Hereditas(Beijing) ›› 2021, Vol. 43 ›› Issue (10): 962-971.doi: 10.16288/j.yczz.21-186
• Research Article • Previous Articles Next Articles
Forensic genomics research on microhaplotypes
Xi Li( ), Haoyu Wang(
), Haoyu Wang( ), Yueyan Cao, Qiang Zhu, Panyin Shu, Tingyun Hou, Yuting Wang, Ji Zhang(
), Yueyan Cao, Qiang Zhu, Panyin Shu, Tingyun Hou, Yuting Wang, Ji Zhang( )
)
- West China School of Basic Medical Sciences & Forensic Medicine, Sichuan University, Chengdu 610041, China
-
Received:2021-05-26Revised:2021-07-29Online:2021-10-20Published:2021-08-10 -
Contact:Zhang Ji E-mail:lixi1105@foxmail.com;wanghy0707@gmail.com;zhangj@scu.edu.cn -
Supported by:Supported by the National Natural Science Foundation of China Nos(81571861);Supported by the National Natural Science Foundation of China Nos(81630054)
Cite this article
Xi Li, Haoyu Wang, Yueyan Cao, Qiang Zhu, Panyin Shu, Tingyun Hou, Yuting Wang, Ji Zhang. Forensic genomics research on microhaplotypes[J]. Hereditas(Beijing), 2021, 43(10): 962-971.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
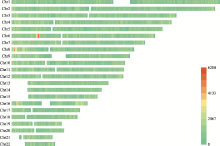
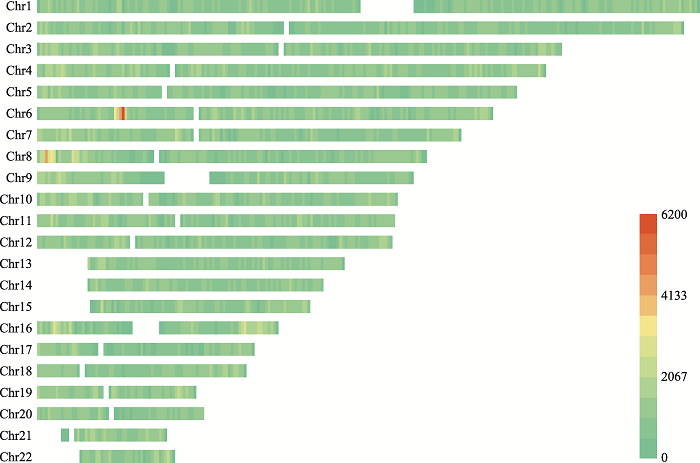
Table 1
The number of SNPs and MHs on different chromosomes"
| 染色体 | #SNPs a | #MHs≤350 bp | #MHs≤150 bp | #MHs≤100 bp | #MHs≤50 bp | ||||
|---|---|---|---|---|---|---|---|---|---|
| A | B | A | B | A | B | A | B | ||
| 1 | 463,261 | 684,624 | 224,137 | 307,923 | 160,070 | 211,609 | 127,357 | 112,562 | 80,220 |
| 2 | 485,172 | 697,551 | 235,330 | 312,548 | 167,213 | 213,273 | 132,583 | 113,234 | 82,973 |
| 3 | 429,260 | 648,976 | 208,260 | 289,892 | 149,992 | 197,993 | 119,687 | 104,404 | 75,230 |
| 4 | 444,134 | 719,321 | 214,966 | 322,127 | 158,179 | 220,652 | 127,643 | 116,157 | 81,220 |
| 5 | 369,559 | 536,677 | 179,135 | 240,372 | 127,576 | 164,475 | 101,232 | 87,636 | 63,535 |
| 6 | 406,810 | 855,491 | 197,195 | 380,915 | 145,792 | 258,670 | 118,720 | 134,514 | 77,942 |
| 7 | 357,021 | 560,129 | 172,793 | 252,329 | 125,618 | 172,901 | 100,917 | 91,761 | 64,047 |
| 8 | 323,908 | 538,902 | 157,309 | 239,578 | 116,180 | 163,404 | 93,911 | 85,902 | 60,124 |
| 9 | 264,750 | 408,354 | 128,051 | 183,076 | 94,273 | 125,067 | 75,556 | 65,695 | 47,502 |
| 10 | 310,578 | 493,463 | 150,444 | 221,083 | 109,703 | 151,560 | 88,309 | 80,883 | 56,820 |
| 11 | 290,938 | 445,661 | 140,840 | 199,579 | 102,589 | 136,071 | 81,970 | 71,787 | 52,025 |
| 12 | 287,513 | 430,602 | 139,153 | 194,229 | 100,173 | 133,241 | 79,814 | 70,791 | 50,312 |
| 13 | 217,352 | 335,132 | 105,264 | 150,042 | 76,429 | 102,775 | 61,026 | 54,319 | 38,801 |
| 14 | 194,482 | 293,076 | 94,194 | 131,568 | 67,706 | 90,024 | 53,957 | 47,739 | 34,160 |
| 15 | 176,222 | 274,166 | 84,933 | 123,892 | 61,570 | 85,058 | 49,433 | 45,461 | 31,845 |
| 16 | 187,593 | 331,113 | 90,597 | 148,132 | 68,613 | 101,035 | 56,023 | 53,349 | 36,743 |
| 17 | 155,592 | 237,431 | 74,611 | 108,076 | 54,056 | 74,898 | 43,371 | 40,510 | 27,684 |
| 18 | 172,512 | 267,593 | 83,271 | 121,279 | 60,482 | 83,014 | 48,506 | 43,974 | 30,786 |
| 19 | 142,771 | 254,075 | 67,813 | 117,069 | 51,542 | 81,348 | 42,076 | 44,002 | 27,689 |
| 20 | 127,126 | 186,986 | 61,427 | 84,197 | 44,252 | 57,717 | 35,181 | 30,649 | 22,301 |
| 21 | 86,049 | 140,790 | 41,396 | 63,550 | 31,033 | 43,662 | 25,070 | 22,955 | 16,084 |
| 22 | 85,052 | 149,962 | 40,808 | 68,111 | 30,586 | 47,028 | 24,982 | 25,065 | 16,427 |
| 总计 | 5,977,655 | 9,490,075 | 2,891,927 | 4,259,567 | 2,103,627 | 2,915,475 | 1,687,324 | 1,543,349 | 1,074,470 |
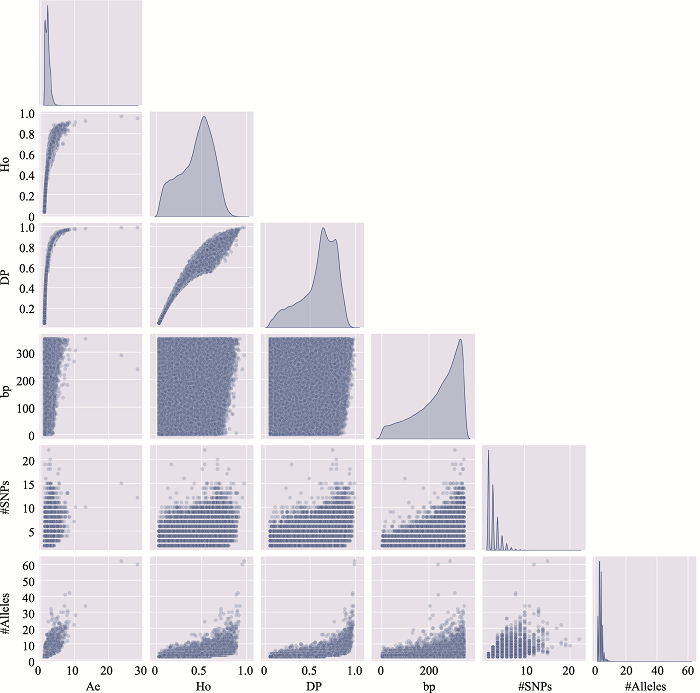
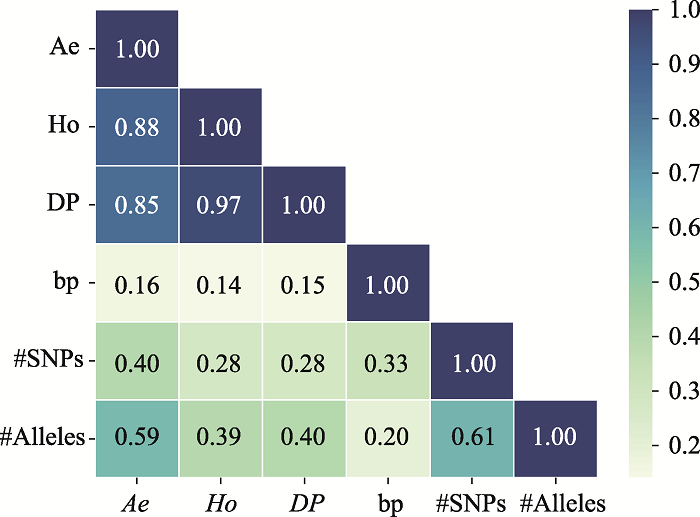
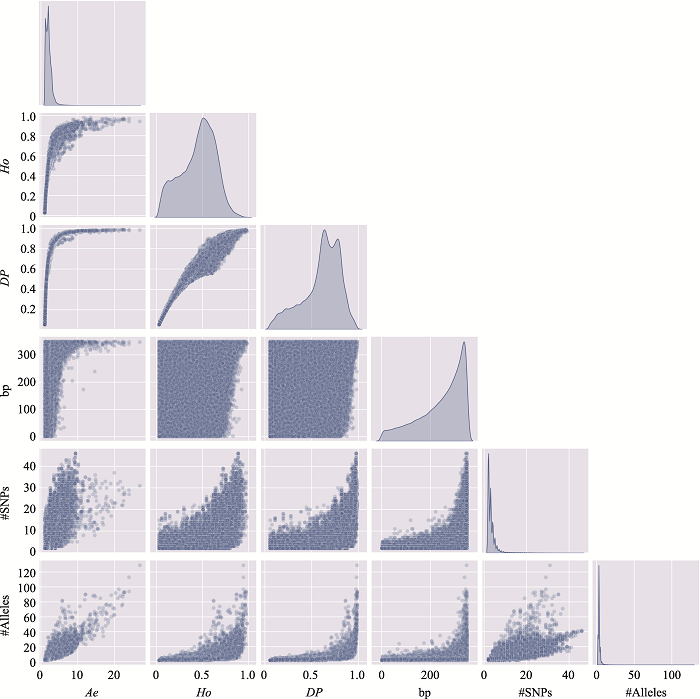
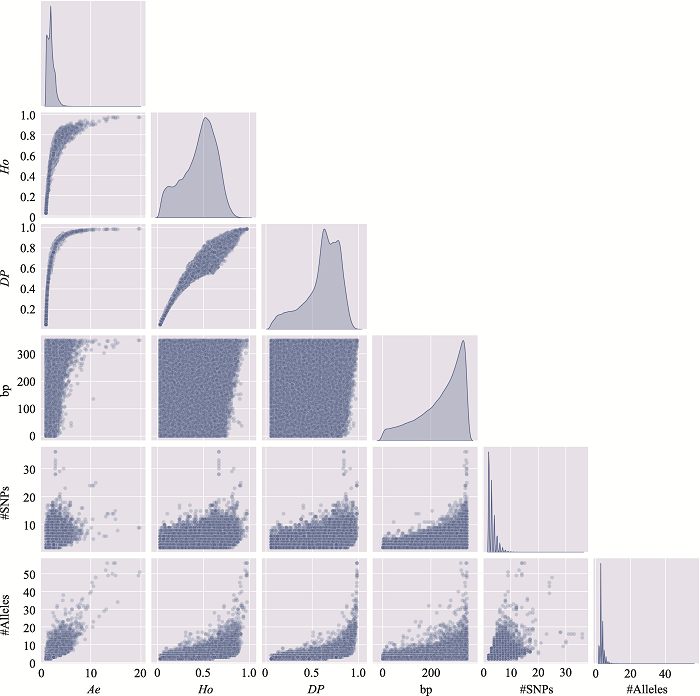
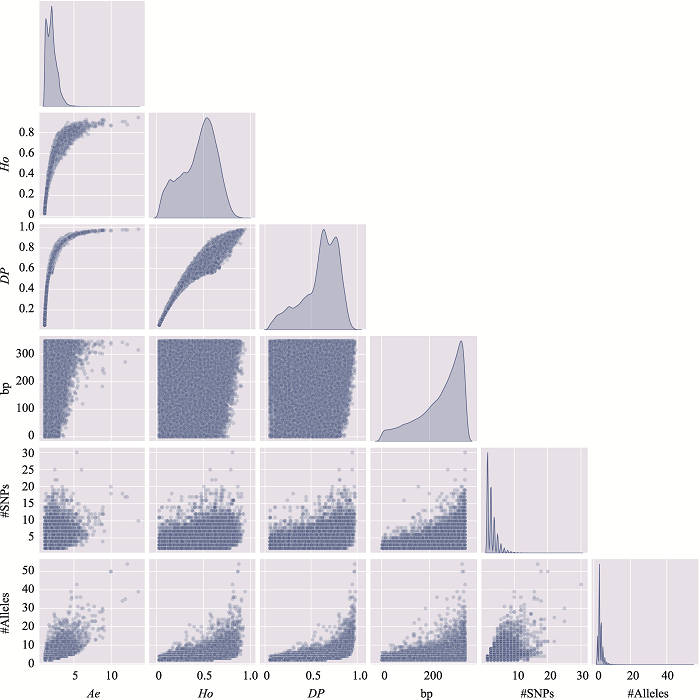
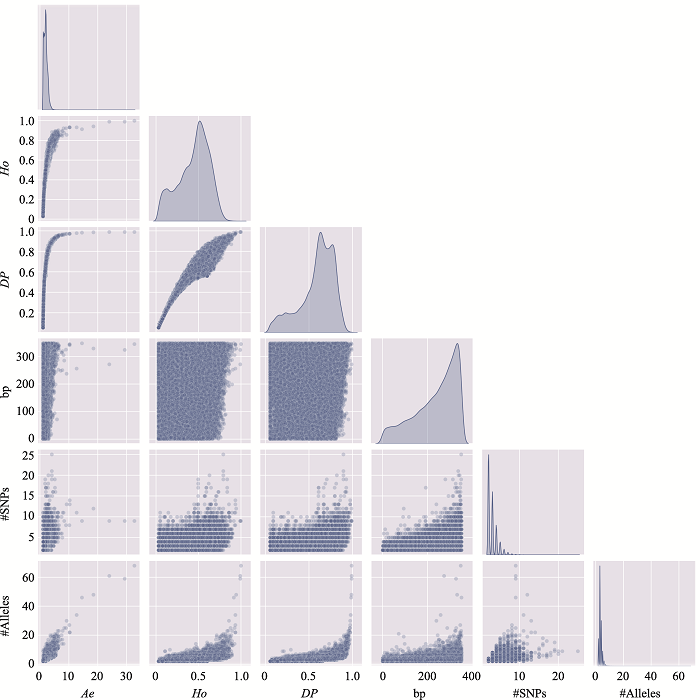
Table 2
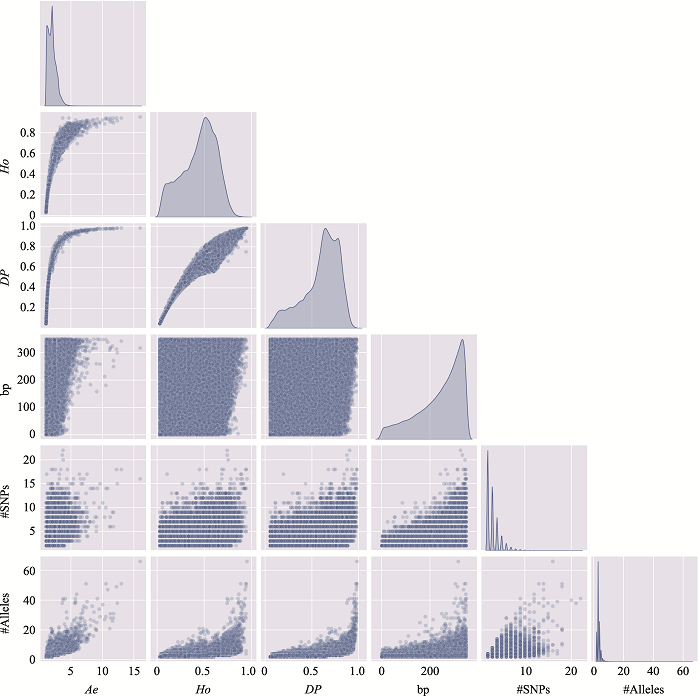
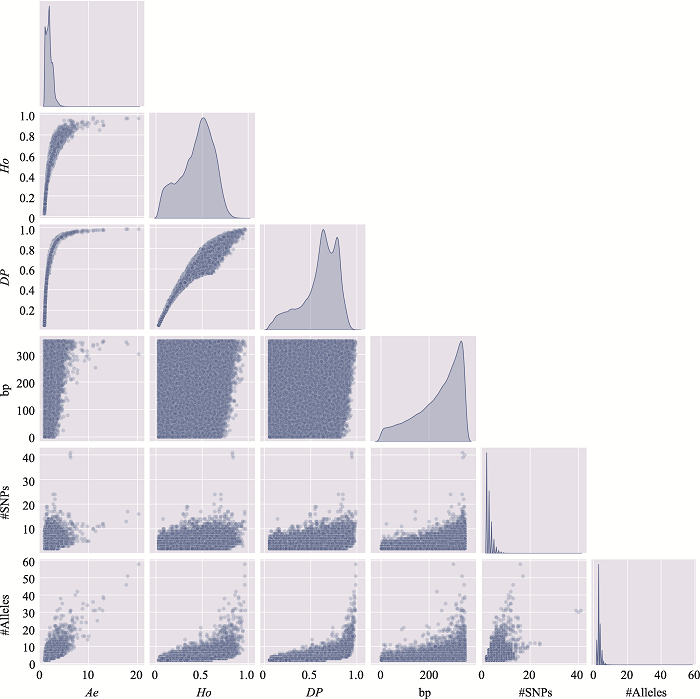
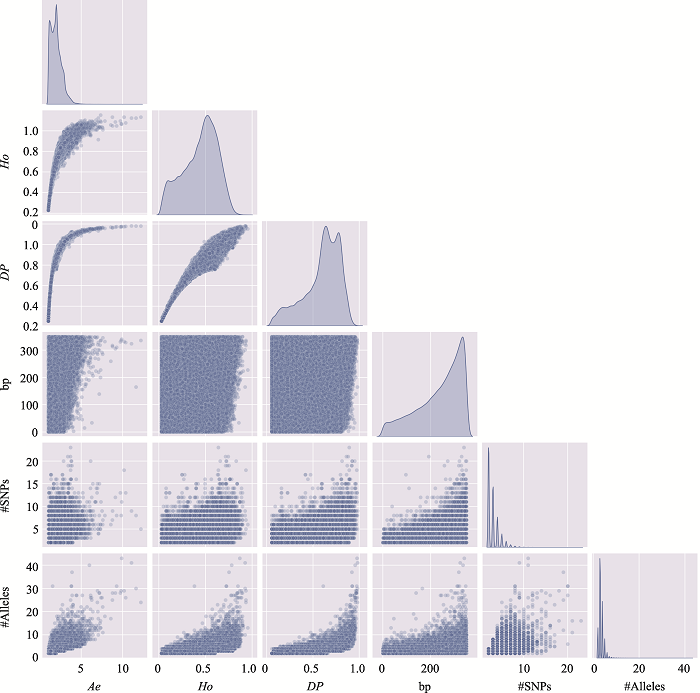
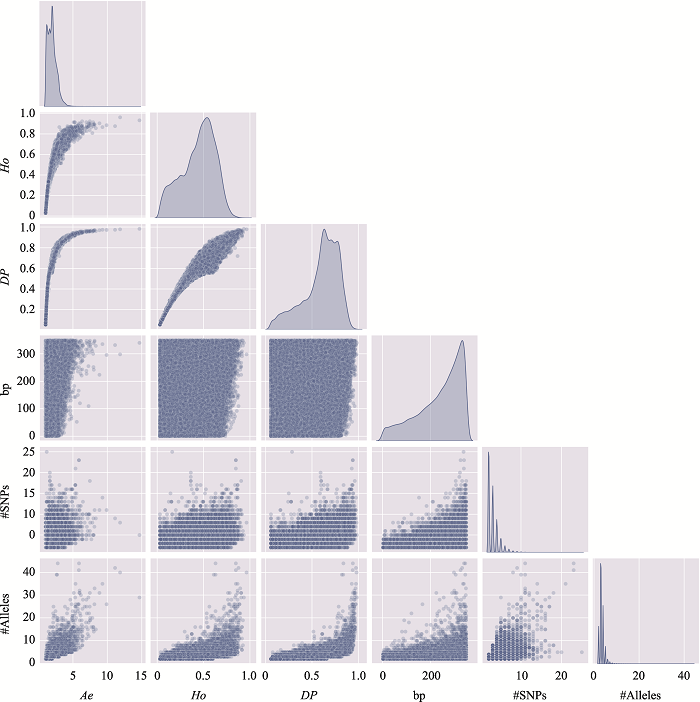
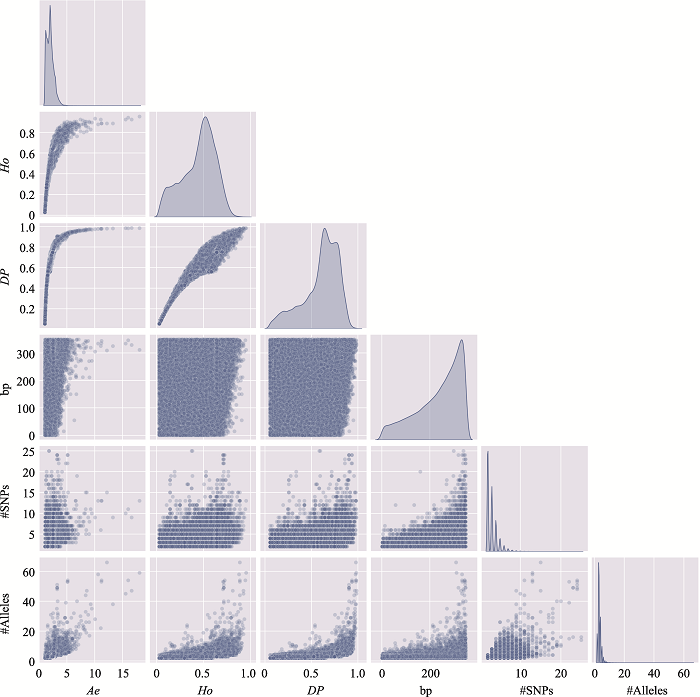
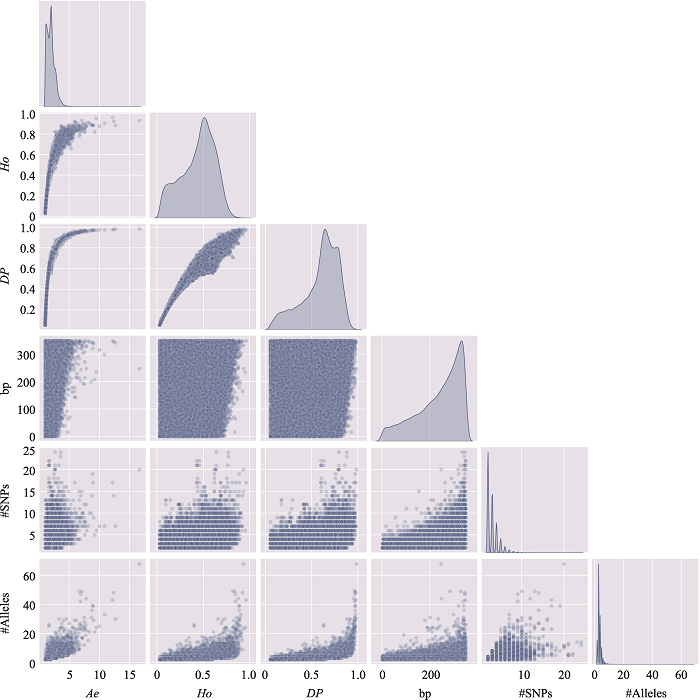
The information of the top 10 Ae values of microhaplotypes"
| MH_ID | bp | Ae | Ho | DP | #SNPs | #Alleles | Position (GRCh37) |
|---|---|---|---|---|---|---|---|
| mh04zj0146583 | 346 | 66.6163 | 0.9905 | 0.9899 | 41 | 134 | Chr4:30279658~30280003 |
| mh01zj0675568 | 248 | 43.2353 | 0.9714 | 0.9905 | 15 | 88 | Chr1:247032193~247032440 |
| mh20zj0185187 | 347 | 32.6183 | 0.9999 | 0.9892 | 9 | 68 | Chr20:62308266~62308612 |
| mh09zj0366544 | 239 | 28.5622 | 0.9524 | 0.9883 | 12 | 60 | Chr9:129479455~129479693 |
| mh07zj0103025 | 323 | 28.3055 | 0.9714 | 0.9892 | 26 | 77 | Chr7:18772264~18772586 |
| mh04zj0352614 | 346 | 27.1218 | 0.9810 | 0.9859 | 22 | 105 | Chr4:88537078~88537423 |
| mh03zj0068937 | 350 | 24.2308 | 0.8762 | 0.9858 | 20 | 75 | Chr3:11955851~11956200 |
| mh02zj0082461 | 320 | 23.7864 | 0.9810 | 0.9874 | 12 | 56 | Chr2:20701112~20701431 |
| mh01zj0508420 | 337 | 21.7028 | 0.9619 | 0.9872 | 37 | 63 | Chr1:200785797~200786133 |
| mh04zj0474307 | 348 | 20.5307 | 0.9714 | 0.9870 | 11 | 51 | Chr4:129682428~129682775 |
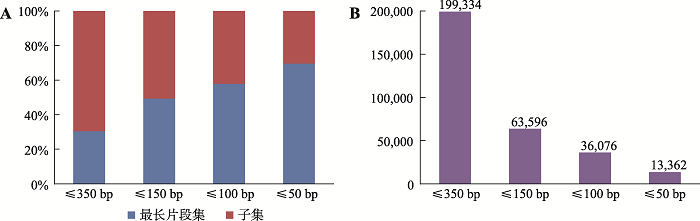
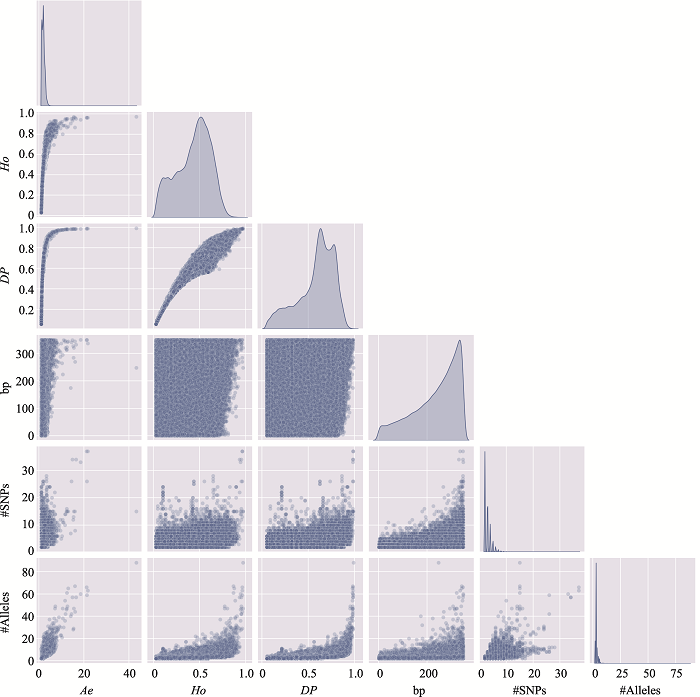
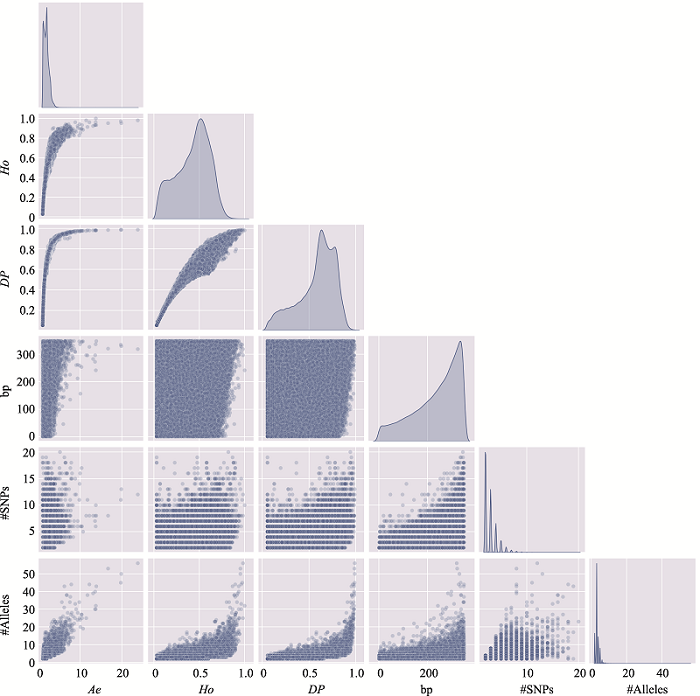
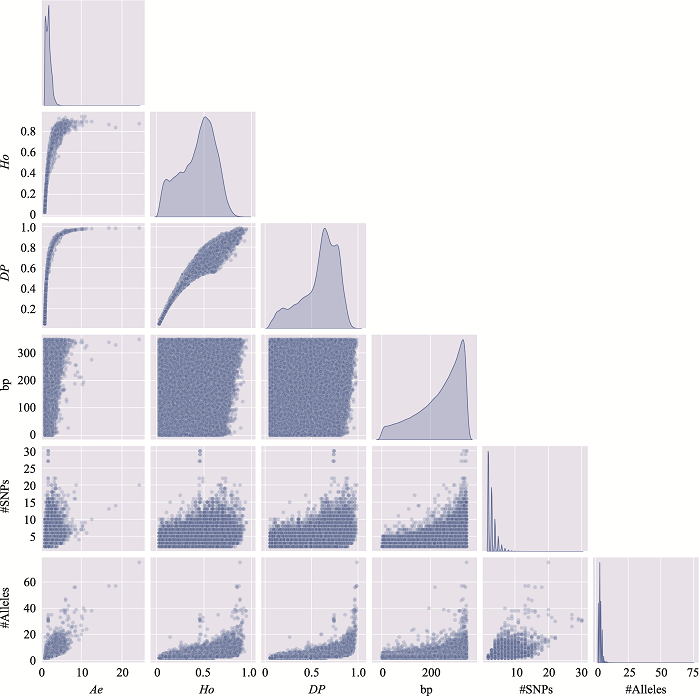
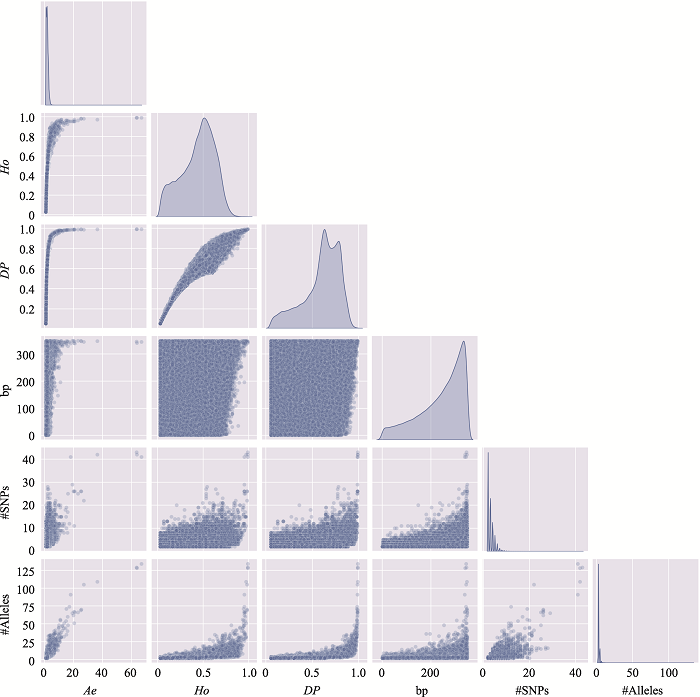
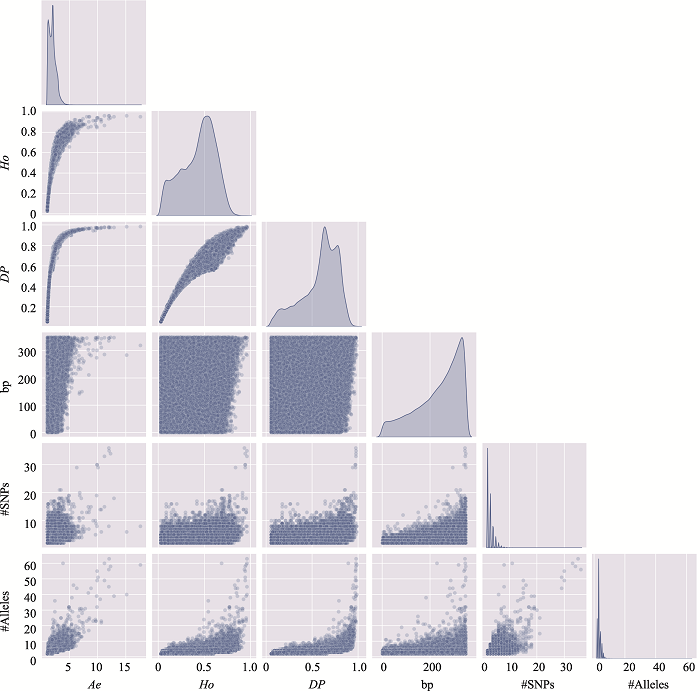
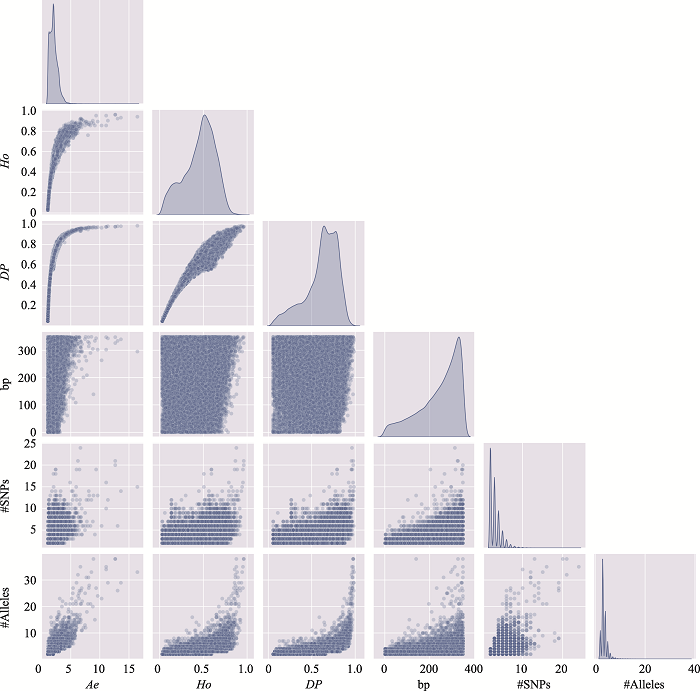
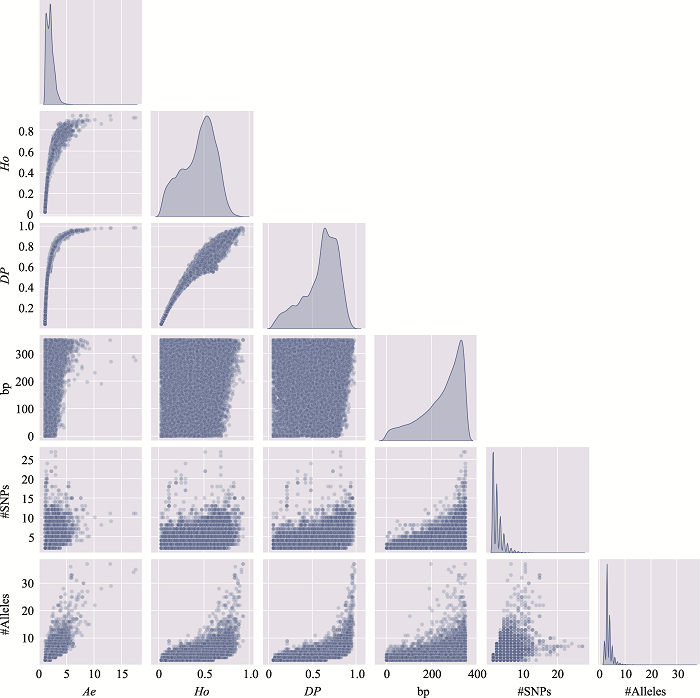
Table 3
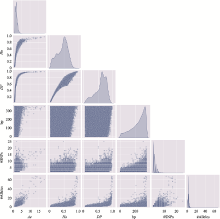
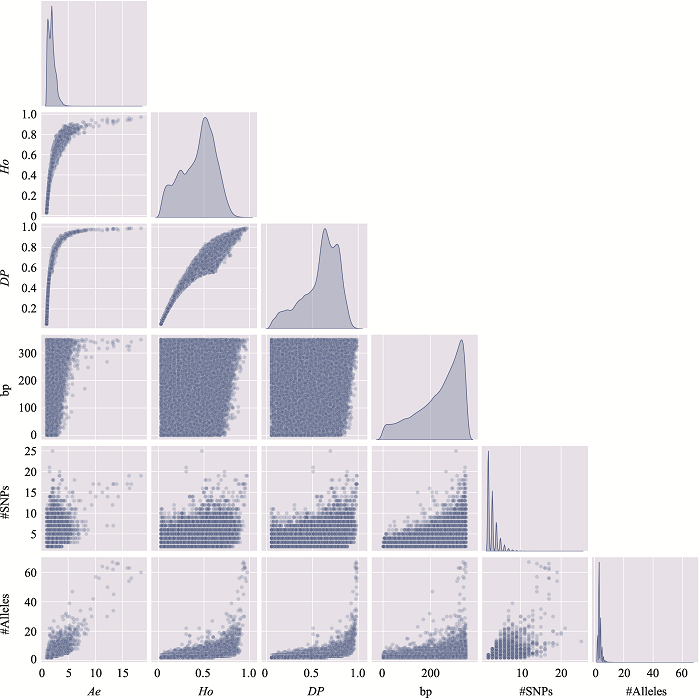
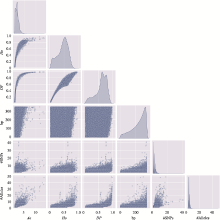
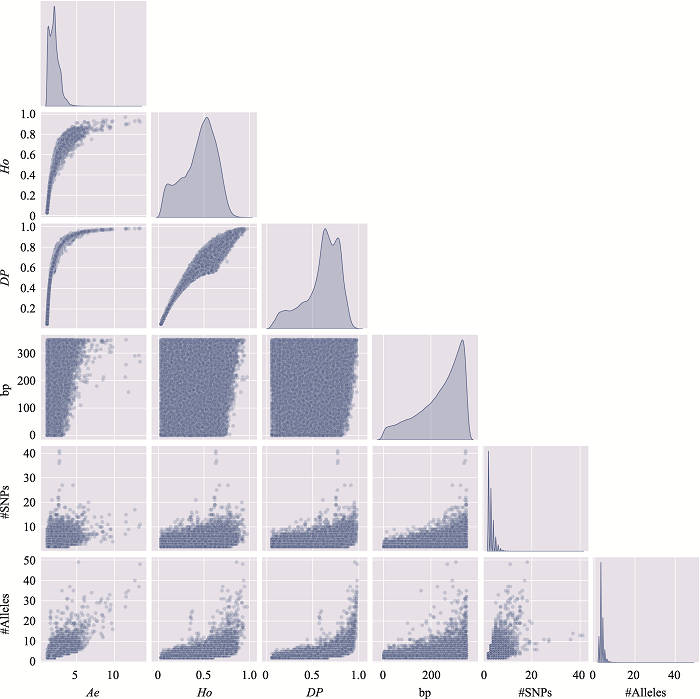
The distribution of microhaplotypes within different lengths of base pairs"
| Ae | #MHs≤350 bp | #MHs≤150 bp | #MHs≤100 bp | #MHs≤50 bp |
|---|---|---|---|---|
| 1~ | 1,634,742 | 1,345,361 | 1,116,786 | 741,819 |
| 2~ | 1,057,851 | 694,670 | 534,462 | 319,289 |
| 3~ | 171,637 | 56,715 | 32,419 | 12,211 |
| 4~ | 20,759 | 4941 | 2683 | 937 |
| 5~ | 6510 | 1875 | 957 | 213 |
| 10~ | 307 | 52 | 11 | 0 |
| 15~ | 80 | 8 | 5 | 1 |
| 20~ | 26 | 0 | 1 | 0 |
| 25~ | 15 | 5 | 0 | 0 |
| [1] | Kidd KK, Pakstis AJ, Speed WC, Lagace R, Chang J, Wootton S, Ihuegbu N . Microhaplotype loci are a powerful new type of forensic marker. Forensic Sci Int Genet Suppl Ser, 2013,4(1):e123-e124. |
| [2] | Oldoni F, Podini D . Forensic molecular biomarkers for mixture analysis. Forensic Sci Int Genet, 2019,41:107-119. |
| [3] | Bennett L, Oldoni F, Long K, Cisana S, Madella K, Wootton S, Chang J, Hasegawa R, Lagacé R, Kidd KK, Podini D. Mixture deconvolution by massively parallel sequencing of microhaplotypes. Int J Legal Med, 2019,133(3):719-729. |
| [4] | Cheung EYY, Phillips C, Eduardoff M, Lareu MV, Mcnevin D . Performance of ancestry-informative SNP and microhaplotype markers. Forensic Sci Int Genet, 2019,43:102141. |
| [5] | Børsting C, Morling N . Next generation sequencing and its applications in forensic genetics. Forensic Sci Int Genet, 2015,18:78-89. |
| [6] | Turchi C, Melchionda F, Pesaresi M, Tagliabracci A . Evaluation of a microhaplotypes panel for forensic genetics using massive parallel sequencing technology. Forensic Sci Int Genet, 2019,41:120-127. |
| [7] | Jin XY, Cui W, Chen C, Guo YX, Zhang XR, Xing GH, Lan JW, Zhu BF . Developing and population analysis of a new multiplex panel of 18 microhaplotypes and compound markers using next generation sequencing and its application in the Shaanxi Han population. Electrophoresis, 2020,41(13-14):1230-1237. |
| [8] | Cao YY, Wang QY, Zhu Q, Huang YG, Hu YH, Zhou YJ, Wang YF, Zhang J . Preliminary exploration of a novel method for the deconvolution of DNA mixtures by pyrosequencing. Forensic Sci Int Genet Suppl Ser, 2019,7(1):843-845. |
| [9] | van der Gaag KJ, de Leeuw RH, Laros J, den Dunnen JT, de Knijff P . Short hypervariable microhaplotypes: A novel set of very short high discriminating power loci without stutter artefacts. Forensic Sci Int Genet, 2018,35:169-175. |
| [10] | de la Puente M, Phillips C, Xavier C, Amigo J, Carracedo A, Parson W, Lareu MV . Building a custom large-scale panel of novel microhaplotypes for forensic identification using MiSeq and Ion S5 massively parallel sequencing systems. Forensic Sci Int Genet, 2020,45:102213. |
| [11] | Oldoni F, Bader D, Fantinato C, Wootton SC, Lagace R, Kidd KK, Podini D . A sequence-based 74plex microhaplotype assay for analysis of forensic DNA mixtures. Forensic Sci Int Genet, 2020,49:102367. |
| [12] | Wu RG, Li HX, Li R, Peng D, Wang NN, Shen XF, Sun HY . Identification and sequencing of 59 highly polymorphic microhaplotypes for analysis of DNA mixtures. Int J Legal Med, 2021,135(4):1137-1149. |
| [13] | Qu N, Lin SB, Gao Y, Liang H, Zhao H, Ou XL . A microhap panel for kinship analysis through massively parallel sequencing technology. Electrophoresis, 2020,41(3-4):246-253. |
| [14] | Sun SL, Liu Y, Li JN, Yang ZD, Wen D, Liang WB, Yan YQ, Yu H, Cai JF, Zha L . Development and application of a nonbinary SNP-based microhaplotype panel for paternity testing involving close relatives. Forensic Sci Int Genet, 2020,46:102255. |
| [15] | Wen D, Sun SL, Liu Y, Li JN, Yang ZD, Kureshi A, Fu Y, Li HN, Jiang BW, Jin C, Cai JF, Zha L . Considering the flanking region variants of nonbinary SNP and phenotype- informative SNP to constitute 30 microhaplotype loci for increasing the discriminative ability of forensic applications. Electrophoresis, 2021,42(9-10):1115-1126. |
| [16] | Chen P, Deng CW, Li Z, Pu Y, Yang JW, Yu YF, Li K, Li D, Liang WB, Zhang L, Chen F . A microhaplotypes panel for massively parallel sequencing analysis of DNA mixtures. Forensic Sci Int Genet, 2019,40:140-149. |
| [17] | Voskoboinik L, Motro U, Darvasi A . Facilitating complex DNA mixture interpretation by sequencing highly polymorphic haplotypes. Forensic Sci Int Genet, 2018,35:136-140. |
| [18] | Kidd KK, Speed WC, Pakstis AJ, Podini DS, Lagace R, Chang J, Wootton S, Haigh E, Soundararajan U . Evaluating 130 microhaplotypes across a global set of 83 populations. Forensic Sci Int Genet, 2017,29:29-37. |
| [19] | Chen P, Yin CY, Li Z, Pu Y, Yu YJ, Zhao P, Chen DX, Liang WB, Zhang L, Chen F . Evaluation of the microhaplotypes panel for DNA mixture analyses. Forensic Sci Int Genet, 2018,35:149-155. |
| [20] | Kureshi A, Li J, Wen D, Sun SL, Yang ZD, Zha L . Construction and forensic application of 20 highly polymorphic microhaplotypes. R Soc Open Sci, 2020,7(5):191937. |
| [21] | 1000 Genomes Project Consortium; Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, Korbel JO, Marchini JL, Mccarthy S, Mcvean GA, Abecasis GR. A global reference for human genetic variation. Nature, 2015,526(7571):68-74. |
| [22] | Kidd KK, Speed WC . Criteria for selecting microhaplotypes: mixture detection and deconvolution. Investig Genet, 2015,6(1):1. |
| [23] | Logsdon GA, Vollger MR, Hsieh P, Mao YF, Liskovykh MA, Koren S, Nurk S, Mercuri L, Dishuck PC, Rhie A, de Lima LG, Dvorkina T, Porubsky D, Harvey WT, Mikheenko A, Bzikadze AV, Kremitzki M, Graves-Lindsay TA, Jain C, Hoekzema K, Murali SC, Munson KM, Baker C, Sorensen M, Lewis AM, Surti U, Gerton JL, Larionov V, Ventura M, Miga KH, Phillippy AM, Eichler EE. The structure, function and evolution of a complete human chromosome 8. Nature, 2021,593(7857):101-107. |
| [24] | Collins JR, Stephens RM, Gold B, Long B, Dean M, Burt SK . An exhaustive DNA micro-satellite map of the human genome using high performance computing. Genomics, 2003,82(1):10-19. |
| [25] | Oldoni F, Kidd KK, Podini D . Microhaplotypes in forensic genetics. Forensic Sci Int Genet, 2019,38:54-69. |
| [1] | Yongqiang Kong, Jinkai Liu, Jiaqi Gu, Jingyi Xu, Yunuo Zheng, Yiliang Wei, Shaoyuan Wu. Optimization scheme of machine learning model for genetic division between northern Han, southern Han, Korean and Japanese [J]. Hereditas(Beijing), 2022, 44(11): 1028-1043. |
| [2] | Xiaoyuan Guo, Changchun Sun, Siyao Xue, Hui Zhao, Li Jiang, Caixia Li. 49AISNP: a study on the ancestry inference of the three ethnic groups in the north of East Asia [J]. Hereditas(Beijing), 2021, 43(9): 880-889. |
| [3] | Zhiyong Liu, Riga Wu, Ran Li, Qiangwei Wang, Hongyu Sun. Ethical issues of the research and practice in forensic genetics [J]. Hereditas(Beijing), 2021, 43(10): 994-1002. |
| [4] | Yang Liu, Changchun Sun, Mi Ma, Ling Wang, Wenting Zhao, Quan Ma, Anquan Ji, Jing Liu, Caixia Li. The ancestry inference of Chinese populations using 74-plex SNPs system [J]. Hereditas(Beijing), 2020, 42(3): 296-308. |
| [5] | Li Jiang,Qifan Sun,Quan Ma,Wenting Zhao,Jing Liu,Lei Zhao,Anquan Ji,Caixia Li. Optimization and validation of analysis method based on 27-plex SNP panel for ancestry inference [J]. Hereditas(Beijing), 2017, 39(2): 166-173. |
| [6] | Xiuyan Ruan, Weini Wang, Yaran Yang, Bingbing Xie, Jing Chen, Yacheng Liu, Jiangwei Yan. Genetic variability and phylogenetic analysis of 39 short tandem repeat loci in Beijing Han population [J]. HEREDITAS(Beijing), 2015, 37(7): 683-691. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||