | G/A | 0.1521 |
| 5 | rs10973829 | 9 | 38446392 | C/T | 0.0781 | 30 | rs3111745 | 3 | 21827159 | G/T | 0.0751 |
| 6 | rs11034709 | 11 | 38428289 | A/G | 0.1083 | 31 | rs3217805 | 12 | 4388084 | C/A/G/T | 0.0844 |
| 7 | rs11841589 | 13 | 73814891 | G/T | 0.0862 | 32 | rs374722 | 2 | 147839830 | G/A | 0.0740 |
| 8 | rs11846710 | 14 | 58343352 | G/A | 0.0768 | 33 | rs3923736 | 7 | 155060730 | A/C/G/T | 0.0714 |
| 9 | rs11959012 | 5 | 167668843 | C/T | 0.0694 | 34 | rs4353835 | 3 | 32446775 | C/T | 0.0849 |
| 10 | rs12006467 | 9 | 35090720 | T/C | 0.0993 | 35 | rs4372441 | 11 | 7657259 | C/T | 0.0787 |
| 11 | rs12039715 | 1 | 242801261 | G/C | 0.1726 | 36 | rs4718412 | 7 | 66286867 | T/C | 0.0751 |
| 12 | rs12483769 | 22 | 47790072 | T/C | 0.0870 | 37 | rs5022079 | 18 | 76478188 | A/G | 0.0931 |
| 13 | rs1256519 | 14 | 65736324 | G/A | 0.1027 | 38 | rs6123723 | 20 | 37082145 | C/T | 0.0891 |
| 14 | rs1322944 | 13 | 53683163 | A/G | 0.0740 | 39 | rs6436971 | 2 | 231582797 | T/C | 0.0815 |
| 15 | rs1333099 | 13 | 73691236 | G/A | 0.0715 | 40 | rs6576127 | 14 | 106194763 | C/T | 0.1076 |
| 16 | rs1678537 | 12 | 57900341 | G/A | 0.0885 | 41 | rs7655849 | 4 | 161867415 | T/A | 0.0759 |
| 17 | rs17121800 | 10 | 108710873 | C/T | 0.1007 | 42 | rs928844 | 21 | 37999799 | C/T | 0.0846 |
| 18 | rs17207681 | 5 | 138168527 | A/G | 0.0823 | 43 | rs929115 | 1 | 171591189 | T/G | 0.0757 |
| 19 | rs17451739 | 5 | 144030993 | C/T | 0.0732 | 44 | rs9321180 | 6 | 130102959 | C/T | 0.0866 |
| 20 | rs174520 | 14 | 88006776 | T/G | 0.0899 | 45 | rs9549212 | 13 | 41022093 | C/T | 0.0708 |
| 21 | rs17599827 | 5 | 89518433 | A/C | 0.0810 | 46 | rs9896443 | 17 | 47125982 | T/C | 0.0738 |
| 22 | rs17631488 | 5 | 18777746 | A/G | 0.1030 | 47 | rs2770310 | 6 | 70165296 | C/A/T | 0.0924 |
| 23 | rs1981370 | 18 | 74070815 | A/G | 0.0722 | 48 | rs3027238 | 17 | 8135061 | T/C | 0.9803 |
| 24 | rs2269275 | 5 | 83261405 | A/G | 0.0688 | 49 | rs4578397 | 11 | 131832284 | G/T | 0.0737 |
| 25 | rs229562 | 22 | 37599065 | G/T | 0.0827 |
Supplementary Table 1
Information of the 428AISNP"
| 序号 | rs号 | 染色体 | 第37版序列位置 | 等位基因 | Fst值 | 序号 | rs号 | 染色体 | 第37版序列位置 | 等位基因 | Fst值 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | rs10018432 | 4 | 189437086 | C/A | 0.0460 | 40 | rs10957985 | 8 | 81789921 | G/A | 0.0645 |
| 2 | rs10058739 | 5 | 144074365 | G/A | 0.0297 | 41 | rs10972006 | 9 | 3430684 | T/C | 0.0266 |
| 3 | rs10088365 | 8 | 10097398 | G/A | 0.2709 | 42 | rs10973829 | 9 | 38446392 | C/T | 0.0781 |
| 4 | rs1009544 | 22 | 42239348 | G/C | 0.0430 | 43 | rs10991718 | 9 | 93716118 | T/C | 0.0048 |
| 5 | rs10107492 | 8 | 80568935 | T/A/C | 0.0503 | 44 | rs11012716 | 10 | 21762267 | A/G | 0.0302 |
| 6 | rs10110194 | 8 | 119479124 | G/C/T | 0.0585 | 45 | rs11023463 | 11 | 15209506 | A/C | 0.0052 |
| 7 | rs10132336 | 14 | 74869017 | A/G | 0.0327 | 46 | rs11032331 | 11 | 33678161 | G/T | 0.0519 |
| 8 | rs10132483 | 14 | 102299420 | T/C | 0.0519 | 47 | rs11034709 | 11 | 38428289 | A/G | 0.1083 |
| 9 | rs10134903 | 14 | 95229777 | G/A | 0.0360 | 48 | rs11042911 | 11 | 10678785 | T/C | 0.0432 |
| 10 | rs10139575 | 14 | 57087118 | A/C/G | 0.0342 | 49 | rs11058961 | 12 | 127544998 | G/A | 0.0438 |
| 11 | rs10166397 | 2 | 44372909 | G/T | 0.0490 | 50 | rs11086012 | 19 | 16032643 | A/C | 0.0537 |
| 12 | rs10171891 | 2 | 195971878 | C/T | 0.0431 | 51 | rs11104947 | 12 | 88942980 | G/A | 0.0782 |
| 13 | rs10177273 | 2 | 122239287 | C/G | 0.0782 | 52 | rs1111212 | 2 | 159312733 | C/G/T | -0.0007 |
| 14 | rs1026975 | 4 | 113971374 | T/G | 0.0229 | 53 | rs11128125 | 3 | 69389614 | T/A/C/G | 0.0035 |
| 15 | rs10277413 | 7 | 55238464 | T/A/G | 0.0294 | 54 | rs11133144 | 4 | 177229719 | A/C/T | 0.0446 |
| 16 | rs10431079 | 11 | 116440678 | A/G | 0.0728 | 55 | rs11151527 | 18 | 67347133 | A/G | 0.0506 |
| 17 | rs10459664 | 15 | 35064934 | C/T | 0.0505 | 56 | rs11159882 | 14 | 89525994 | G/T | 0.0479 |
| 18 | rs10483991 | 14 | 88682616 | G/A | 0.0607 | 57 | rs11161614 | 1 | 85898160 | T/G | 0.0299 |
| 19 | rs10486260 | 7 | 8859616 | C/T | 0.0210 | 58 | rs11164354 | 1 | 102439329 | G/A | 0.0436 |
| 20 | rs10489744 | 1 | 165380623 | G/A | 0.0690 | 59 | rs11174261 | 12 | 62362843 | C/T | 0.0473 |
| 21 | rs10506294 | 12 | 51070554 | T/C | 0.0019 | 60 | rs11177698 | 12 | 69906287 | A/G | 0.0534 |
| 22 | rs10506725 | 12 | 77449514 | T/C | 0.0444 | 61 | rs11222851 | 11 | 131831335 | G/A | 0.0683 |
| 23 | rs10521076 | 9 | 108878562 | C/T | 0.0775 | 62 | rs11223550 | 11 | 133532372 | A/G | 0.0527 |
| 24 | rs10756234 | 9 | 11316481 | C/T | -0.0004 | 63 | rs11224765 | 11 | 101310590 | C/T | 0.0981 |
| 25 | rs10768017 | 11 | 33681025 | G/T | 0.0571 | 64 | rs11257349 | 10 | 6260717 | T/G | 0.0065 |
| 26 | rs10771923 | 12 | 32320912 | C/T | 0.0332 | 65 | rs1125832 | 16 | 52421140 | T/A | 0.0337 |
| 27 | rs10778125 | 12 | 101962634 | C/A | 0.0513 | 66 | rs11466640 | 4 | 38778903 | G/A/T | 0.0090 |
| 28 | rs10806975 | 6 | 23766367 | A/G/T | 0.0372 | 67 | rs11611033 | 12 | 63771136 | C/T | 0.0488 |
| 29 | rs10819066 | 9 | 101356091 | C/G | 0.0262 | 68 | rs11621121 | 14 | 65822493 | C/T | 0.0990 |
| 30 | rs10833071 | 11 | 19209050 | T/C | 0.0328 | 69 | rs11625485 | 14 | 80242132 | T/C | 0.0538 |
| 31 | rs10836092 | 11 | 33676946 | G/A/C | 0.0515 | 70 | rs11625876 | 14 | 35248467 | C/T | 0.0559 |
| 32 | rs10836093 | 11 | 33678526 | A/G | 0.0530 | 71 | rs11680024 | 2 | 49535856 | A/C | -0.0040 |
| 33 | rs10847616 | 12 | 128910954 | A/C/T | 0.0308 | 72 | rs1178148 | 7 | 18778113 | A/C | 0.0795 |
| 34 | rs10849181 | 12 | 638166 | C/T | 0.0525 | 73 | rs11841589 | 13 | 73814891 | G/T | 0.0862 |
| 35 | rs10858883 | 12 | 89893348 | T/C | -0.0004 | 74 | rs11846710 | 14 | 58343352 | G/A | 0.0768 |
| 36 | rs10883736 | 10 | 104320029 | G/T | 0.0365 | 75 | rs11847263 | 14 | 65775695 | T/A/G | 0.0635 |
| 37 | rs10890510 | 1 | 48334490 | A/C | 0.0495 | 76 | rs11874960 | 18 | 1948608 | A/C/G/T | 0.0049 |
| 38 | rs10894034 | 11 | 129255618 | C/T | 0.0521 | 77 | rs11959012 | 5 | 167668843 | C/T | 0.0694 |
| 39 | rs10894724 | 11 | 133547942 | T/A/G | 0.0812 | 78 | rs12006467 | 9 | 35090720 | T/C | 0.0993 |
| 79 | rs12039715 | 1 | 242801261 | G/C | 0.1726 | 119 | rs1391099 | 4 | 143373910 | A/G/T | 0.0025 |
| 80 | rs12045644 | 1 | 40084488 | G/T | 0.0393 | 120 | rs1420288 | 16 | 54477881 | T/C | 0.0519 |
| 81 | rs12095503 | 1 | 171653329 | C/T | 0.0346 | 121 | rs1420528 | 16 | 52417241 | C/G | 0.0374 |
| 82 | rs12134013 | 1 | 164459913 | C/T | 0.0586 | 122 | rs1420638 | 3 | 181497374 | G/A | 0.0407 |
| 83 | rs12141436 | 1 | 68463212 | G/A | 0.0418 | 123 | rs1422931 | 5 | 167397540 | C/T | 0.0462 |
| 84 | rs12151767 | 2 | 198274929 | G/A | 0.0548 | 124 | rs1428150 | 5 | 132490807 | A/G | 0.0666 |
| 85 | rs12231617 | 12 | 124930127 | C/G | 0.0458 | 125 | rs1442502 | 1 | 166548317 | G/A | 0.0277 |
| 86 | rs12351269 | 9 | 16806521 | T/C | 0.0558 | 126 | rs1453054 | 2 | 181359907 | T/C/G | 0.0119 |
| 87 | rs12459941 | 19 | 10666112 | G/A | 0.0631 | 127 | rs1465306 | 7 | 66699784 | T/C | 0.0672 |
| 88 | rs12483769 | 22 | 47790072 | T/C | 0.0870 | 128 | rs1465759 | 2 | 142576915 | T/C | 0.0365 |
| 89 | rs12488690 | 3 | 150490463 | G/A | 0.0502 | 129 | rs1488485 | 3 | 7951364 | T/C | 0.0125 |
| 90 | rs12522710 | 5 | 65051804 | G/A | 0.0125 | 130 | rs1522340 | 3 | 70369504 | G/A | 0.0320 |
| 91 | rs1256519 | 14 | 65736324 | G/A | 0.1027 | 131 | rs1524930 | 21 | 40968460 | A/G | 0.0649 |
| 92 | rs1256520 | 14 | 65737193 | C/A | 0.0958 | 132 | rs1537523 | 9 | 3649097 | A/C | 0.0512 |
| 93 | rs12567990 | 1 | 207681685 | C/T | -0.0038 | 133 | rs1538374 | 10 | 72718336 | G/A | 0.0663 |
| 94 | rs12571942 | 10 | 127954919 | G/A | 0.0056 | 134 | rs1551653 | 8 | 34324240 | A/G | 0.0084 |
| 95 | rs12586912 | 14 | 34023334 | G/T | 0.0422 | 135 | rs155872 | 3 | 1454063 | C/T | 0.0227 |
| 96 | rs12588061 | 14 | 50481926 | T/C | 0.0703 | 136 | rs1561201 | 18 | 38205375 | G/A | 0.0330 |
| 97 | rs12589835 | 14 | 30848932 | C/T | 0.0615 | 137 | rs1566857 | 4 | 46048880 | A/T | 0.0468 |
| 98 | rs12598852 | 16 | 13329469 | G/A | 0.0500 | 138 | rs160539 | 16 | 63353953 | C/G | 0.0641 |
| 99 | rs12630087 | 3 | 80935171 | T/C | 0.0429 | 139 | rs1609763 | 5 | 142018424 | T/G | 0.0232 |
| 100 | rs12632233 | 3 | 94033744 | G/T | 0.0334 | 140 | rs1613215 | 3 | 77738317 | T/C | 0.0509 |
| 101 | rs12676684 | 8 | 118857704 | G/C/T | 0.0584 | 141 | rs1652519 | 15 | 58612113 | G/A/C | 0.0219 |
| 102 | rs12691557 | 2 | 141469911 | G/A/T | 0.0461 | 142 | rs1678537 | 12 | 57900341 | G/A | 0.0885 |
| 103 | rs12768145 | 10 | 17610277 | G/A | 0.0532 | 143 | rs16834705 | 2 | 193385209 | A/G | 0.0168 |
| 104 | rs12776442 | 10 | 59297413 | A/G | 0.0075 | 144 | rs16850913 | 2 | 166421763 | A/G | 0.0450 |
| 105 | rs1284605 | 12 | 57921188 | C/T | 0.0863 | 145 | rs16862627 | 3 | 150114701 | G/A | -0.0022 |
| 106 | rs13076655 | 3 | 80920612 | C/T | 0.0412 | 146 | rs16893074 | 5 | 24074021 | T/A | 0.0042 |
| 107 | rs13094402 | 3 | 80710455 | A/G | 0.0467 | 147 | rs16895073 | 5 | 65596821 | T/C | 0.0302 |
| 108 | rs13160399 | 5 | 165711426 | G/A | 0.0550 | 148 | rs16944149 | 17 | 10938783 | T/G | 0.0223 |
| 109 | rs1317548 | 10 | 12245520 | G/A | 0.0575 | 149 | rs16951472 | 13 | 97073977 | C/T | 0.0503 |
| 110 | rs131864 | 22 | 47271217 | T/G | 0.0450 | 150 | rs16986850 | 20 | 41048928 | T/C | 0.0307 |
| 111 | rs1322944 | 13 | 53683163 | A/G | 0.0740 | 151 | rs16991180 | 20 | 10148468 | G/A | 0.0655 |
| 112 | rs13249584 | 8 | 9710943 | A/G | 0.0786 | 152 | rs16997770 | 4 | 124693346 | T/C | 0.0611 |
| 113 | rs1325192 | 1 | 199244248 | G/C/T | 0.0625 | 153 | rs17002737 | 22 | 42282012 | C/T | 0.0454 |
| 114 | rs1333099 | 13 | 73691236 | G/A | 0.0715 | 154 | rs17016175 | 4 | 143391299 | A/G | 0.0018 |
| 115 | rs13419695 | 2 | 7347211 | A/T | 0.0808 | 155 | rs17029405 | 3 | 32451697 | C/A | 0.0508 |
| 116 | rs1362553 | 16 | 52438954 | C/G | 0.0329 | 156 | rs17034099 | 3 | 11220006 | C/A | 0.0412 |
| 117 | rs1371566 | 2 | 119738187 | T/G | 0.0337 | 157 | rs17072984 | 18 | 62051788 | G/A | 0.0312 |
| 118 | rs1378365 | 4 | 61775645 | G/A | 0.0587 | 158 | rs17076328 | 5 | 173143508 | G/A | 0.0164 |
| 159 | rs17121800 | 10 | 108710873 | C/T | 0.1007 | 199 | rs2175743 | 1 | 70295901 | C/T | 0.0413 |
| 160 | rs17129041 | 1 | 66996037 | T/C | 0.0529 | 200 | rs2194757 | 2 | 21706029 | G/A | 0.0108 |
| 161 | rs17207681 | 5 | 138168527 | A/G | 0.0823 | 201 | rs2195856 | 1 | 187922876 | A/C | 0.0548 |
| 162 | rs17239258 | 4 | 183263055 | A/G | 0.0500 | 202 | rs2253974 | 21 | 23231832 | C/T | 0.0187 |
| 163 | rs17372695 | 4 | 155060317 | A/G | -0.0030 | 203 | rs2255957 | 22 | 42241372 | G/A/T | 0.0440 |
| 164 | rs17377643 | 22 | 42152988 | A/C | 0.0565 | 204 | rs2269275 | 5 | 83261405 | A/G | 0.0688 |
| 165 | rs17451739 | 5 | 144030993 | C/T | 0.0732 | 205 | rs2269658 | 22 | 42280618 | T/C | 0.0375 |
| 166 | rs174520 | 14 | 88006776 | T/G | 0.0899 | 206 | rs2278339 | 18 | 50866195 | T/G | 0.0298 |
| 167 | rs174534 | 11 | 61549458 | A/G | -0.0023 | 207 | rs229562 | 22 | 37599065 | G/T | 0.0827 |
| 168 | rs174583 | 11 | 61609750 | C/T | -0.0046 | 208 | rs2295756 | 11 | 35241229 | T/C | 0.0000 |
| 169 | rs17469810 | 3 | 80807568 | G/A | 0.0390 | 209 | rs2313427 | 16 | 8815000 | G/A | 0.0470 |
| 170 | rs17582830 | 4 | 38867427 | A/G | 0.0004 | 210 | rs2330015 | 22 | 22536956 | C/T | 0.0678 |
| 171 | rs17583068 | 4 | 38908616 | G/C/T | 0.0558 | 211 | rs2333656 | 17 | 58184540 | T/C | 0.0576 |
| 172 | rs17599827 | 5 | 89518433 | A/C | 0.0810 | 212 | rs2349093 | 12 | 128520953 | T/A | 0.0004 |
| 173 | rs17631488 | 5 | 18777746 | A/G | 0.1030 | 213 | rs2352181 | 10 | 87653766 | G/A | 0.0257 |
| 174 | rs17717717 | 3 | 71404405 | G/A | 0.0320 | 214 | rs2353686 | 16 | 86354605 | T/C | 0.0566 |
| 175 | rs17766621 | 14 | 71262932 | T/C | 0.0349 | 215 | rs2377962 | 18 | 9101722 | G/T | 0.0522 |
| 176 | rs17823795 | 14 | 63384260 | G/A | 0.0411 | 216 | rs2430184 | 1 | 187786661 | G/T | 0.0482 |
| 177 | rs1794072 | 11 | 61303803 | C/T | 0.0446 | 217 | rs2526371 | 17 | 56443545 | G/A | 0.0501 |
| 178 | rs1795704 | 12 | 58739693 | C/A | 0.0502 | 218 | rs2547353 | 19 | 58582797 | G/C/T | 0.0666 |
| 179 | rs1835298 | 11 | 112057620 | G/A | 0.0428 | 219 | rs255630 | 5 | 127677188 | T/C | 0.0133 |
| 180 | rs1841305 | 18 | 50163950 | A/T | 0.0556 | 220 | rs258728 | 7 | 81663275 | C/A | 0.0337 |
| 181 | rs1861693 | 12 | 107603157 | C/T | 0.0471 | 221 | rs2587694 | 2 | 120213497 | A/G | 0.0026 |
| 182 | rs1864307 | 18 | 57332158 | T/G | 0.0440 | 222 | rs2589787 | 5 | 38260303 | G/A | 0.0374 |
| 183 | rs1875174 | 10 | 2971488 | C/G/T | 0.0806 | 223 | rs2622637 | 8 | 106505971 | G/A/T | 0.0951 |
| 184 | rs1877696 | 8 | 9712822 | C/G/T | 0.0956 | 224 | rs2642066 | 17 | 65639014 | G/T | 0.0910 |
| 185 | rs189062 | 2 | 137349278 | G/A | 0.0429 | 225 | rs2645645 | 4 | 77859067 | A/G/T | 0.0469 |
| 186 | rs1898227 | 16 | 63282080 | A/G/T | 0.0546 | 226 | rs26461 | 5 | 11202649 | C/T | 0.0211 |
| 187 | rs1901786 | 3 | 130021945 | G/A | 0.0411 | 227 | rs27061 | 5 | 1362793 | T/C | 0.0761 |
| 188 | rs1944975 | 18 | 57276748 | T/C | 0.0369 | 228 | rs2770310 | 6 | 70165296 | C/A/T | 0.0924 |
| 189 | rs1981370 | 18 | 74070815 | A/G | 0.0722 | 229 | rs2793438 | 6 | 24461427 | G/C | 0.0382 |
| 190 | rs198464 | 11 | 61521621 | G/A | -0.0050 | 230 | rs2836749 | 21 | 40287238 | A/C/T | 0.0548 |
| 191 | rs2008819 | 7 | 39662268 | C/T | 0.0459 | 231 | rs2837352 | 21 | 41363778 | C/T | 0.0312 |
| 192 | rs2011071 | 5 | 88741077 | A/G | 0.0583 | 232 | rs2838408 | 21 | 45246422 | A/G | 0.0750 |
| 193 | rs2035023 | 4 | 137753683 | T/C | 0.0549 | 233 | rs2853668 | 5 | 1300025 | G/T | 0.0580 |
| 194 | rs2047297 | 4 | 121511851 | G/C/T | 0.0271 | 234 | rs2901142 | 2 | 124764392 | C/T | 0.0727 |
| 195 | rs2068746 | 17 | 11389480 | T/C | 0.0464 | 235 | rs2945733 | 8 | 134615750 | T/G | 0.0602 |
| 196 | rs2099563 | 3 | 80740366 | G/A | 0.0447 | 236 | rs2972336 | 5 | 76504150 | G/C | -0.0028 |
| 197 | rs2160913 | 18 | 8716090 | A/C/G | 0.0216 | 237 | rs2976396 | 8 | 143764001 | G/A | 0.1521 |
| 198 | rs2174739 | 6 | 79659170 | A/G | 0.0042 | 238 | rs3027238 | 17 | 8135061 | T/C | 0.9803 |
| 239 | rs3104517 | 15 | 27940339 | A/G | 0.0288 | 279 | rs4690508 | 4 | 178295766 | G/T | 0.0504 |
| 240 | rs3111745 | 3 | 21827159 | G/T | 0.0751 | 280 | rs4718412 | 7 | 66286867 | T/C | 0.0751 |
| 241 | rs315808 | 5 | 169750158 | G/T | 0.0626 | 281 | rs475235 | 18 | 10029988 | G/A/T | 0.0289 |
| 242 | rs3217805 | 12 | 4388084 | C/A/G/T | 0.0844 | 282 | rs4756305 | 11 | 36292065 | T/A/C | 0.0532 |
| 243 | rs321967 | 7 | 78314549 | G/A/C | 0.0454 | 283 | rs4790409 | 17 | 1243941 | C/T | 0.0600 |
| 244 | rs34052939 | 3 | 80765431 | C/G/T | 0.0447 | 284 | rs480631 | 6 | 138406328 | C/A/T | 0.0620 |
| 245 | rs34254286 | 4 | 164411734 | A/G | 0 | 285 | rs4820428 | 22 | 41537589 | A/G | 0.0762 |
| 246 | rs374722 | 2 | 147839830 | G/A | 0.0740 | 286 | rs4834226 | 4 | 128986874 | C/T | -0.0008 |
| 247 | rs3757419 | 7 | 66029429 | C/T | 0.0718 | 287 | rs4839523 | 1 | 116800016 | T/C | 0.0284 |
| 248 | rs3757425 | 7 | 150067640 | A/C | 0.0706 | 288 | rs4846992 | 1 | 230137518 | A/G | 0.0426 |
| 249 | rs3758229 | 9 | 95078852 | C/A/G | 0.0574 | 289 | rs4865142 | 4 | 57549583 | C/T | 0.0372 |
| 250 | rs3775539 | 4 | 99304600 | A/G | 0.0452 | 290 | rs4865290 | 4 | 53278689 | G/A | 0.0307 |
| 251 | rs3910142 | 20 | 12921966 | T/C | 0.0613 | 291 | rs4883926 | 13 | 73843655 | T/C | 0.0583 |
| 252 | rs3923084 | 2 | 227948726 | A/G/T | 0.0586 | 292 | rs4902391 | 14 | 65815979 | G/T | 0.0929 |
| 253 | rs3923736 | 7 | 155060730 | A/C/G/T | 0.0714 | 293 | rs4903754 | 14 | 40865770 | G/A | 0.0492 |
| 254 | rs3924308 | 9 | 130185385 | C/A | 0.0129 | 294 | rs4912933 | 5 | 143038150 | G/A/C/T | -0.0034 |
| 255 | rs4133446 | 5 | 86155113 | T/C | 0.0645 | 295 | rs4918000 | 10 | 94837743 | C/T | 0.0399 |
| 256 | rs4144800 | 4 | 38006916 | G/A | 0.0228 | 296 | rs4922234 | 8 | 16639271 | G/A/C/T | 0.0433 |
| 257 | rs4254643 | 3 | 792207 | G/A | 0.0307 | 297 | rs4938285 | 11 | 116440011 | T/C | 0.0697 |
| 258 | rs4263026 | 18 | 73309989 | G/C | 0.0466 | 298 | rs5022079 | 18 | 76478188 | A/G | 0.0931 |
| 259 | rs4280278 | 16 | 29439227 | C/G | 0.0841 | 299 | rs5030883 | 10 | 70480868 | A/C/T | 0.0759 |
| 260 | rs4325622 | 17 | 28526475 | T/C | 0.3617 | 300 | rs520605 | 1 | 184795779 | C/T | 0.0581 |
| 261 | rs4328700 | 20 | 23709545 | A/G | 0.0344 | 301 | rs536430 | 10 | 79001643 | C/T | 0.0107 |
| 262 | rs4353835 | 3 | 32446775 | C/T | 0.0849 | 302 | rs5770018 | 22 | 49595560 | C/T | 0.0411 |
| 263 | rs4355871 | 9 | 29653331 | G/A/C | 0.0665 | 303 | rs5996064 | 22 | 42172080 | C/T | 0.0476 |
| 264 | rs4372441 | 11 | 7657259 | C/T | 0.0787 | 304 | rs6000401 | 22 | 37149336 | C/T | 0.0426 |
| 265 | rs4393669 | 18 | 67296561 | T/C | 0.0330 | 305 | rs6007756 | 22 | 48282309 | G/A/C | 0.0571 |
| 266 | rs4414069 | 1 | 200391561 | T/C | 0.0478 | 306 | rs6016226 | 20 | 38661387 | C/T | 0.0181 |
| 267 | rs4466495 | 9 | 17496114 | C/A/T | 0.0489 | 307 | rs6030932 | 20 | 42146320 | T/G | 0.0659 |
| 268 | rs4486887 | 16 | 50677571 | C/T | 0.0579 | 308 | rs6031579 | 20 | 43015108 | C/G | 0.0654 |
| 269 | rs4491175 | 11 | 90956214 | T/C | 0.0485 | 309 | rs6049064 | 20 | 23727145 | C/T | 0.0360 |
| 270 | rs4533076 | 12 | 56069231 | C/A | 0.0410 | 310 | rs6074520 | 20 | 12905188 | T/A/C | 0.0559 |
| 271 | rs4543050 | 3 | 74954560 | A/C/T | 0.0194 | 311 | rs6117562 | 20 | 753310 | G/A | -0.0011 |
| 272 | rs4578397 | 11 | 131832284 | G/T | 0.0737 | 312 | rs6123723 | 20 | 37082145 | C/T | 0.0891 |
| 273 | rs4603782 | 2 | 102735809 | T/A | 0.0642 | 313 | rs6137010 | 20 | 2090118 | C/T | 0.0110 |
| 274 | rs4668680 | 2 | 10529961 | A/G | 0.0199 | 314 | rs6138046 | 20 | 23704388 | G/C/T | 0.0342 |
| 275 | rs4674759 | 2 | 224068666 | G/A | 0.0059 | 315 | rs616022 | 3 | 80711349 | G/C/T | 0.0253 |
| 276 | rs4677399 | 3 | 74519384 | T/C | 0.0307 | 316 | rs6436971 | 2 | 231582797 | T/C | 0.0815 |
| 277 | rs4678169 | 3 | 124543103 | C/A | 0.0557 | 317 | rs6450876 | 5 | 31925423 | A/G | -0.0024 |
| 278 | rs4681869 | 3 | 58573534 | C/T | 0.0583 | 318 | rs6465469 | 7 | 95179593 | G/A | 0.0611 |
| 319 | rs6478966 | 9 | 101776125 | G/A | 0.0192 | 359 | rs760873 | 20 | 45393072 | T/G | 0.0582 |
| 320 | rs6502840 | 17 | 4989857 | C/T | 0.0515 | 360 | rs7630111 | 3 | 114165901 | C/A | 0.0499 |
| 321 | rs6549596 | 3 | 74507710 | T/C | 0.0271 | 361 | rs7642488 | 3 | 84688723 | C/T | 0.0406 |
| 322 | rs6549601 | 3 | 74525187 | C/T | 0.0307 | 362 | rs7655849 | 4 | 161867415 | T/A | 0.0759 |
| 323 | rs6559935 | 9 | 89140339 | G/A | 0.0425 | 363 | rs7666030 | 4 | 24728480 | G/A | 0.0492 |
| 324 | rs6561294 | 13 | 46695350 | T/C | 0.0215 | 364 | rs7674135 | 4 | 84577025 | T/C | 0.0399 |
| 325 | rs6576127 | 14 | 106194763 | C/T | 0.1076 | 365 | rs769411 | 2 | 171693639 | C/G/T | 0.0537 |
| 326 | rs6589072 | 11 | 109322914 | C/G | 0.0501 | 366 | rs7698051 | 4 | 60278484 | A/G | 0.0445 |
| 327 | rs6599390 | 4 | 956047 | A/G | 0.0584 | 367 | rs770576 | 11 | 99884272 | A/G | 0.0565 |
| 328 | rs6707773 | 2 | 191344132 | C/T | 0.0279 | 368 | rs7802058 | 7 | 81697666 | A/C | 0.0512 |
| 329 | rs6717406 | 2 | 187743228 | T/G | 0.0326 | 369 | rs7845383 | 8 | 88289450 | G/T | 0.0361 |
| 330 | rs6738590 | 2 | 65059603 | A/G | 0.0429 | 370 | rs789378 | 3 | 154079645 | C/T | 0.0205 |
| 331 | rs6743998 | 2 | 196676489 | T/A/C/G | 0.0612 | 371 | rs8014475 | 14 | 65811537 | T/C | 0.0937 |
| 332 | rs6760967 | 2 | 4669324 | C/T | 0.0400 | 372 | rs8015594 | 14 | 40942142 | C/G | 0.0688 |
| 333 | rs6767410 | 3 | 80699208 | C/T | 0.0505 | 373 | rs8049660 | 16 | 89857700 | A/G | 0.0398 |
| 334 | rs6768622 | 3 | 74508075 | C/T | 0.0271 | 374 | rs8050932 | 16 | 50687673 | C/T | 0.0545 |
| 335 | rs6833150 | 4 | 61659236 | A/G | 0.0235 | 375 | rs8060207 | 16 | 82273372 | G/A | 0.0287 |
| 336 | rs6898653 | 5 | 115975656 | A/G | 0.0654 | 376 | rs807959 | 19 | 22115835 | A/G | 0.0561 |
| 337 | rs6924957 | 6 | 96853579 | G/T | 0.0527 | 377 | rs8084884 | 18 | 75513922 | A/T | 0.0597 |
| 338 | rs6955018 | 7 | 125115418 | A/G | 0.0474 | 378 | rs8107011 | 19 | 30841000 | A/C | 0.0341 |
| 339 | rs7006443 | 8 | 9290753 | T/C | 0.0383 | 379 | rs827287 | 10 | 72708276 | A/T | 0.0435 |
| 340 | rs7022178 | 9 | 29780195 | G/A | 0.0720 | 380 | rs883433 | 19 | 39206288 | C/T | 0.0580 |
| 341 | rs7032231 | 9 | 117025771 | C/A | 0.0706 | 381 | rs928844 | 21 | 37999799 | C/T | 0.0846 |
| 342 | rs7038964 | 9 | 104423907 | C/T | 0.0419 | 382 | rs929115 | 1 | 171591189 | T/G | 0.0757 |
| 343 | rs7097617 | 10 | 77504287 | A/G | 0.0447 | 383 | rs9302550 | 16 | 52405521 | C/T | 0.0374 |
| 344 | rs7117447 | 11 | 101351383 | G/A | 0.1118 | 384 | rs9303660 | 17 | 31420730 | C/T | 0.0614 |
| 345 | rs712645 | 5 | 110585627 | C/T | 0.0604 | 385 | rs9321180 | 6 | 130102959 | C/T | 0.0866 |
| 346 | rs713278 | 11 | 133563491 | G/A | 0.0619 | 386 | rs933199 | 6 | 26112893 | T/C | 0.0107 |
| 347 | rs7173716 | 15 | 70123826 | T/G | 0.0723 | 387 | rs9384981 | 6 | 116803328 | T/C | 0.0259 |
| 348 | rs7173982 | 15 | 36685718 | C/A/T | -0.0013 | 388 | rs9398434 | 6 | 116787017 | T/G | 0.0229 |
| 349 | rs7193505 | 16 | 52401477 | C/G | 0.0394 | 389 | rs943327 | 9 | 113599422 | T/C | 0.0695 |
| 350 | rs7195477 | 16 | 29288634 | T/C | 0.0020 | 390 | rs9532080 | 13 | 38126537 | G/A | 0.0395 |
| 351 | rs7211426 | 17 | 53654548 | G/A/C | 0.0037 | 391 | rs9549212 | 13 | 41022093 | C/T | 0.0708 |
| 352 | rs7268940 | 20 | 9961532 | C/T | 0.0461 | 392 | rs9590216 | 13 | 95905305 | T/C | -0.0035 |
| 353 | rs7271913 | 20 | 39263421 | C/A | 0.0155 | 393 | rs9663367 | 10 | 19776910 | A/C/T | 0.0370 |
| 354 | rs7317643 | 13 | 108536028 | C/T | 0.1665 | 394 | rs978605 | 10 | 94903693 | A/G | 0.0208 |
| 355 | rs738745 | 22 | 48246398 | A/G | 0.0255 | 395 | rs9825713 | 3 | 100936248 | C/A | 0.0450 |
| 356 | rs741245 | 12 | 4227248 | G/A | 0.0535 | 396 | rs9826148 | 3 | 114464858 | C/A/T | 0.0677 |
| 357 | rs7555405 | 1 | 31183486 | C/T | 0.0420 | 397 | rs9852677 | 3 | 50291617 | G/A/T | 0.0412 |
| 358 | rs758219 | 19 | 40294953 | T/C | 0.0461 | 398 | rs9857773 | 3 | 149344615 | C/A | 0.0547 |
| 399 | rs9860483 | 3 | 138730737 | C/T | 0.0985 | 414 | rs1991955 | 6 | 29884066 | A/C | 0.0938 |
| 400 | rs9896443 | 17 | 47125982 | T/C | 0.0738 | 415 | rs2182268 | 13 | 113170683 | G/A | 0.0573 |
| 401 | rs9941426 | 18 | 487281 | T/G | 0.0689 | 416 | rs2261033 | 6 | 31603591 | A/G | 0.1538 |
| 402 | rs9947737 | 18 | 455128 | A/G | 0.0477 | 417 | rs2524095 | 6 | 31266117 | A/C | 0.0456 |
| 403 | rs9979724 | 21 | 22447717 | A/G | 0.0383 | 418 | rs284784 | 4 | 100335874 | C/A | 0.0255 |
| 404 | rs10901244 | 9 | 136079463 | C/T | 0.0077 | 419 | rs3015224 | 9 | 72728388 | A/G | 0.1419 |
| 405 | rs11078951 | 17 | 38831415 | T/A/G | 0.1249 | 420 | rs3130688 | 6 | 31210216 | C/A/T | 0.0991 |
| 406 | rs11673276 | 19 | 55347963 | C/T | 0.0264 | 421 | rs3135402 | 6 | 33024654 | A/C | 0.1028 |
| 407 | rs12896399 | 14 | 92773663 | G/T | 0.0615 | 422 | rs376877 | 6 | 33024606 | C/G | 0.0902 |
| 408 | rs1414241 | 9 | 21744392 | G/A/T | 0.0756 | 423 | rs4800105 | 18 | 19651982 | C/G/T | 0.0906 |
| 409 | rs1431403 | 6 | 33047031 | T/C | -0.0048 | 424 | rs5758649 | 22 | 42605548 | C/T | 0.0059 |
| 410 | rs1496279 | 5 | 18680420 | A/T | - | 425 | rs6062275 | 20 | 61146909 | C/G/T | 0.1420 |
| 411 | rs1500127 | 5 | 165739982 | C/T | 0.0416 | 426 | rs7207346 | 17 | 75668619 | C/A/G/T | 0.0105 |
| 412 | rs174592 | 11 | 61618608 | A/G | -0.0049 | 427 | rs8074124 | 17 | 36531565 | C/T | 0.0457 |
| 413 | rs1927568 | 13 | 99740117 | T/C | 0.0137 | 428 | rs901134 | 3 | 84688173 | G/A | 0.0574 |
Supplementary Table 2
r2 of AISNP between paired LD"
| Block | rs号-1 | rs号-2 | r2值 | Block | rs号-1 | rs号-2 | r2值 |
|---|---|---|---|---|---|---|---|
| Block1 | rs2853668 | rs27061 | 0.243 | rs5996064 | rs2255957 | 0.869 | |
| Block2 | rs13249584 | rs1877696 | 0.885 | rs5996064 | rs2269658 | 0.695 | |
| Block3 | rs6074520 | rs3910142 | 0.771 | rs5996064 | rs17002737 | 0.574 | |
| Block4 | rs6138046 | rs4328700 | 1 | rs1009544 | rs2255957 | 0.991 | |
| rs6138046 | rs6049064 | 0.993 | rs1009544 | rs2269658 | 0.795 | ||
| rs4328700 | rs6049064 | 0.993 | rs1009544 | rs17002737 | 0.679 | ||
| Block5 | rs4355871 | rs7022178 | 0.221 | rs2255957 | rs2269658 | 0.802 | |
| Block6 | rs10836092 | rs11032331 | 0.98 | rs2255957 | rs17002737 | 0.686 | |
| rs10836092 | rs10836093 | 0.987 | rs2269658 | rs17002737 | 0.816 | ||
| rs10836092 | rs10768017 | 0.906 | Block10 | rs4486887 | rs8050932 | 0.371 | |
| rs11032331 | rs10836093 | 0.981 | Block11 | rs7193505 | rs9302550 | 0.959 | |
| rs11032331 | rs10768017 | 0.899 | rs7193505 | rs1420528 | 0.959 | ||
| rs10836093 | rs10768017 | 0.919 | rs7193505 | rs1125832 | 0.93 | ||
| Block7 | rs11466640 | rs17582830 | 0.449 | rs7193505 | rs1362553 | 0.911 | |
| Block8 | rs4903754 | rs8015594 | 0.285 | rs9302550 | rs1420528 | 1 | |
| Block9 | rs17377643 | rs5996064 | 0.849 | rs9302550 | rs1125832 | 0.95 | |
| rs17377643 | rs1009544 | 0.735 | rs9302550 | rs1362553 | 0.912 | ||
| rs17377643 | rs2255957 | 0.742 | rs1420528 | rs1125832 | 0.95 | ||
| rs17377643 | rs2269658 | 0.62 | rs1420528 | rs1362553 | 0.912 | ||
| rs17377643 | rs17002737 | 0.66 | rs1125832 | rs1362553 | 0.961 | ||
| rs5996064 | rs1009544 | 0.861 | Block12 | rs1678537 | rs1284605 | 0.965 | |
| Block13 | rs198464 | rs174534 | 0.388 | rs6767410 | rs34052939 | 0.96 | |
| Block14 | rs6833150 | rs1378365 | 0.278 | rs6767410 | rs17469810 | 0.944 | |
| Block15 | rs1898227 | rs160539 | 0.326 | rs6767410 | rs13076655 | 0.898 | |
| Block16 | rs1256519 | rs1256520 | 0.898 | rs6767410 | rs12630087 | 0.905 | |
| rs1256519 | rs11847263 | 0.565 | rs13094402 | rs616022 | 0.468 | ||
| rs1256519 | rs8014475 | 0.61 | rs13094402 | rs2099563 | 0.992 | ||
| rs1256519 | rs4902391 | 0.61 | rs13094402 | rs34052939 | 0.992 | ||
| rs1256519 | rs11621121 | 0.606 | rs13094402 | rs17469810 | 0.976 | ||
| rs1256520 | rs11847263 | 0.528 | rs13094402 | rs13076655 | 0.929 | ||
| rs1256520 | rs8014475 | 0.596 | rs13094402 | rs12630087 | 0.937 | ||
| rs1256520 | rs4902391 | 0.575 | rs616022 | rs2099563 | 0.472 | ||
| rs1256520 | rs11621121 | 0.572 | rs616022 | rs34052939 | 0.472 | ||
| rs11847263 | rs8014475 | 0.814 | rs616022 | rs17469810 | 0.466 | ||
| rs11847263 | rs4902391 | 0.791 | rs616022 | rs13076655 | 0.468 | ||
| rs11847263 | rs11621121 | 0.791 | rs616022 | rs12630087 | 0.476 | ||
| rs8014475 | rs4902391 | 0.95 | rs2099563 | rs34052939 | 1 | ||
| rs8014475 | rs11621121 | 0.925 | rs2099563 | rs17469810 | 0.984 | ||
| rs4902391 | rs11621121 | 0.975 | rs2099563 | rs13076655 | 0.937 | ||
| Block17 | rs3757419 | rs4718412 | 0.312 | rs2099563 | rs12630087 | 0.944 | |
| Block18 | rs827287 | rs1538374 | 0.258 | rs34052939 | rs17469810 | 0.984 | |
| Block19 | rs6549596 | rs6768622 | 1 | rs34052939 | rs13076655 | 0.937 | |
| rs6549596 | rs4677399 | 0.923 | rs34052939 | rs12630087 | 0.944 | ||
| rs6549596 | rs6549601 | 0.975 | rs17469810 | rs13076655 | 0.921 | ||
| rs6768622 | rs4677399 | 0.923 | rs17469810 | rs12630087 | 0.929 | ||
| rs6768622 | rs6549601 | 0.975 | rs13076655 | rs12630087 | 0.992 | ||
| rs4677399 | rs6549601 | 0.916 | Block21 | rs11224765 | rs7117447 | 0.442 | |
| Block20 | rs6767410 | rs13094402 | 0.952 | Block22 | rs4938285 | rs10431079 | 0.876 |
| rs6767410 | rs616022 | 0.468 | Block23 | rs9398434 | rs9384981 | 0.949 | |
| rs6767410 | rs2099563 | 0.96 | Block24 | rs1391099 | rs17016175 | 0.593 |
| [1] |
Budowle B, van Daal A. Forensically relevant SNP classes. Biotechniques, 2008, 44(5):603-608, 610.
doi: 10.2144/000112806 pmid: 18474034 |
| [2] |
Phillips C. Forensic genetic analysis of bio-geographical ancestry. Forensic Sci Int Genet, 2015, 18:49-65.
doi: 10.1016/j.fsigen.2015.05.012 |
| [3] | Phillips C. Ancestry informative markers. Encycl Forensic Sci, 2013: 323-331. |
| [4] |
Kidd KK, Speed WC, Pakstis AJ, Furtado MR, Fang RX, Madbouly A, Maiers M, Middha M, Friedlaender FR, Kidd JR. Progress toward an efficient panel of SNPs for ancestry inference. Forensic Sci Int Genet, 2014, 10:23-32.
doi: 10.1016/j.fsigen.2014.01.002 |
| [5] |
Phillips C, Salas A, Sánchez JJ, Fondevila M, Gómez-Tato A, Alvarez-Dios J, Calaza M, de Cal MC, Ballard D, Lareu MV, Carracedo A, SNPfor ID Consortium. Inferring ancestral origin using a single multiplex assay of ancestry-informative marker SNPs. Forensic Sci Int Genet, 2007, 1(3-4):273-280.
doi: 10.1016/j.fsigen.2007.06.008 |
| [6] |
Kidd JR, Friedlaender FR, Speed WC, Pakstis AJ, De La Vega FM, Kidd KK. Analyses of a set of 128 ancestry informative single-nucleotide polymorphisms in a global set of 119 population samples. Investig Genet, 2011, 2(1):1.
doi: 10.1186/2041-2223-2-1 |
| [7] | Wei L, Wei YL, Jiang L, Sun QF, Wang YY, Li CX. The development of a 27-plex SNP multiplex system. Chin J Foren Med, 2016, 31(1):13-17. |
| 魏丽, 魏以梁, 江丽, 孙启凡, 王英元, 李彩霞. 27-plex SNPs 复合扩增检测体系构建与应用评价. 中国法医学杂志, 2016, 31(1):13-17. | |
| [8] | Hao WQ, Liu J, Jiang L, Huang MS, Li JL, Ma Q, Liu C, Li CX, Wang HJ. The study of a SNP-multiplex for the ancestry inference of five continental populations. Acta Univ Med Nanjing, 2018, 38(3):331-337. |
| 郝伟琪, 刘京, 江丽, 黄美莎, 李玖玲, 马泉, 刘超, 李彩霞, 王慧君. 用于五大洲际人群区分的SNP体系研究. 南京医科大学学报(自然科学版), 2018, 38(3):331-337. | |
| [9] |
Li CX, Pakstis AJ, Jiang L, Wei YL, Sun QF, Wu H, Bulbul O, Wang P, Kang LL, Kidd JR, Kidd KK. A panel of 74 AISNPs: Improved ancestry inference within Eastern Asia. Forensic Sci Int Genet, 2016, 23:101-110.
doi: 10.1016/j.fsigen.2016.04.002 |
| [10] |
Qu SQ, Zhu J, Wang YJ, Yin L, Lv ML, Wang L, Jian H, Tan Y, Zhang RR, Liu YQ, Li F, Huang SC, Liang WB, Zhang L. Establishing a second-tier panel of 18 ancestry informative markers to improve ancestry distinctions among Asian populations. Forensic Sci Int Genet, 2019, 41:159-167.
doi: 10.1016/j.fsigen.2019.05.001 |
| [11] |
Hwa HL, Wu MY, Lin CP, Hsieh WH, Yin HI, Lee TT, Lee JC. A single nucleotide polymorphism panel for individual identification and ancestry assignment in Caucasians and four East and Southeast Asian populations using a machine learning classifier. Forensic Sci Med Pathol, 2019, 15(1):67-74.
doi: 10.1007/s12024-018-0071-y |
| [12] |
Zhang F, Su B, Zhang YP, Jin L. Genetic studies of human diversity in East Asia. Philos Trans R Soc Lond B Biol Sci, 2007, 362(1482):987-995.
doi: 10.1098/rstb.2007.2028 |
| [13] | Tian JY, Li YC, Kong QP, Zhang YP. The origin and evolution history of East Asian populations from genetic perspectives. Hereditas(Beijing), 2018, 40(10):814-824. |
| 田娇阳, 李玉春, 孔庆鹏, 张亚平. 遗传学视角下东亚人群的起源和演化. 遗传, 2018, 40(10):814-824. | |
| [14] |
Suo C, Xu HY, Khor CC, Ong RT, Sim XL, Chen JM, Tay WT, Sim KS, Zeng YX, Zhang XJ, Liu JJ, Tai ES, Wong TY, Chia KS, Teo YY. Natural positive selection and north-south genetic diversity in East Asia. Eur J Hum Genet, 2012, 20(1):102-110.
doi: 10.1038/ejhg.2011.139 pmid: 21792231 |
| [15] |
HUGO Pan-Asian SNP Consortium, Abdulla MA, Ahmed I, Assawamakin A, Bhak J, Brahmachari SK, Calacal GC, Chaurasia A, Chen CH, Chen JM, Chen YT, Chu JY, Cutiongco-de la Paz EMC, De Ungria MCA, Delfin FC, Edo J, Fuchareon S, Ghang H, Gojobori T, Han JS, Ho SF, Hoh BP, Huang W, Inoko H, Jha P, Jinam TA, Jin L, Jung JS, Kangwanpong D, Kampuansai J, Kennedy GC, Khurana P, Kim HL, Kim K, Kim S, Kim WY, Kimm K, Kimura R, Koike T, Kulawonganunchai S, Kumar V, Lai PS, Lee JY, Lee S, Liu ET, Majumder PP, Mandapati KK, Marzuki S, Mitchell W, Mukerji M, Naritomi K, Ngamphiw C, Niikawa N, Nishida N, Oh B, Oh S, Ohashi J, Oka A, Ong R, Padilla CD, Palittapongarnpim P, Perdigon HB, Phipps ME, Png E, Sakaki Y, Salvador JM, Sandraling Y, Scaria V, Seielstad M, Sidek MR, Sinha A, Srikummool M, Sudoyo H, Sugano S, Suryadi H, Suzuki Y, Tabbada KA, Tan A, Tokunaga K, Tongsima S, Villamor LP, Wang E, Wang Y, Wang HF, Wu JY, Xiao HS, Xu SH, Yang JO, Shugart YY, Yoo HS, Yuan WT, Zhao GP, Zilfalil BA; Indian Genome Variation Consortium. Mapping human genetic diversity in Asia. Science, 2009, 326(5959):1541-1545.
doi: 10.1126/science.1177074 |
| [16] |
Shi CM, Liu Q, Zhao SL, Chen H. Ancestry informative SNP panels for discriminating the major East Asian populations: Han Chinese, Japanese and Korean. Ann Hum Genet, 2019, 83(5):348-354.
doi: 10.1111/ahg.v83.5 |
| [17] |
Wang YC, Lu DS, Chung YJ, Xu SH. Genetic structure, divergence and admixture of Han Chinese, Japanese and Korean populations. Hereditas, 2018, 155:19.
doi: 10.1186/s41065-018-0057-5 |
| [18] |
Kim W, Shin DJ, Harihara S, Kim YJ. Y chromosomal DNA variation in east Asian populations and its potential for inferring the peopling of Korea. J Hum Genet, 2000, 45(2):76-83.
pmid: 10721667 |
| [19] |
Ding QL, Wang CC, Farina SE, Li H. Mapping human genetic diversity on the Japanese archipelago. Advances in Anthropology, 2011, 1(2):19-25.
doi: 10.4236/aa.2011.12004 |
| [20] |
Harihara S, Saitou N, Hirai M, Gojobori T, Park KS, Misawa S, Ellepola SB, Ishida T, Omoto K. Mitochondrial DNA polymorphism among five Asian populations. Am J Hum Genet, 1988, 43(2):134-143.
pmid: 2840820 |
| [21] |
Horai S, Murayama K, Hayasaka K, Matsubayashi S, Hattori Y, Fucharoen G, Harihara S, Park KS, Omoto K, Pan IH. mtDNA polymorphism in East Asian populations, with special reference to the peopling of Japan. Am J Hum Genet, 1996, 59(3):579-590.
pmid: 8751859 |
| [22] |
Rolf B, Horst B, Eigel A, Sagansermsri T, Brinkmann B, Horst J. Microsatellite profiles reveal an unexpected genetic relationship between Asian populations. Hum Genet, 1998, 102(6):647-652.
pmid: 9703426 |
| [23] |
Jeon S, Bhak Y, Choi Y, Jeon Y, Kim S, Jang J, Jang J, Blazyte A, Kim C, Kim Y, Shim J, Kim N, Kim YJ, Park SG, Kim J, Cho YS, Park Y, Kim HM, Kim BC, Park NH, Shin ES, Kim BC, Bolser D, Manica A, Edwards JS, Church G, Lee S, Bhak J. Korean Genome Project: 1094 Korean personal genomes with clinical information. Sci Adv, 2020, 6(22): eaaz7835.
doi: 10.1126/sciadv.aaz7835 |
| [24] |
1000 Genomes Project Consortium, Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, Korbel JO, Marchini JL, McCarthy S, McVean GA, Abecasis GR. A global reference for human genetic variation. Nature, 2015, 526(7571):68-74.
doi: 10.1038/nature15393 |
| [25] |
Clarke L, Fairley S, Zheng-Bradley XQ, Streeter I, Perry E, Lowy E, Tassé AM, Flicek P. The international Genome sample resource (IGSR): A worldwide collection of genome variation incorporating the 1000 Genomes Project data. Nucleic Acids Res, 2017, 45(D1):D854-D859.
doi: 10.1093/nar/gkw829 |
| [26] |
Xu SH, Yin XY, Li SL, Jin WF, Lou HY, Yang L, Gong XH, Wang HY, Shen YP, Pan XD, He YG, Yang YJ, Wang Y, Fu WQ, An Y, Wang JC, Tan JZ, Qian J, Chen XL, Zhang X, Sun YF, Zhang XJ, Wu BL, Jin L. Genomic dissection of population substructure of Han Chinese and its implication in association studies. Am J Hum Genet, 2009, 85(6):762-774.
doi: 10.1016/j.ajhg.2009.10.015 |
| [27] |
Jinam TA, Kanzawa-Kiriyama H, Inoue I, Tokunaga K, Omoto K, Saitou N. Unique characteristics of the Ainu population in Northern Japan. J Hum Genet, 2015, 60(10):565-571.
doi: 10.1038/jhg.2015.79 |
| [28] |
Kim JJ, Verdu P, Pakstis AJ, Speed WC, Kidd JR, Kidd KK. Use of autosomal loci for clustering individuals and populations of East Asian origin. Hum Genet, 2005, 117(6):511-519.
doi: 10.1007/s00439-005-1334-8 |
| [29] |
Qin PF, Li ZQ, Jin WF, Lu DS, Lou HY, Shen JW, Jin L, Shi YY, Xu SH. A panel of ancestry informative markers to estimate and correct potential effects of population stratification in Han Chinese. Eur J Hum Genet, 2014, 22(2):248-253.
doi: 10.1038/ejhg.2013.111 |
| [30] |
Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics, 2005, 21(2):263-265.
pmid: 15297300 |
| [31] |
Carlson CS, Eberle MA, Rieder MJ, Yi Q, Kruglyak L, Nickerson DA. Selecting a maximally informative set of single-nucleotide polymorphisms for association analyses using linkage disequilibrium. Am J Hum Genet, 2004, 74(1):106-120.
doi: 10.1086/381000 |
| [32] |
Wei YL, Wei L, Zhao L, Sun QF, Jiang L, Zhang T, Liu HB, Chen JG, Ye J, Hu L, Li CX. A single-tube 27-plex SNP assay for estimating individual ancestry and admixture from three continents. Int J Legal Med, 2015, 130(1):27-37.
doi: 10.1007/s00414-015-1183-5 |
| [33] |
Nei M. Analysis of gene diversity in subdivided populations. Proc Natl Acad Sci USA, 1973, 70(12):3321-3323.
doi: 10.1073/pnas.70.12.3321 |
| [34] |
Rousset F. genepop'007: a complete re-implementation of the genepop software for Windows and Linux. Mol Ecol Resour, 2008, 8(1):103-106.
doi: 10.1111/j.1471-8286.2007.01931.x |
| [35] |
de la Puente M, Santos C, Fondevila M, Manzo L, EUROFORGEN-NoE Consortium, Carracedo Á, Lareu MV, Phillips C. The Global AIMs Nano set: A 31-plex SNaPshot assay of ancestry-informative SNPs. Forensic Sci Int Genet, 2016, 22:81-88.
doi: 10.1016/j.fsigen.2016.01.015 |
| [36] | Liu J, Li S, Jiang L, Zhao L, Zhao WT, Feng L, Liu HB, Ji AQ, Li CX. DNA Ancestry Analyzer: an automatic program for ancestry inference of unknown individuals. Life Sci Res, 2018, 22(1):3-7, 41. |
| 刘京, 李盛, 江丽, 赵蕾, 赵雯婷, 丰蕾, 刘海渤, 季安全, 李彩霞. 对于未知来源个体进行族群推断的自动分析系统. 生命科学研究, 2018, 22(1):3-7, 41. | |
| [37] |
Geck C. The world factbook. Charleston Adv, 2017, 19(1):58-60.
doi: 10.5260/chara.19.1.58 |
| [38] |
Siska V, Jones ER, Jeon S, Bhak Y, Kim HM, Cho YS, Kim H, Lee K, Veselovskaya E, Balueva T, Gallego- Llorente M, Hofreiter M, Bradley DG, Eriksson A, Pinhasi R, Bhak J, Manica A. Genome-wide data from two early Neolithic East Asian individuals dating to 7700 years ago. Sci Adv, 2017, 3(2):e1601877.
doi: 10.1126/sciadv.1601877 |
| [39] |
Pan ZQ, Xu SH. Population genomics of East Asian ethnic groups. Hereditas, 2020, 157(1):49.
doi: 10.1186/s41065-020-00162-w |
| [40] | Wen H, Wei YL, Guo XY, Sun CC, Xue SY, Liu J, Fan H, Jiang L. High-resolution SNP ancestry inference model and efficiency evaluation in three East Asian populations. Progress in Biochemistry and Biophysics, 2021, 1-11. |
| 文豪, 魏以梁, 郭晓媛, 孙昌春, 薛思瑶, 刘京, 范虹, 江丽. 东亚三族群SNP高分辨推断模型构建与效能评估. 生物化学与生物物理进展, 2021, 1-11. | |
| [41] | Jiang L, Sun QF, Ma Q, Zhao WT, Liu J, Zhao L, Ji AQ, Li CX. Optimization and validation of analysis method based on 27-plex SNP panel for ancestry inference. Hereditas (Beijing), 2017, 39(2):166-173. |
| 江丽, 孙启凡, 马泉, 赵雯婷, 刘京, 赵蕾, 季安全, 李彩霞. 27-plex SNP 种族推断方法的优化及验证. 遗传, 2017, 39(2):166-173. |
| [1] | Yongqiang Kong, Jinkai Liu, Jiaqi Gu, Jingyi Xu, Yunuo Zheng, Yiliang Wei, Shaoyuan Wu. Optimization scheme of machine learning model for genetic division between northern Han, southern Han, Korean and Japanese [J]. Hereditas(Beijing), 2022, 44(11): 1028-1043. |
| [2] | Xi Li, Haoyu Wang, Yueyan Cao, Qiang Zhu, Panyin Shu, Tingyun Hou, Yuting Wang, Ji Zhang. Forensic genomics research on microhaplotypes [J]. Hereditas(Beijing), 2021, 43(10): 962-971. |
| [3] | Zhiyong Liu, Riga Wu, Ran Li, Qiangwei Wang, Hongyu Sun. Ethical issues of the research and practice in forensic genetics [J]. Hereditas(Beijing), 2021, 43(10): 994-1002. |
| [4] | Yang Liu, Changchun Sun, Mi Ma, Ling Wang, Wenting Zhao, Quan Ma, Anquan Ji, Jing Liu, Caixia Li. The ancestry inference of Chinese populations using 74-plex SNPs system [J]. Hereditas(Beijing), 2020, 42(3): 296-308. |
| [5] | Li Jiang,Qifan Sun,Quan Ma,Wenting Zhao,Jing Liu,Lei Zhao,Anquan Ji,Caixia Li. Optimization and validation of analysis method based on 27-plex SNP panel for ancestry inference [J]. Hereditas(Beijing), 2017, 39(2): 166-173. |
| [6] | Xiuyan Ruan, Weini Wang, Yaran Yang, Bingbing Xie, Jing Chen, Yacheng Liu, Jiangwei Yan. Genetic variability and phylogenetic analysis of 39 short tandem repeat loci in Beijing Han population [J]. HEREDITAS(Beijing), 2015, 37(7): 683-691. |
| [7] | . The selection of 30 ancestry informative markers and its application in ancestry inference [J]. HEREDITAS(Beijing), 2014, 36(8): 779-785. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||