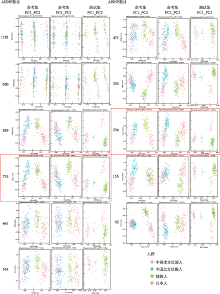
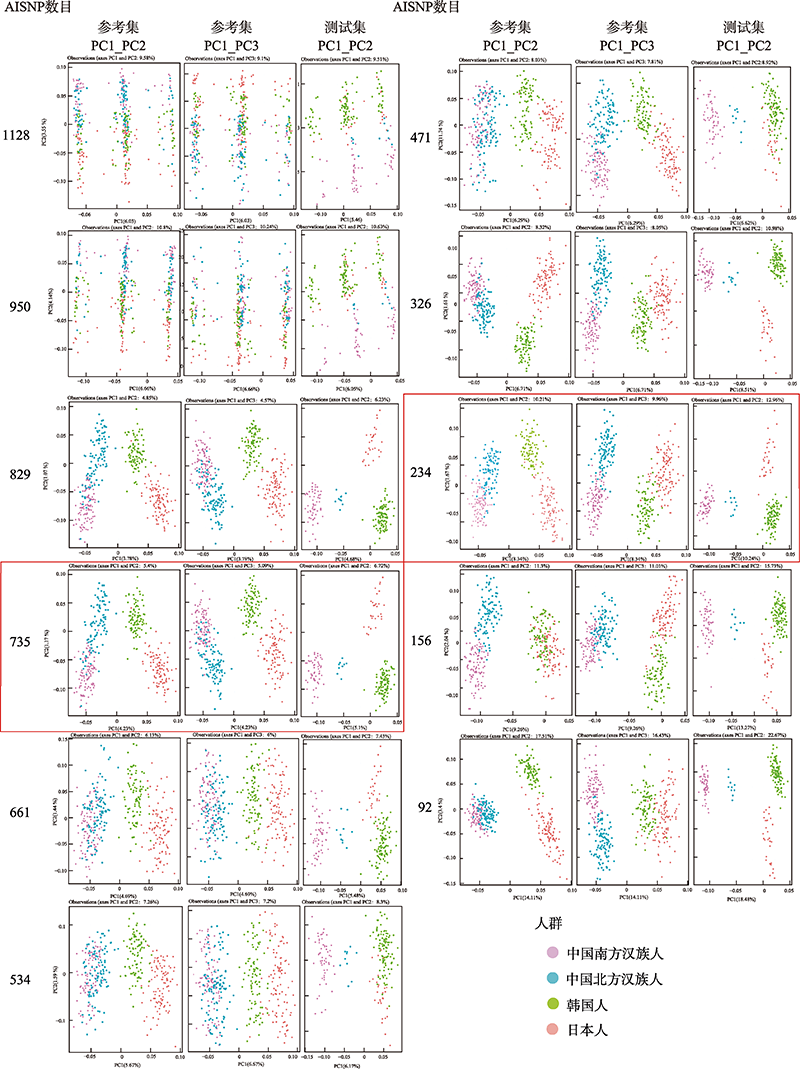
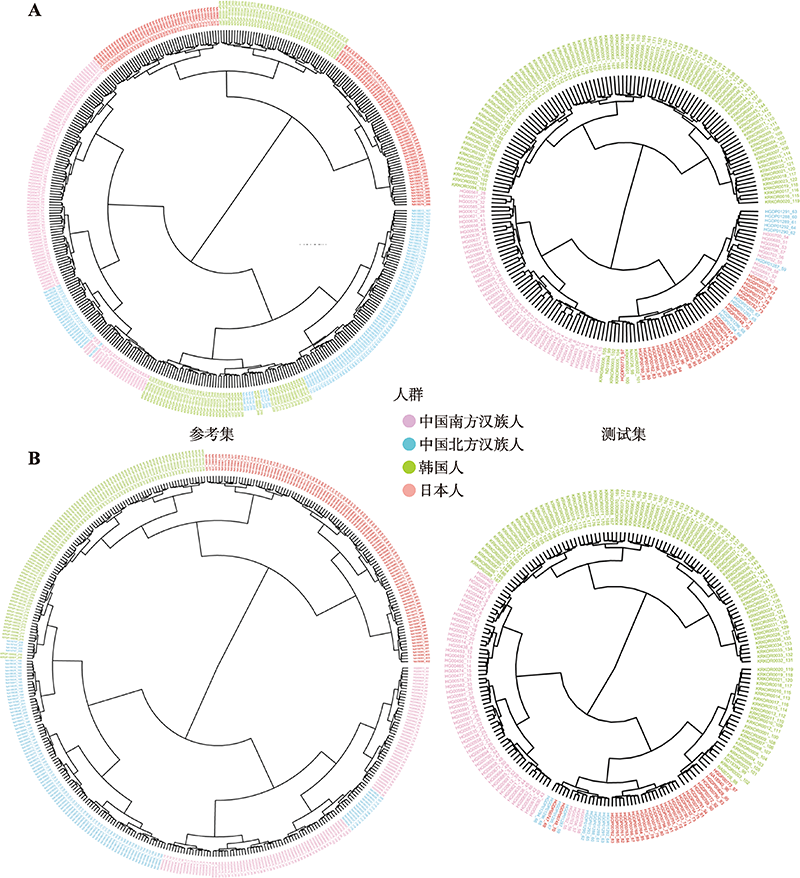
Hereditas(Beijing) ›› 2022, Vol. 44 ›› Issue (11): 1028-1043.doi: 10.16288/j.yczz.22-073
• Research Article • Previous Articles Next Articles
Optimization scheme of machine learning model for genetic division between northern Han, southern Han, Korean and Japanese
Yongqiang Kong1( ), Jinkai Liu1, Jiaqi Gu2, Jingyi Xu1, Yunuo Zheng2, Yiliang Wei2(
), Jinkai Liu1, Jiaqi Gu2, Jingyi Xu1, Yunuo Zheng2, Yiliang Wei2( ), Shaoyuan Wu1,2(
), Shaoyuan Wu1,2( )
)
- 1. Key Laboratory of Tianjin for Epigenetics, Department of Biochemistry and Molecular Biology, School of Basic Medical Sciences, Tianjin Medical University, Tianjin 300070, China
2. Key Laboratory of Phylogeny and Comparative Genomics of Jiangsu Province, Jiangsu Normal University, Xuzhou 221116, China
-
Received:2022-05-03Revised:2022-07-13Online:2022-11-20Published:2022-08-11 -
Contact:Wei Yiliang,Wu Shaoyuan E-mail:kongyongqiang@tmu.edu.cn;weiyiliang.2013@tsinghua.org.cn;shaoyuan5@gmail.com -
Supported by:Supported by the Key Laboratory of Forensic Genetics of China No(2020FGKFKT01);the Graduate Research and Practice Innovation Program of Jiangsu Normal University Nos(KYCX20_2286);the Graduate Research and Practice Innovation Program of Jiangsu Normal University Nos(KYCX21_2597)
Cite this article
Yongqiang Kong, Jinkai Liu, Jiaqi Gu, Jingyi Xu, Yunuo Zheng, Yiliang Wei, Shaoyuan Wu. Optimization scheme of machine learning model for genetic division between northern Han, southern Han, Korean and Japanese[J]. Hereditas(Beijing), 2022, 44(11): 1028-1043.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
Table 3
The collection and references of AISNP"
| 目标区分人群 | 样本量 | 参考来源 |
|---|---|---|
| 中国汉族人 | 461 | Qin等(2014)[ |
| 韩国人 | 141 | Jeon等(2020)[ |
| 东亚人(韩国人) | 100 | Jung等(2019)[ |
| 日本人 | 111 | Aklyma等(2017)[ |
| 中国汉族人、韩国人和日本人 | 341 | Shi等(2019)[ |
| 东亚人 | 8 | Kim等(2005)[ |
| 中国北方汉族人和日本人 | 23 | 本实验室[ |
Table 4
Performance assessment of training and testing set in softmax and randomForest model"
| AISNP数 | 准确率(95%置信区间) | kappa系数 | ||
|---|---|---|---|---|
| softmax | 随机森林 | softmax | 随机森林 | |
| 92 | 0.7538(0.6879~0.8119) | 0.7337(0.6665~0.7937) | 0.6339 | 0.6155 |
| 156 | 0.8844(0.8316~0.9253) | 0.7889(0.7256~0.8435) | 0.8156 | 0.6925 |
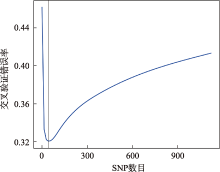
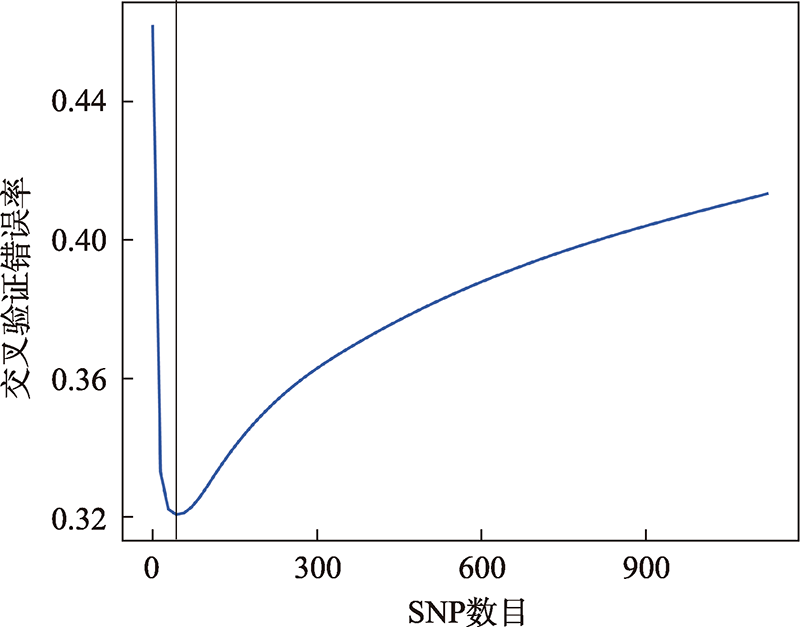
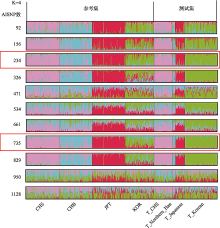
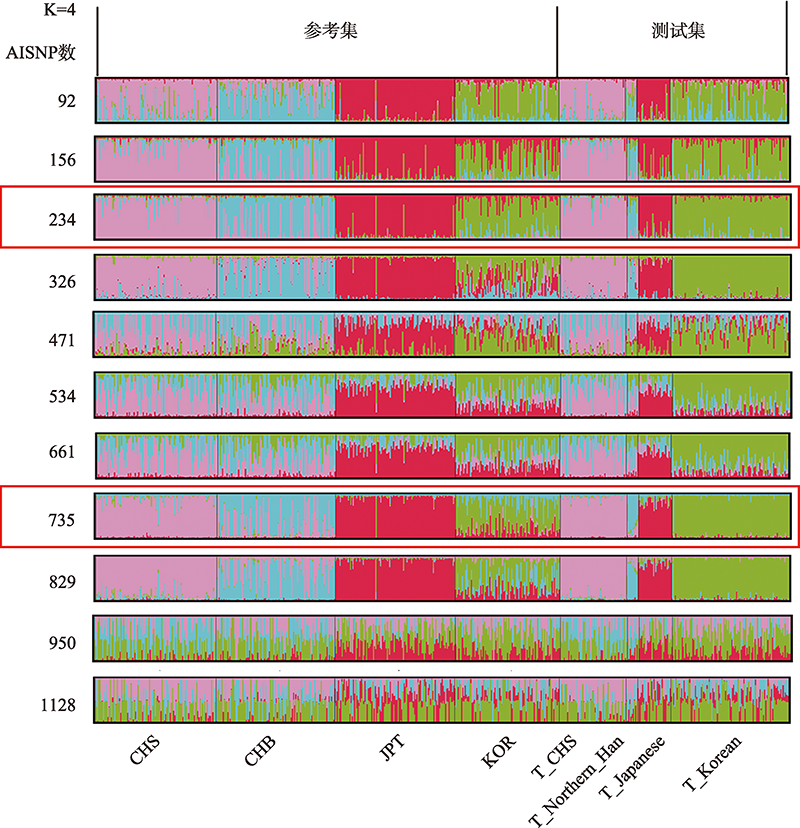
| 234 | 0.9196(0.8727~0.9533) | 0.8342(0.7751~0.883) | 0.8713 | 0.7554 |
| 326 | 0.9095(0.8608~0.9455) | 0.7839(0.7202~0.839) | 0.8532 | 0.6564 |
| 471 | 0.9146(0.8667~0.9494) | 0.8241(0.764~0.8743) | 0.8605 | 0.7285 |
| 534 | 0.9196(0.8727~0.9533) | 0.7889(0.7256~0.8435) | 0.8693 | 0.6778 |
| 661 | 0.4573(0.3867~0.5292) | 0.8191(0.7585~0.87) | 0.358 | 0.719 |
| 735 | 0.1457 (0.0998~0.2025) | 0.9447(0.9032~0.9721) | 0 | 0.914 |
| 829 | 0.4422(0.372~0.5141) | 0.9146(0.8667~0.9494) | 0.3022 | 0.8679 |
| 950 | 0.4824(0.4112~0.5542) | 0.8693(0.8144~0.9128) | 0.3651 | 0.7935 |
| 1128 | 0.4724(0.4014~0.5442) | 0.8794(0.8259~0.9212) | 0.3316 | 0.8076 |
Table 5
The information of 234 AISNPs"
| rs号 | 染色体 | 第37版序列位置 | 等位基因 | Fst | In | rs号 | 染色体 | 第37版序列位置 | 等位基因 | Fst | In |
|---|---|---|---|---|---|---|---|---|---|---|---|
| rs11119385 | 1 | 205317503 | C/T | 0.024 | 0.005 | rs6478966 | 9 | 93716118 | T/C | 0.029 | 0.006 |
| rs11161614 | 1 | 48334490 | A/C | 0.051 | 0.011 | rs7022178 | 9 | 16806521 | T/A/C | 0.073 | 0.015 |
| rs11164354 | 1 | 70295901 | C/T | 0.022 | 0.005 | rs7032231 | 9 | 108878562 | C/T | 0.062 | 0.014 |
| rs11805598 | 1 | 151830098 | C/T | 0.016 | 0.004 | rs7038964 | 9 | 101776125 | G/A | 0.052 | 0.012 |
| rs1371566 | 1 | 230137518 | A/G | 0.032 | 0.007 | rs943327 | 9 | 101356091 | C/G | 0.015 | 0.003 |
| rs1442502 | 1 | 165380623 | G/A | 0.032 | 0.007 | rs998410 | 9 | 100556109 | A/G/T | 0.054 | 0.012 |
| rs1564032 | 1 | 244136216 | G/A | 0.014 | 0.003 | rs10900048 | 10 | 25401705 | G/A | 0.016 | 0.004 |
| rs2430184 | 1 | 184795779 | C/T | 0.031 | 0.007 | rs1341667 | 10 | 64415184 | A/C/G | 0.028 | 0.006 |
| rs2488457 | 1 | 114377568 | A/G | 0.011 | 0.002 | rs17121800 | 10 | 104320029 | G/T | 0.068 | 0.013 |
| rs4414069 | 1 | 199244248 | G/C/T | 0.020 | 0.004 | rs2295756 | 10 | 77504287 | A/G | 0.032 | 0.007 |
| rs520605 | 1 | 166548317 | G/A | 0.013 | 0.003 | rs4372441 | 10 | 127954919 | G/A | 0.053 | 0.012 |
| rs6549596 | 1 | 31183486 | C/T | 0.022 | 0.005 | rs4918000 | 10 | 21762267 | A/G | 0.044 | 0.010 |
| rs723848 | 1 | 172862939 | C/T | 0.043 | 0.009 | rs703989 | 10 | 44987921 | T/A/C | 0.017 | 0.004 |
| rs929115 | 1 | 164459913 | C/G/T | 0.052 | 0.011 | rs7909976 | 10 | 13914443 | C/A/T | 0.020 | 0.005 |
| rs10929660 | 2 | 1985248 | G/A | 0.034 | 0.007 | rs978605 | 10 | 94837743 | C/T | 0.031 | 0.008 |
| rs12151767 | 2 | 193385209 | A/G | 0.055 | 0.012 | rs10431079 | 11 | 101351383 | G/A | 0.053 | 0.011 |
| rs12691557 | 2 | 124764392 | C/T | 0.013 | 0.003 | rs10894034 | 11 | 116440011 | T/C | 0.010 | 0.002 |
| rs13390103 | 2 | 142291181 | T/A/C | 0.019 | 0.004 | rs11034709 | 11 | 19209050 | T/C | 0.079 | 0.015 |
| rs1453054 | 2 | 171693639 | C/G/T | 0.020 | 0.004 | rs11222851 | 11 | 129255618 | C/T | 0.024 | 0.005 |
| rs16834705 | 2 | 187743228 | T/G | 0.034 | 0.008 | rs11223550 | 11 | 131832284 | G/T | 0.046 | 0.010 |
| rs16850913 | 2 | 137349278 | G/A | 0.069 | 0.016 | rs11224765 | 11 | 99884272 | A/G | 0.063 | 0.013 |
| rs2042020 | 2 | 5702577 | C/T | 0.031 | 0.007 | rs12574415 | 11 | 68091265 | C/T | 0.024 | 0.005 |
| rs2194757 | 2 | 10529961 | A/C/G | 0.011 | 0.003 | rs1566734 | 11 | 18281916 | C/T | 0.019 | 0.004 |
| rs4254643 | 2 | 231582797 | T/C | 0.024 | 0.007 | rs174583 | 11 | 48145375 | A/C | 0.039 | 0.008 |
| rs4611596 | 2 | 130255181 | A/G | 0.024 | 0.005 | rs1794072 | 11 | 38428289 | A/G | 0.019 | 0.004 |
| rs4675874 | 2 | 234858801 | G/A | 0.013 | 0.003 | rs1835298 | 11 | 109322914 | C/G | 0.017 | 0.004 |
| rs6436971 | 2 | 198274929 | G/A | 0.066 | 0.015 | rs4491175 | 11 | 61303803 | C/G/T | 0.053 | 0.012 |
| rs6717406 | 2 | 166421763 | A/G | 0.037 | 0.010 | rs4578397 | 11 | 116440678 | A/G | 0.071 | 0.014 |
| rs7569376 | 2 | 48955683 | A/G | 0.022 | 0.005 | rs4938285 | 11 | 112057620 | G/A | 0.043 | 0.009 |
| rs7630111 | 2 | 191344132 | C/T | 0.040 | 0.009 | rs659366 | 11 | 69462910 | G/A | 0.016 | 0.004 |
| rs11128125 | 3 | 61814273 | G/T | 0.040 | 0.008 | rs7117447 | 11 | 101310590 | C/T | 0.082 | 0.017 |
| rs1225051 | 3 | 49971514 | C/A/G/T | 0.012 | 0.003 | rs713278 | 11 | 133532372 | A/G | 0.048 | 0.011 |
| rs12488690 | 3 | 149344615 | C/A | 0.048 | 0.012 | rs741245 | 11 | 133547942 | T/A/G | 0.023 | 0.005 |
| rs1488485 | 3 | 1454063 | C/T | 0.027 | 0.006 | rs770576 | 11 | 90956214 | T/A/C | 0.053 | 0.011 |
| rs1613215 | 3 | 74954560 | A/C/T | 0.039 | 0.009 | rs10858883 | 12 | 41753811 | C/G/T | 0.037 | 0.008 |
| rs16862627 | 3 | 69389614 | T/A/C/G | 0.013 | 0.003 | rs11104947 | 12 | 69906287 | A/G/T | 0.084 | 0.019 |
| rs17008485 | 3 | 17194071 | G/A | 0.014 | 0.003 | rs3217805 | 12 | 4227248 | G/A | 0.073 | 0.017 |
| rs1709621 | 3 | 172389873 | G/C | 0.022 | 0.005 | rs4533076 | 12 | 638166 | C/T | 0.045 | 0.010 |
| rs17582830 | 3 | 114165901 | C/A/T | 0.018 | 0.004 | rs9532080 | 12 | 88942980 | G/A | 0.052 | 0.012 |
| rs4353835 | 3 | 11220006 | C/A | 0.070 | 0.013 | rs9549212 | 12 | 128910954 | A/C/T | 0.038 | 0.009 |
| rs4678169 | 3 | 94033744 | G/C/T | 0.030 | 0.007 | rs10492574 | 13 | 90173515 | A/C/G | 0.055 | 0.012 |
| rs6599390 | 3 | 181497374 | G/A | 0.025 | 0.005 | rs11841589 | 13 | 46695350 | T/C | 0.087 | 0.016 |
| rs9826148 | 3 | 100936248 | C/A/G | 0.039 | 0.009 | rs11841652 | 13 | 43086351 | A/G | 0.011 | 0.002 |
| rs9852677 | 3 | 32451697 | C/A | 0.028 | 0.006 | rs11846710 | 13 | 73814891 | G/T | 0.052 | 0.011 |
| rs9857773 | 3 | 138730737 | C/T | 0.042 | 0.009 | rs12589835 | 13 | 108536028 | C/T | 0.031 | 0.007 |
| rs9860483 | 3 | 130021945 | G/A | 0.081 | 0.016 | rs1322944 | 13 | 41022093 | C/T | 0.048 | 0.011 |
| rs9869826 | 3 | 45900369 | G/A/T | 0.015 | 0.003 | rs1333099 | 13 | 53683163 | A/G | 0.053 | 0.011 |
| rs1026975 | 4 | 99304600 | A/G | 0.025 | 0.006 | rs1751034 | 13 | 47411985 | A/G | 0.019 | 0.004 |
| rs11466640 | 4 | 38138661 | C/T | 0.026 | 0.006 | rs452748 | 13 | 99683635 | T/C | 0.019 | 0.005 |
| rs1522221 | 4 | 71229896 | G/A/T | 0.026 | 0.006 | rs4941430 | 13 | 27534715 | G/A | 0.026 | 0.005 |
| rs16997770 | 4 | 113971374 | T/G | 0.058 | 0.013 | rs9513535 | 13 | 94969796 | T/C | 0.052 | 0.011 |
| rs17016175 | 4 | 143373910 | A/G/T | 0.022 | 0.005 | rs10132336 | 14 | 71262932 | T/C | 0.038 | 0.009 |
| rs17239258 | 4 | 178295766 | G/A/T | 0.027 | 0.007 | rs10134903 | 14 | 88682616 | G/A | 0.023 | 0.005 |
| rs17579988 | 4 | 19044103 | G/A | 0.018 | 0.004 | rs10139575 | 14 | 50481926 | T/C | 0.020 | 0.004 |
| rs17583068 | 4 | 24728480 | G/A | 0.028 | 0.006 | rs10483991 | 14 | 80242132 | T/C/G | 0.050 | 0.011 |
| rs2035023 | 4 | 124693346 | T/C | 0.052 | 0.010 | rs11159882 | 14 | 88006776 | T/G | 0.029 | 0.007 |
| rs2622637 | 4 | 100335874 | C/A | 0.076 | 0.017 | rs11625485 | 14 | 74869017 | A/G/T | 0.054 | 0.011 |
| rs279845 | 4 | 46305733 | T/C | 0.013 | 0.003 | rs1256520 | 14 | 57087118 | A/C/G | 0.049 | 0.011 |
| rs2972336 | 4 | 143391299 | A/G | 0.032 | 0.007 | rs12588061 | 14 | 40942142 | C/G | 0.042 | 0.010 |
| rs3775539 | 4 | 84577025 | T/C | 0.039 | 0.009 | rs174520 | 14 | 65737193 | C/A | 0.053 | 0.012 |
| rs4865142 | 4 | 53278689 | G/A | 0.043 | 0.010 | rs17823795 | 14 | 58343352 | G/A | 0.033 | 0.008 |
| rs639617 | 4 | 178655568 | C/T | 0.030 | 0.006 | rs4902391 | 14 | 65811537 | T/C | 0.045 | 0.010 |
| rs7666030 | 4 | 956047 | A/G | 0.015 | 0.003 | rs8005568 | 14 | 101151012 | C/G/T | 0.040 | 0.009 |
| rs7676014 | 4 | 4346373 | C/T | 0.046 | 0.009 | rs8015594 | 14 | 40865770 | G/A | 0.054 | 0.011 |
| rs10058739 | 5 | 127677188 | T/C | 0.014 | 0.003 | rs10459664 | 15 | 27940339 | A/G | 0.052 | 0.011 |
| rs10806975 | 5 | 167397540 | C/T | 0.011 | 0.002 | rs17822931 | 15 | 84586464 | T/C | 0.037 | 0.009 |
| rs11745587 | 5 | 88188058 | A/C/G | 0.021 | 0.005 | rs2313427 | 15 | 70123826 | T/G | 0.028 | 0.006 |
| rs11959012 | 5 | 165711426 | G/A | 0.035 | 0.008 | rs4486887 | 15 | 35064934 | C/T | 0.033 | 0.007 |
| rs12654905 | 5 | 11984496 | T/A/G | 0.024 | 0.005 | rs11639903 | 16 | 9222411 | A/G | 0.011 | 0.002 |
| rs13160399 | 5 | 144030993 | C/A/T | 0.040 | 0.009 | rs1420288 | 16 | 50677571 | C/T | 0.041 | 0.009 |
| rs1422931 | 5 | 144074365 | G/A | 0.019 | 0.004 | rs160539 | 16 | 63282080 | A/G/T | 0.036 | 0.008 |
| rs1428150 | 5 | 89518433 | A/C | 0.040 | 0.008 | rs2297514 | 16 | 89972345 | T/C | 0.026 | 0.006 |
| rs1468722 | 5 | 143038150 | G/A/C/T | 0.013 | 0.003 | rs2353686 | 16 | 63353953 | C/G | 0.032 | 0.008 |
| rs17076328 | 5 | 169750158 | G/T | 0.013 | 0.003 | rs4280278 | 16 | 29288634 | T/C | 0.047 | 0.010 |
| rs17207681 | 5 | 132490807 | A/G | 0.051 | 0.012 | rs6502840 | 16 | 82273372 | G/A | 0.042 | 0.009 |
| rs17451739 | 5 | 142018424 | T/G | 0.056 | 0.013 | rs8049660 | 16 | 86354605 | T/C | 0.043 | 0.009 |
| rs17599827 | 5 | 65596821 | T/C | 0.059 | 0.014 | rs8060207 | 16 | 54477881 | T/C | 0.039 | 0.009 |
| rs17631488 | 5 | 18680420 | A/T | 0.072 | 0.014 | rs16944149 | 17 | 8135061 | T/C | 0.025 | 0.006 |
| rs2589787 | 5 | 11202649 | C/A/T | 0.035 | 0.008 | rs16966855 | 17 | 26093315 | T/C/G | 0.010 | 0.002 |
| rs304141 | 5 | 79020426 | A/G | 0.076 | 0.016 | rs2068746 | 17 | 1243941 | C/T | 0.021 | 0.005 |
| rs315808 | 5 | 167668843 | C/T | 0.066 | 0.013 | rs2642066 | 17 | 58184540 | T/C | 0.067 | 0.014 |
| rs4912933 | 5 | 132561277 | G/A/C/T | 0.028 | 0.006 | rs9303660 | 17 | 28526475 | T/C | 0.021 | 0.005 |
| rs6898653 | 5 | 110585627 | C/T | 0.045 | 0.009 | rs9941426 | 17 | 75668619 | C/A/G/T | 0.044 | 0.010 |
| rs11042911 | 6 | 33024654 | A/C | 0.036 | 0.007 | rs11086012 | 18 | 76478188 | A/G | 0.015 | 0.003 |
| rs1178148 | 6 | 138406328 | C/A/T | 0.039 | 0.009 | rs1561201 | 18 | 1948608 | A/C/G/T | 0.014 | 0.003 |
| rs12660882 | 6 | 108906200 | C/T | 0.015 | 0.003 | rs17072984 | 18 | 57332158 | T/G | 0.048 | 0.011 |
| rs1539348 | 6 | 63640369 | C/T | 0.011 | 0.002 | rs1708698 | 18 | 28158452 | C/A | 0.014 | 0.003 |
| rs210798 | 6 | 134423175 | A/T | 0.013 | 0.003 | rs2278339 | 18 | 10029988 | G/A/T | 0.031 | 0.009 |
| rs480631 | 6 | 130102959 | C/T | 0.024 | 0.005 | rs2377962 | 18 | 455128 | A/G | 0.076 | 0.017 |
| rs6924957 | 6 | 79659170 | A/G | 0.039 | 0.009 | rs475235 | 18 | 9101722 | G/T | 0.042 | 0.009 |
| rs761798 | 6 | 33047031 | T/C | 0.015 | 0.003 | rs6117562 | 18 | 46454048 | A/G/T | 0.032 | 0.007 |
| rs7740161 | 6 | 120775626 | G/A | 0.016 | 0.004 | rs9964230 | 18 | 852562 | G/A | 0.022 | 0.005 |
| rs258728 | 7 | 8859616 | C/T | 0.023 | 0.005 | rs7268940 | 19 | 39206288 | C/T | 0.037 | 0.008 |
| rs321967 | 7 | 66699784 | T/C | 0.017 | 0.004 | rs8107011 | 19 | 10666112 | G/A/T | 0.030 | 0.007 |
| rs3757419 | 7 | 55238464 | T/A/G | 0.032 | 0.007 | rs16986850 | 20 | 39263421 | C/A | 0.033 | 0.008 |
| rs3757425 | 7 | 125115418 | A/G | 0.034 | 0.007 | rs16991180 | 20 | 2090118 | C/A/T | 0.055 | 0.012 |
| rs3923736 | 7 | 81663275 | C/A | 0.050 | 0.011 | rs17377643 | 20 | 23727145 | C/T | 0.052 | 0.011 |
| rs4718412 | 7 | 66029429 | C/T | 0.060 | 0.013 | rs2837352 | 20 | 12905188 | T/A/C | 0.023 | 0.005 |
| rs6465469 | 7 | 81697666 | A/C | 0.011 | 0.002 | rs6030932 | 20 | 41048928 | T/C | 0.051 | 0.011 |
| rs7006443 | 7 | 155060730 | A/C/G/T | 0.041 | 0.012 | rs6123723 | 20 | 12921966 | T/C | 0.047 | 0.012 |
| rs7794745 | 7 | 141673345 | C/G | 0.012 | 0.003 | rs760873 | 20 | 42146320 | T/G | 0.054 | 0.012 |
| rs7802058 | 7 | 78314549 | G/A/C | 0.014 | 0.003 | rs1524930 | 21 | 37999799 | C/T | 0.033 | 0.007 |
| rs10504726 | 8 | 72774349 | G/A | 0.020 | 0.004 | rs2330015 | 21 | 40287238 | A/C/T | 0.043 | 0.010 |
| rs10957985 | 8 | 80568935 | T/A/C | 0.041 | 0.009 | rs2836749 | 21 | 22447717 | A/G | 0.030 | 0.007 |
| rs12676684 | 8 | 88289450 | G/T | 0.053 | 0.012 | rs6000401 | 21 | 45246422 | A/G | 0.037 | 0.008 |
| rs1517114 | 8 | 18258316 | G/A/T | 0.013 | 0.003 | rs928844 | 21 | 23231832 | C/T | 0.055 | 0.012 |
| rs1537523 | 8 | 119479124 | G/C/T | 0.026 | 0.007 | rs12483769 | 22 | 47271217 | T/C/G | 0.047 | 0.009 |
| rs2945733 | 8 | 10097398 | G/A | 0.044 | 0.010 | rs131864 | 22 | 41537589 | A/G | 0.037 | 0.008 |
| rs2976396 | 8 | 134615750 | T/G | 0.116 | 0.026 | rs17002737 | 22 | 42280618 | T/C | 0.053 | 0.012 |
| rs4922234 | 8 | 9712822 | C/G/T | 0.013 | 0.003 | rs17129041 | 22 | 24825511 | C/A/T | 0.022 | 0.005 |
| rs10521076 | 9 | 104423907 | C/T | 0.054 | 0.012 | rs198464 | 22 | 49595560 | C/T | 0.028 | 0.006 |
| rs10991718 | 9 | 89140339 | G/A | 0.025 | 0.006 | rs2269658 | 22 | 42241372 | G/A/T | 0.038 | 0.008 |
| rs12006467 | 9 | 29780195 | G/A | 0.071 | 0.014 | rs229562 | 22 | 22536956 | C/T | 0.056 | 0.012 |
| rs12351269 | 9 | 3430684 | T/C | 0.037 | 0.008 | rs4820428 | 22 | 37149336 | C/T | 0.029 | 0.008 |
| rs1875174 | 9 | 136079463 | C/T | 0.025 | 0.005 | rs5770018 | 22 | 48282309 | G/A/C | 0.014 | 0.003 |
| rs1981500 | 9 | 117032448 | T/C | 0.028 | 0.007 | rs6007756 | 22 | 37599065 | G/T | 0.030 | 0.007 |
| rs2636864 | 9 | 92800165 | G/C | 0.038 | 0.008 | rs860236 | 22 | 42605548 | C/T | 0.016 | 0.006 |
| [1] |
Phillips C. Forensic genetic analysis of bio-geographical ancestry. Forensic Sci Int Genet, 2015, 18: 49-65.
doi: 10.1016/j.fsigen.2015.05.012 |
| [2] |
Tishkoff SA, Kidd KK. Implications of biogeography of human populations for 'race' and medicine. Nat Genet, 2004, 36(< W>11 Suppl): S21-S27.
doi: 10.1038/ng1438 |
| [3] |
Marchini J, Cardon LR, Phillips MS, Donnelly P. The effects of human population structure on large genetic association studies. Nat Genet, 2004, 36(5): 512-517.
pmid: 15052271 |
| [4] |
Paschou P, Lewis J, Javed A, Drineas P. Ancestry informative markers for fine-scale individual assignment to worldwide populations. J Med Genet, 2010, 47(12): 835-847.
doi: 10.1136/jmg.2010.078212 |
| [5] |
Phillips C, Salas A, Sánchez JJ, Fondevila M, Gómez-Tato A, Alvarez-Dios J, Calaza M, de Cal MC, Ballard D, Lareu MV, Carracedo A. Inferring ancestral origin using a single multiplex assay of ancestry-informative marker SNPs. Forensic Sci Int Genet, 2007, 1(3-4): 273-280.
doi: 10.1016/j.fsigen.2007.06.008 |
| [6] |
Kidd KK, Speed WC, Pakstis AJ, Furtado MR, Fang RX, Madbouly A, Maiers M, Middha M, Friedlaender FR, Kidd JR. Progress toward an efficient panel of SNPs for ancestry inference. Forensic Sci Int Genet, 2014, 10: 23-32.
doi: 10.1016/j.fsigen.2014.01.002 |
| [7] | Jiang L, Sun QF, Ma Q, Zhao WT, Liu J, Zhao L, Ji AQ, Li CX. Optimization and validation of analysis method based on 27-plex SNP panel for ancestry inference. Hereditas (Beijing), 2017, 39(2): 166-173. |
| 江丽, 孙启凡, 马泉, 赵雯婷, 刘京, 赵蕾, 季安全, 李彩霞. 27-plex SNP 种族推断方法的优化及验证. 遗传, 2017, 39(2): 166-173. | |
| [8] |
Qin PF, Li ZQ, Jin WF, Lu DS, Lou HY, Shen JW, Jin L, Shi YY, Xu SH. A panel of ancestry informative markers to estimate and correct potential effects of population stratification in Han Chinese. Eur J Hum Genet, 2014, 22(2): 248-253.
doi: 10.1038/ejhg.2013.111 pmid: 23714748 |
| [9] |
Wang YC, Lu DS, Chung YJ, Xu SH. Genetic structure, divergence and admixture of Han Chinese, Japanese and Korean populations. Hereditas, 2018, 155: 19.
doi: 10.1186/s41065-018-0057-5 pmid: 29636655 |
| [10] |
Shi CM, Liu Q, Zhao SL, Chen H. Ancestry informative SNP panels for discriminating the major East Asian populations: Han Chinese, Japanese and Korean. Ann Hum Genet, 2019, 83(5): 348-354.
doi: 10.1111/ahg.12320 |
| [11] |
Wang CC, Yeh HY, Popov AN, Zhang HQ, Matsumura H, Sirak K, Cheronet O, Kovalev A, Rohland N, Kim AM, Mallick S, Bernardos R, Tumen D, Zhao J, Liu YC, Liu JY, Mah M, Wang K, Zhang Z, Adamski N, Broomandkhoshbacht N, Callan K, Candilio F, Carlson KSD, Culleton BJ, Eccles L, Freilich S, Keating D, Lawson AM, Mandl K, Michel M, Oppenheimer J, Özdoğan KT, Stewardson K, Wen SQ, Yan S, Zalzala F, Chuang R, Huang CJ, Looh H, Shiung CC, Nikitin YG, Tabarev AV, Tishkin AA, Lin S, Sun ZY, Wu XM, Yang TL, Hu X, Chen L, Du H, Bayarsaikhan J, Mijiddorj E, Erdenebaatar D, Iderkhangai TO, Myagmar E, Kanzawa-Kiriyama H, Nishino M, Shinoda KI, Shubina OA, Guo J, Cai WW, Deng QY, Kang LL, Li D, Li DW, Lin RM, Nini, Shrestha R, Wang LX, Wei LW, Xie GM, Yao HB, Zhang MF, He GL, Yang XM, Hu R, Robbeets M, Schiffels S, Kennett DJ, Jin L, Li H, Krause J, Pinhasi R, Reich D. Genomic insights into the formation of human populations in East Asia. Nature, 2021, 591(7850): 413-419.
doi: 10.1038/s41586-021-03336-2 |
| [12] |
Jung JY, Kang PW, Kim E, Chacon D, Beck D, McNevin D. Ancestry informative markers (AIMs) for Korean and other East Asian and South East Asian populations. Int J Legal Med, 2019, 133(6): 1711-1719.
doi: 10.1007/s00414-019-02129-7 pmid: 31388795 |
| [13] |
Okada Y, Momozawa Y, Sakaue S, Kanai M, Ishigaki K, Akiyama M, Kishikawa T, Arai Y, Sasaki T, Kosaki K, Suematsu M, Matsuda K, Yamamoto K, Kubo M, Hirose N, Kamatani Y. Deep whole-genome sequencing reveals recent selection signatures linked to evolution and disease risk of Japanese. Nat Commun, 2018, 9(1): 1631.
doi: 10.1038/s41467-018-03274-0 pmid: 29691385 |
| [14] |
Akiyama M, Okada Y, Kanai M, Takahashi A, Momozawa Y, Ikeda M, Iwata N, Ikegawa S, Hirata M, Matsuda K, Iwasaki M, Yamaji T, Sawada N, Hachiya T, Tanno K, Shimizu A, Hozawa A, Minegishi N, Tsugane S, Yamamoto M, Kubo M, Kamatani Y. Genome-wide association study identifies 112 new loci for body mass index in the Japanese population. Nat Genet, 2017, 49(10): 1458-1467.
doi: 10.1038/ng.3951 pmid: 28892062 |
| [15] |
Liu SY, Huang SJ, Chen F, Zhao LJ, Yuan YY, Francis SS, Fang L, Li ZL, Lin L, Liu R, Zhang Y, Xu HX, Li SK, Zhou YW, Davies RW, Liu Q, Walters RG, Lin K, Ju J, Korneliussen T, Yang MA, Fu QM, Wang J, Zhou LJ, Krogh A, Zhang HY, Wang W, Chen ZM, Cai ZM, Yin Y, Yang HM, Mao M, Shendure J, Wang J, Albrechtsen A, Jin X, Nielsen R, Xu X. Genomic analyses from non-invasive prenatal testing reveal genetic associations, patterns of viral infections, and Chinese population history. Cell, 2018, 175(2): 347-359.
doi: S0092-8674(18)31032-8 pmid: 30290141 |
| [16] |
Xu SH, Yin XY, Li SL, Jin WF, Lou HY, Yang L, Gong XH, Wang HY, Shen YP, Pan XD, He YG, Yang YJ, Wang Y, Fu WQ, An Y, Wang JC, Tan JZ, Qian J, Chen XL, Zhang X, Sun YF, Zhang XJ, Wu BL, Jin L. Genomic dissection of population substructure of Han Chinese and its implication in association studies. Am J Hum Genet, 2009, 85(6): 762-774.
doi: 10.1016/j.ajhg.2009.10.015 |
| [17] |
Jeon S, Bhak Y, Choi Y, Jeon Y, Kim S, Jang J, Jang J, Blazyte A, Kim C, Kim Y, Shim J, Kim N, Kim YJ, Park SG, Kim J, Cho YS, Park Y, Kim HM, Kim BC, Park NH, Shin ES, Kim BC, Bolser D, Manica A, Edwards JS, Church G, Lee S, Bhak J. Korean genome project: 1094 Korean personal genomes with clinical information. Sci Adv, 2020, 6(22): eaaz7835.
doi: 10.1126/sciadv.aaz7835 |
| [18] |
Cao YN, Li L, Xu M, Feng ZM, Sun XH, Lu JL, Xu Y, Du PN, Wang TG, Hu RY, Ye Z, Shi LX, Tang XL, Yan L, Gao ZN, Chen G, Zhang YF, Chen LL, Ning G, Bi YF, Wang WQ, Consortium C. The ChinaMAP analytics of deep whole genome sequences in 10,588 individuals. Cell Res, 2020, 30(9): 717-731.
doi: 10.1038/s41422-020-0322-9 pmid: 32355288 |
| [19] |
Jinam TA, Kanzawa-Kiriyama H, Inoue I, Tokunaga K, Omoto K, Saitou N. Unique characteristics of the Ainu population in Northern Japan. J Hum Genet, 2015, 60(10): 565-571.
doi: 10.1038/jhg.2015.79 pmid: 26178428 |
| [20] |
Kim JJ, Verdu P, Pakstis AJ, Speed WC, Kidd JR, Kidd KK. Use of autosomal loci for clustering individuals and populations of East Asian origin. Hum Genet, 2005, 117(6): 511-519.
doi: 10.1007/s00439-005-1334-8 |
| [21] | Clarke L, Fairley S, Zheng-Bradley X, Streeter I, Perry E, Lowy E, Tassé A-M, and Flicek P. The international genome sample resource (IGSR): a worldwide collection of genome variation incorporating the 1000 genomes project data. Nucleic Acids Res, 2016, 45(1): 854-859. |
| [22] |
Zhang WQ, Meehan J, Su ZQ, Ng HW, Shu M, Luo H, Ge WG, Perkins R, Tong WD, Hong HX. Whole genome sequencing of 35 individuals provides insights into the genetic architecture of Korean population. BMC Bioinformatics, 2014, 15(11): 6-18.
doi: 10.1186/1471-2105-15-6 |
| [23] |
Byrska-Bishop M, Evani US, Zhao XF, Basile AO, Abel HJ, Regier AA, Corvelo A, Clarke WE, Musunuri R, Nagulapalli K, Fairley S, Runnels A, Winterkorn L, Lowy E, Flicek P, Germer S, Brand H, Hall IM, Talkowski ME, Narzisi G, Zody MC, The Human Genome Structural Variation Consortium. High coverage whole genome sequencing of the expanded 1000 Genomes Project cohort including 602 trios. bioRxiv, 2021, doi: 10.1101/2021.02.06.430068.
doi: 10.1101/2021.02.06.430068 |
| [24] |
Bergström A, McCarthy SA, Hui RY, Almarri MA, Ayub Q, Danecek P, Chen Y, Felkel S, Hallast P, Kamm J, Blanché H, Deleuze JF, Cann H, Mallick S, Reich D, Sandhu MS, Skoglund P, Scally A, Xue YL, Durbin R, Tyler-Smith C. Insights into human genetic variation and population history from 929 diverse genomes. Science, 2020, 367(6484): eaay5012.
doi: 10.1126/science.aay5012 |
| [25] | Mallick S, Li H, Lipson M, Mathieson I, Gymrek M, Racimo F, Zhao MY, Chennagiri N, Nordenfelt S, Tandon A, Skoglund P, Lazaridis I, Sankararaman S, Fu QM, Rohland N, Renaud G, Erlich Y, Willems T, Gallo C, Spence JP, Song YS, Poletti G, Balloux F, van Driem G, de Knijff P, Romero IG, Jha AR, Behar DM, Bravi CM, Capelli C, Hervig T, Moreno-Estrada A, Posukh OL, Balanovska E, Balanovsky O, Karachanak-Yankova S, Sahakyan H, Toncheva D, Yepiskoposyan L, Tyler-Smith C, Xue YL, Abdullah MS, Ruiz-Linares A, Beall CM, Di Rienzo A, Jeong C, Starikovskaya EB, Metspalu E, Parik J, Villems R, Henn BM, Hodoglugil U, Mahley R, Sajantila A, Stamatoyannopoulos G, Wee JTS, Khusainova R, Khusnutdinova E, Litvinov S, Ayodo G, Comas D, Hammer MF, Kivisild T, Klitz W, Winkler CA, Labuda D, Bamshad M, Jorde LB, Tishkoff SA, Watkins WS, Metspalu M, Dryomov S, Sukernik R, Singh L, Thangaraj K, Pääbo S, Kelso J, Patterson N, Reich D. The Simons genome diversity project: 300 genomes from 142 diverse populations. Nature, 2016, 538(7624): 201-206. |
| [26] |
Liu XY, Lu DS, Saw WY, Shaw PJ, Wangkumhang P, Ngamphiw C, Fucharoen S, Lert-Itthiporn W, Chin- Inmanu K, Chau TNB, Anders K, Kasturiratne A, de Silva HJ, Katsuya T, Kimura R, Nabika T, Ohkubo T, Tabara Y, Takeuchi F, Yamamoto K, Yokota M, Mamatyusupu D, Yang WJ, Chung YJ, Jin L, Hoh BP, Wickremasinghe AR, Ong RH, Khor CC, Dunstan SJ, Simmons C, Tongsima S, Suriyaphol P, Kato N, Xu SH, Teo YY. Characterising private and shared signatures of positive selection in 37 Asian populations. Eur J Hum Genet, 2017, 25(4): 499-508.
doi: 10.1038/ejhg.2016.181 pmid: 28098149 |
| [27] | Wen H, Wei YL, Guo XY, Sun CC, Xue SY, Liu J, Fan H, Jiang L. High-resolution SNP ancestry inference model and efficiency evaluation in three East Asian populations. Prog Biochem Biophys, 2021, 48(8): 973-981. |
| 文豪, 魏以梁, 郭晓媛, 孙昌春, 薛思瑶, 刘京, 范虹, 江丽. 东亚三族群SNP高分辨推断模型构建与效能评估. 生物化学与生物物理进展, 2021, 48(8): 973-981. | |
| [28] | Guo XY, Sun CC, Xue SY, Zhao H, Jiang L, Li CX. 49AISNP: a study on the ancestry inference of the three ethnic groups in the north of East Asia. Hereditas (Beijing), 2021, 43(9): 880-889. |
| 郭晓媛, 孙昌春, 薛思瑶, 赵慧, 江丽, 李彩霞. 49AISNP:东亚北方三个族群遗传来源推断. 遗传, 2021, 43(9): 880-889. | |
| [29] |
Kim T, Seo HD, Hennighausen L, Lee D, Kang K. Octopus-toolkit: a workflow to automate mining of public epigenomic and transcriptomic next-generation sequencing data. Nucleic Acids Res, 2018, 46(9): 53-58.
doi: 10.1093/nar/gky083 pmid: 29420797 |
| [30] | 1000 Genomes Project Consortium, Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, Korbel JO, Marchini JL, McCarthy S, McVean GA, Abecasis GR. A global reference for human genetic variation. Nature, 2015, 526(7571): 68-74. |
| [31] |
Sudmant PH, Rausch T, Gardner EJ, Handsaker RE, Abyzov A, Huddleston J, Zhang Y, Ye K, Jun G, Fritz MHY, Konkel MK, Malhotra A, Stütz AM, Shi XH, Casale FP, Chen JM, Hormozdiari F, Dayama G, Chen K, Malig M, Chaisson MJP, Walter K, Meiers S, Kashin S, Garrison E, Auton A, Lam HYK, Mu XJ, Alkan C, Antaki D, Bae T, Cerveira E, Chines P, Chong ZC, Clarke L, Dal E, Ding L, Emery S, Fan X, Gujral M, Kahveci F, Kidd JM, Kong Y, Lameijer EW, McCarthy S, Flicek P, Gibbs RA, Marth G, Mason CE, Menelaou A, Muzny DM, Nelson BJ, Noor A, Parrish NF, Pendleton M, Quitadamo A, Raeder B, Schadt EE, Romanovitch M, Schlattl A, Sebra R, Shabalin AA, Untergasser A, Walker JA, Wang M, Yu FL, Zhang C, Zhang J, Zheng-Bradley XQ, Zhou WD, Zichner T, Sebat J, Batzer MA, McCarroll SA, 1000 Genomes Project Consortium, Mills RE, Gerstein MB, Bashir A, Stegle O, Devine SE, Lee C, Eichler EE, Korbel JO. An integrated map of structural variation in 2,504 human genomes. Nature, 2015, 526(7571): 75-81.
doi: 10.1038/nature15394 |
| [32] |
Korn JM, Kuruvilla FG, McCarroll SA, Wysoker A, Nemesh J, Cawley S, Hubbell E, Veitch J, Collins PJ, Darvishi K, Lee C, Nizzari MM, Gabriel SB, Purcell S, Daly MJ, Altshuler D. Integrated genotype calling and association analysis of SNPs, common copy number polymorphisms and rare CNVs. Nat Genet, 2008, 40(10): 1253-1260.
doi: 10.1038/ng.237 pmid: 18776909 |
| [33] | Van der Auwera GA, O'Connor BD. Genomics in the Cloud: Using Docker, GATK, and WDL in Terra. 2020: O'Reilly Media, Incorporated. |
| [34] | Meire M, Ballings M, Van den Poel D. imputeMissings: impute missing values in a predictive context. 2016. |
| [35] | Rustowicz R. Crop classification with multi-temporal satellite imagery. 2017. |
| [36] | Breiman L, Cutler A, Liaw A, Wiener M. Package ‘randomForest’. 2018. |
| [37] |
Yu GC, Smith DK, Zhu HC, Guan Y, Lam TTY. ggtree: an R package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol Evol, 2017, 8(1): 28-36.
doi: 10.1111/2041-210X.12628 |
| [38] |
Hao W, Storey JD. Extending tests of Hardy-Weinberg equilibrium to structured populations. Genetics, 2019, 213(3): 759-770.
doi: 10.1534/genetics.119.302370 |
| [39] |
Pritchard JK, Przeworski M. Linkage disequilibrium in humans: models and data. Am J Hum Genet, 2001, 69(1): 1-14.
doi: 10.1086/321275 |
| [40] |
Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics, 2005, 21(2): 263-265.
pmid: 15297300 |
| [41] |
Armstrong RA. When to use the Bonferroni correction. Ophthalmic Physiol Opt, 2014, 34(5): 502-508.
doi: 10.1111/opo.12131 |
| [42] |
Boca SM, Rosenberg NA. Mathematical properties of Fst between admixed populations and their parental source populations. Theor Popul Biol, 2011, 80(3): 208-216.
doi: 10.1016/j.tpb.2011.05.003 pmid: 21640742 |
| [43] |
Rosenberg NA, Li LM, Ward R, Pritchard JK. Informativeness of genetic markers for inference of ancestry. Am J Hum Genet, 2003, 73(6): 1402-1422.
doi: 10.1086/380416 |
| [44] | Hui SB, Wang WJ. Improvement of multi-variable's redundant attributes in classification algorithm of support vector machines. Computer Engineering and Design, 2006, 27(8): 1385-138. |
| 惠守博, 王文杰. 支持向量机分类算法中多元变量共线性问题的改进. 计算机工程与设计, 2006, 27(8): 1385- 1388. | |
| [45] | Zhao YD, Liu R, Liu YL, Xiao F, Zhang Y. Multivariate logistic regression collinearity diagnosis analysis. Chinese Journal of Health Statistics, 2000, (5): 3-5. |
| 赵宇东, 刘嵘, 刘延龄, 肖峰, 张扬. 多元logistic回归的共线性分析. 中国卫生统计, 2000, (5): 3-5. | |
| [46] | Wang L, Tong X, Sheng MW, Qin HD, Tang QS. Review of image classification based on softmax classifier in deep learning. Navigation and Control, 2019, 18(6): 1-9+47. |
| 万磊, 佟鑫, 盛明伟, 秦洪德, 唐松奇. Softmax分类器深度学习图像分类方法应用综述. 导航与控制, 2019, 18(6): 1-9+47. | |
| [47] |
Rigatti SJ. Random Forest. J Insur Med, 2017, 47(1): 31-39.
doi: 10.17849/insm-47-01-31-39.1 pmid: 28836909 |
| [48] |
Heo J, Yoon JG, Park H, Kim YD, Nam HS, Heo JH. Machine learning-based model for prediction of outcomes in acute stroke. Stroke, 2019, 50(5): 1263-1265.
doi: 10.1161/STROKEAHA.118.024293 pmid: 30890116 |
| [49] |
Che DS, Liu Q, Rasheed K, Tao XP. Decision tree and ensemble learning algorithms with their applications in bioinformatics. Adv Exp Med Biol, 2011, 696: 191-199.
doi: 10.1007/978-1-4419-7046-6_19 pmid: 21431559 |
| [50] |
Connor CW. Artificial intelligence and machine learning in anesthesiology. Anesthesiology, 2019, 131(6): 1346-1359.
doi: 10.1097/ALN.0000000000002694 pmid: 30973516 |
| [51] |
Pandis N. Linear regression. Am J Orthod Dentofacial Orthop, 2016, 149(3): 431-434.
doi: 10.1016/j.ajodo.2015.11.019 |
| [52] |
LaValley MP. Logistic regression. Circulation, 2008, 117(18): 2395-2399.
doi: 10.1161/CIRCULATIONAHA.106.682658 pmid: 18458181 |
| [53] | Huang SJ, Cai NG, Pacheco PP, Narrandes S, Wang Y, Xu W. Applications of support vector machine (SVM) learning in cancer genomics. Cancer Genomics Proteomics, 2018, 15(1): 41-51. |
| [54] |
Karalis G. Decision trees and applications. Adv Exp Med Biol, 2020, 1194: 239-242.
doi: 10.1007/978-3-030-32622-7_21 pmid: 32468539 |
| [55] |
Hatwell J, Gaber MM, Atif Azad RM. Ada-WHIPS: explaining AdaBoost classification with applications in the health sciences. BMC Med Inform Decis Mak, 2020, 20(1): 250.
doi: 10.1186/s12911-020-01201-2 |
| [56] |
Wen J, Xu Y, Li ZY, Ma ZL, Xu YR. Inter-class sparsity based discriminative least square regression. Neural Netw, 2018, 102: 36-47.
doi: 10.1016/j.neunet.2018.02.002 |
| [57] |
Kloumann IM, Ugander J, Kleinberg J. Block models and personalized PageRank. Proc Natl Acad Sci USA, 2017, 114(1): 33-38.
doi: 10.1073/pnas.1611275114 |
| [58] |
Jung Y, Hu JH. A k-fold averaging cross-validation procedure. J Nonparametr Stat, 2015, 27(2): 167-179.
doi: 10.1080/10485252.2015.1010532 |
| [59] | Liu J, Li S, Jiang L, Zhao L, Zhao WT, Feng L, Liu HB, Ji AQ, Li CX. DNA ancestry analyzer: an automatic program for ancestry inference of unknown individuals. Life Sci Res, 2018, 22(1): 3-7+41. |
| 刘京, 李盛, 江丽, 赵蕾, 赵雯婷, 丰蕾, 刘海渤, 季安全, 李彩霞. 对于未知来源个体进行族群推断的自动分析系统. 生命科学研究, 2018, 22(1): 3-7+41. | |
| [60] |
Ringnér M. What is principal component analysis? Nat Biotechnol, 2008, 26(3): 303-304.
doi: 10.1038/nbt0308-303 pmid: 18327243 |
| [61] |
Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics, 2000, 155(2): 945-959.
doi: 10.1093/genetics/155.2.945 |
| [62] | Cai LJ. The technical means of forensic material evidence identification in criminal investigation cases-DNA identification technology. Legality Vision, 2018, (34): 177. |
| 蔡立君. 刑侦案件中法医物证鉴定的技术手段——DNA鉴定技术. 法制博览, 2018, (34): 177. | |
| [63] | Jiang L, Zhao L, Liu J, Zhao WT, Ma Q, Zhao H, Ji AQ, Li CX. DNA ancestry inference assisting to have a case solved. Forensic Science and Technology, 2019, 44(4): 371-373. |
| 江丽, 赵蕾, 刘京, 赵雯婷, 马泉, 赵慧, 季安全, 李彩霞. DNA供者族群推断技术在案件中的应用. 刑事技术, 2019, 44(4): 371-373. | |
| [64] |
Charilaou P, Battat R. Machine learning models and over-fitting considerations. World J Gastroenterol, 2022, 28(5): 605-607.
doi: 10.3748/wjg.v28.i5.605 |
| [65] |
Dizaji KG, Chen W, Huang H. Deep large-scale multitask learning network for gene expression inference. J Comput Biol, 2021, 28(5): 485-500.
doi: 10.1089/cmb.2020.0438 pmid: 34014778 |
| [1] | Xiaoyuan Guo, Changchun Sun, Siyao Xue, Hui Zhao, Li Jiang, Caixia Li. 49AISNP: a study on the ancestry inference of the three ethnic groups in the north of East Asia [J]. Hereditas(Beijing), 2021, 43(9): 880-889. |
| [2] | Haoyu Wang, Yuhan Hu, Yueyan Cao, Qiang Zhu, Yuguo Huang, Xi Li, Ji Zhang. AI-SNPs screening based on the whole genome data and research on genetic structure differences of subcontinent populations [J]. Hereditas(Beijing), 2021, 43(10): 938-948. |
| [3] | Xi Li, Haoyu Wang, Yueyan Cao, Qiang Zhu, Panyin Shu, Tingyun Hou, Yuting Wang, Ji Zhang. Forensic genomics research on microhaplotypes [J]. Hereditas(Beijing), 2021, 43(10): 962-971. |
| [4] | Zhiyong Liu, Riga Wu, Ran Li, Qiangwei Wang, Hongyu Sun. Ethical issues of the research and practice in forensic genetics [J]. Hereditas(Beijing), 2021, 43(10): 994-1002. |
| [5] | Yali Hu, Rui Dai, Yongxin Liu, Jingying Zhang, Bin Hu, Chengcai Chu, Huaibo Yuan, Yang Bai. Analysis of rice root bacterial microbiota of Nipponbare and IR24 [J]. Hereditas(Beijing), 2020, 42(5): 506-518. |
| [6] | Yang Liu, Changchun Sun, Mi Ma, Ling Wang, Wenting Zhao, Quan Ma, Anquan Ji, Jing Liu, Caixia Li. The ancestry inference of Chinese populations using 74-plex SNPs system [J]. Hereditas(Beijing), 2020, 42(3): 296-308. |
| [7] | Zhang Guishan, Yang Yong, Zhang Lingmin, Dai Xianhua. Application of machine learning in the CRISPR/Cas9 system [J]. Hereditas(Beijing), 2018, 40(9): 704-723. |
| [8] | Zhao Xuetong, Yang Yadong, Qu Hongzhu, Fang Xiangdong. Applications of machine learning in clinical decision support in the omic era [J]. Hereditas(Beijing), 2018, 40(9): 693-703. |
| [9] | Zhe-ye Peng,Zi-jun Tang,Min-zhu Xie. Research progress in machine learning methods for gene-gene interaction detection [J]. Hereditas(Beijing), 2018, 40(3): 218-226. |
| [10] | Jiaoyang Tian, Yuchun Li, Qingpeng Kong, Yaping Zhang. The origin and evolution history of East Asian populations from genetic perspectives [J]. Hereditas(Beijing), 2018, 40(10): 814-824. |
| [11] | Li Jiang,Qifan Sun,Quan Ma,Wenting Zhao,Jing Liu,Lei Zhao,Anquan Ji,Caixia Li. Optimization and validation of analysis method based on 27-plex SNP panel for ancestry inference [J]. Hereditas(Beijing), 2017, 39(2): 166-173. |
| [12] | Xiuyan Ruan, Weini Wang, Yaran Yang, Bingbing Xie, Jing Chen, Yacheng Liu, Jiangwei Yan. Genetic variability and phylogenetic analysis of 39 short tandem repeat loci in Beijing Han population [J]. HEREDITAS(Beijing), 2015, 37(7): 683-691. |
| [13] | HOU Yan-Yan, YING Xiao-Min, LI Wu-Ju . Computational approaches to microRNA discovery [J]. HEREDITAS, 2008, 30(6): 687-696. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||