Hereditas(Beijing) ›› 2021, Vol. 43 ›› Issue (9): 910-920.doi: 10.16288/j.yczz.21-171
• Research Article • Previous Articles
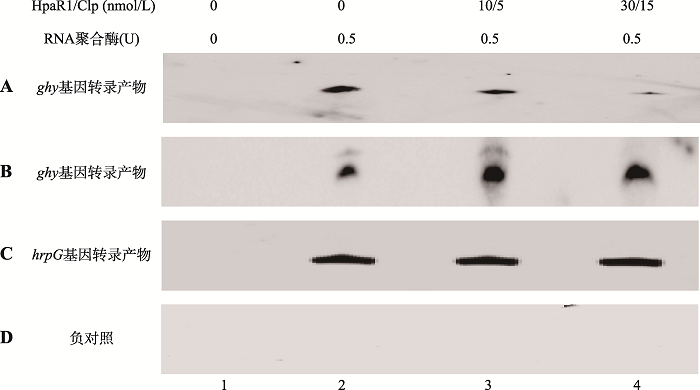
Analysis of transcriptional regulators HpaR1 and Clp regulating the expression of glycoside hydrolase-encoding gene in the Xanthomonas campestris pv. campestris
Guofang Liu1( ), Peidong Ren2, Wenxin Ye2, Guangtao Lu2(
), Peidong Ren2, Wenxin Ye2, Guangtao Lu2( )
)
- 1. Guangxi Key Laboratory for Polysaccharide Materials and Modifications,School of Marine Sciences and Biotechnology, Guangxi University for Nationalities, Nanning 530008, China
2. College of Life Science and Technology, Guangxi University, Nanning 530004, China
-
Received:2021-05-11Revised:2021-07-05Online:2021-09-20Published:2021-07-14 -
Contact:Lu Guangtao E-mail:lgf8411@126.com;lugt@gxu.edu.cn -
Supported by:Supported by Guangxi University for Nationalities Scientific Research Fund No(2019KJQN004);the National Natural Science Foundation of China No(31860021);Guangxi Natural Science Foundation of China No(2020GXNSFBA297140);Science and Technology Major Project of Guangxi No(AA18242026)
Cite this article
Guofang Liu, Peidong Ren, Wenxin Ye, Guangtao Lu. Analysis of transcriptional regulators HpaR1 and Clp regulating the expression of glycoside hydrolase-encoding gene in the Xanthomonas campestris pv. campestris[J]. Hereditas(Beijing), 2021, 43(9): 910-920.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
Table 1
The strains and plasmids used in this study"
| 菌株和质粒 | 相关特征 |
|---|---|
| 大肠杆菌 | |
| DH5α | F-,φ80d/lacZ△M15,Δ(lacZYA-argF)U169,deoR,recA1,endA1,hsdR17(rk-, mk+),phoA,supE44, λ-,thi-1,gyrA96,relA1 |
| M15 | Nals,trs,Rifs,Thi-,Lac-,Ar+a,Gal+,Mtl-,F-,RecA+,Uvr+,Lon+harboring pREP4 plasmid,Kmr |
| ED8767 | RecA,met,含有帮助质粒 pRK2073,Spcr |
| pQE-HpaR1/M15 | M15中含有质粒pQE-HpaR1,Kmr,Ampr |
| pQE-Clp/M15 | M15中含有质粒 pQE-Clp,Kmr,Ampr |
| pL3-HpaR1::3×Flg/ DH5α | DH5α中含有质粒pL3-HpaR1:: 3×Flg,Tcr |
| pL3-Clp:: 3×Flg/ DH5α | DH5α中含有质粒pL3-Clp:: 3×Flg,Tcr |
| 十字花科黑腐病菌 | |
| 8004 | 野生型,Rifr |
| ΔhpaR1 | hpaR1基因的缺失突变体,Rifr |
| Δclp | Clp基因的缺失突变体,Rifr |
| 8004/pLAFR3 | 8004中含有质粒pLAFR3,Rifr,Tcr |
| ΔhpaR1/pL3-HpaR1::3×Flg | ΔhpaR1中含有质粒 pL3-HpaR1::3×Flg,Rifr,Tcr |
| Δclp/pL3-Clp:: 3×Flg | Δclp中含有质粒 pL3-Clp:: 3×Flg,Rifr,Tcr |
| 质粒 | |
| pK18mob | 黄单胞菌属中的自杀质粒,Kmr |
| pRK2073 | 帮助质粒,Tra+,Mob+,ColE1,Spcr |
| pQE30 | 表达载体,Ampr |
| pQE30-HpaR1 | pQE30上含有hpaR1基因的360 bp ORF,Ampr |
| pQE30-Clp | pQE30上含有 clp基因的690 bp ORF,Ampr |
| pLAFR3 | PK2 replicon;Mob+,Tra-;cos+,Tcr |
| pL3-HpaR1::3×Flg | pLAFR3上含有 hpaR1::3×Flg,Tcr |
| pL3-Clp::3×Flg | pLAFR3上含有 clp::3×Flg,Tcr |
Table 2
Primers used in this study"
| 用途 | 引物名称 | 引物序列(5′→3′) |
|---|---|---|
| 蛋白表达 | Clp-OF | ACAGTTGGATCC ATGAGCCTAGGGAACAGCAC |
| Clp-OR | ACAGTTAAGCTT TTAGCGCGTGCCGTACAACA | |
| 凝胶阻滞分析 | 0026 F | ACGGCAATCGATCAGTTCGC |
| 0026 R | CATGGAAACCACTCCTTCGACG | |
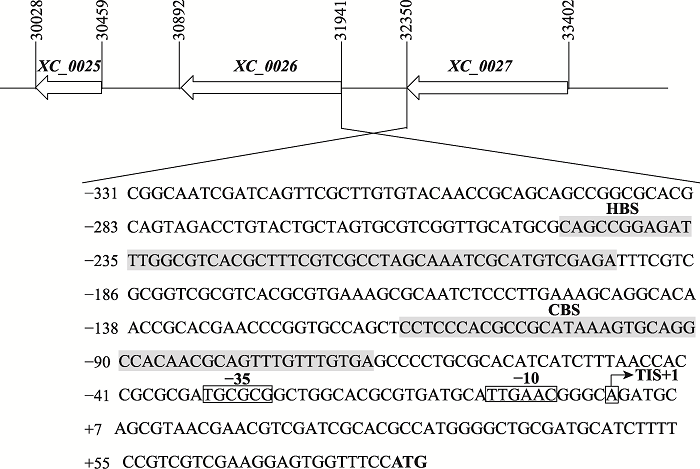
| DNase I保护实验 | 0026(HpaR1)F | GACAAGCCGCAGATGAAAACCC |
| 0026(HpaR1)R | GTTACGCTGCATCTGCCCGT | |
| 0026(Clp)F | GCAGCCGGCGCACGCAGTAGACCTGTACT | |
| 0026(Clp)R | CGACGGAAAAGATGCATCGCAGCCC | |
| 5ʹ-RACE | AAP | GGCCACGCGTCGACTAGTACGGGGGGGGGG |
| AUAP | GGCCACGCGTCGACTAGTAC | |
| 0026GSP1 | GTTCCCACAGGATCGGCAGG | |
| 0026GSP2 | TCCAGTCCCAGCGAAATAGC | |
| 0026GSP3 | TGTTGAGGACGCCAGGTTTT | |
| 0026GSPF | ATGTCGTGTTCTCTGTGGTTGC | |
| 体外转录 | 0026ivtF | GCGACAAGCCGCAGATGAAAACCCT |
| 0026ivtR | GATGACGCAAATTCCGCACCCGCC | |
| 3077ivtF | TCTCACTCTGTCTTGCAAACTGCGA | |
| 3077ivtR | AATGGGCATCGAAAACCAGAAGC | |
| 染色质免疫共沉淀 | Clp(ChIP)F | CGGGATCCGACTACAAAGACCATGACGGTGATTATAAAGATCATGATATCGACTACAA-AGATGACGACGATAAAATGAGCCTAGGGAACACGACG |
| Clp(ChIP)R | CCAAGCTTTTAGCGCGTGCCGTACAACA | |
| HpaR1(ChIP)F | CGGGATCCGACTACAAAGACCATGACGGTGATTATAAAGATCATGATATCGACTACAA-AGATGACGACGATAAAATGACTGACATCCAGTGGAGC | |
| HpaR1(ChIP)R | CCAAGCTTTCATGGTGTTTTCCCCTGTG | |
| 0026(ChIP)F | CGTCACGCTTTCGTCGCCTA | |
| 0026(ChIP)R | AACTGCGTTGTGGCCTGCAC | |
| 实时荧光定量PCR | 0026qRT-F | ATGTCGTGTTCTCTGTGGTTGC |
| 0026qRT-R | TGTTGAGGACGCCAGGTTTT |
| [1] |
Haydon DJ, Guest JR. A new family of bacterial regulatory proteins. FEMS Microbiol Lett, 1991, 63(2-3):291-295.
pmid: 2060763 |
| [2] |
Hoskisson PA, Rigali S. Chapter 1: Variation in form and function the helix-turn-helix regulators of the GntR superfamily. Adv Appl Microbiol, 2009, 69:1-22.
doi: 10.1016/S0065-2164(09)69001-8 pmid: 19729089 |
| [3] |
Rigali S, Derouaux A, Giannotta F, Dusart J. Subdivision of the helix-turn-helix GntR family of bacterial regulators in the FadR, HutC, MocR, and YtrA subfamilies. J Biol Chem, 2002, 277(15):12507-12515.
doi: 10.1074/jbc.M110968200 |
| [4] |
Vindal V, Suma K, Ranjan A. GntR family of regulators in Mycobacterium smegmatis: a sequence and structure based characterization. BMC Genomics, 2007, 8:289.
doi: 10.1186/1471-2164-8-289 |
| [5] |
Suvorova IA, Korostelev YD, Gelfand MS. GntR family of bacterial transcription factors and their DNA binding motifs: structure, positioning and co-evolution. PLoS One, 2015, 10(7):e0132618.
doi: 10.1371/journal.pone.0132618 |
| [6] |
Simpson RB, Johnson LJ. Arabidopsis thaliana as a host for Xanthomonas campestris pv.campestris. Mol Plant Microbe In, 1990, 3(4):233-237.
doi: 10.1094/MPMI-3-233 |
| [7] |
Sun WX, Liu LJ, Bent AF. Type III secretion-dependent host defence elicitation and type III secretion-independent growth within leaves by Xanthomonas campestris pv. campestris. Mol Plant Pathol, 2011, 12(8):731-745.
doi: 10.1111/mpp.2011.12.issue-8 |
| [8] | Schroth MN, Hildebrand DC, Panopoulos N. Phytopathogenic Pseudomonads and related plant-associated Pseudomonads. In: Balows A, Truper HG, Dworkin M, Harder W, Schleifer KH, eds. The Prokaryotes, 2nd edn. New York: Springer Verlag, 2021, 714-740 |
| [9] |
Hugouvieux V, Barber CE, Daniels MJ. Entry of Xanthomonas campestris pv. campestris into hydathodes of Arabidopsis thaliana leaves: a system for studying early infections events in bacterial pathogenesis. Mol Plant Microbe Interact, 1998, 11(6):537-543.
doi: 10.1094/MPMI.1998.11.6.537 |
| [10] | Alvarez AM. Black rot of crucifers. In A J Slusarenko, R. S. S. Fraser, and L. C. van Loon (ed.), Mechanisms of resistance to plant diseases. Kluwer Academic Publishers, Dordrecht, The Netherlands, 2000, 21-52. |
| [11] |
Lu H, Patil P, Van Sluys MA, White FF, Ryan RP, Dow JM, Rabinowicz P, Salzberg SL, Leach JE, Sonti R, Brendel V, Bogdanove AJ. Acquisition and evolution of plant pathogenesis associated gene clusters and candidate determinants of tissue-specificity in Xanthomonas. PLoS One, 2008, 3(11):e3828.
doi: 10.1371/journal.pone.0003828 |
| [12] |
An SQ, Lu GT, Su HZ, Li RF, He YQ, Jiang BL, Tang DJ, Tang JL. Systematic mutagenesis of all predicted gntR genes in Xanthomonas campestris pv. campestris reveals a GntR family transcriptional regulator controlling hypersensitive response and virulence. Mol Plant Microbe Interact, 2011, 24(9):1027-1039.
doi: 10.1094/MPMI-08-10-0180 |
| [13] |
Su HZ, Wu L, Qi YH, Liu GF, Lu GT, Tang JL. Characterization of the GntR family regulator HpaR1 of the crucifer black rot pathogen Xanthomonas campestris pathovar campestris. Sci Rep, 2016, 6:19862.
doi: 10.1038/srep19862 |
| [14] |
Tao F, He YW, Wu DH, Swarup S, Zhang LH. The cyclic nucleotide monophosphate domain of Xanthomonas campestris global regulator Clp defines a new class of cyclic di-GMP effectors. J Bacteriol, 2010, 192(4):1020-1029.
doi: 10.1128/JB.01253-09 |
| [15] |
Hsiao YM, Liao HY, Lee MC, Yang TC, Tseng YH. Clp upregulates transcription of engA gene encoding a virulence factor in Xanthomonas campestris by direct binding to the upstream tandem Clp sites. FEBS Lett, 2005, 579(17):3525-3533.
doi: 10.1016/j.febslet.2005.05.023 |
| [16] |
Liu GF, Su HZ, Sun HY, Lu GT, Tang JL. Competitive control of endoglucanase gene engXCA expression in the plant pathogen Xanthomonas campestris by the global transcriptional regulators HpaR1 and Clp. Mol Plant Pathol, 2019, 20(1):51-68.
doi: 10.1111/mpp.2019.20.issue-1 |
| [17] |
Berg OG, von Hippel PH. Selection of DNA binding sites by regulatory proteins. II. The binding specificity of cyclic AMP receptor protein to recognition sites. J Mol Biol, 1988, 200(4):709-723.
pmid: 3045325 |
| [18] |
Ge C, He C. Regulation of the type II secretion structural gene xpsE in Xanthomonas campestris Pathovar campestris by the global transcription regulator Clp. Curr Microbiol, 2008, 56(2):122-127.
doi: 10.1007/s00284-007-9081-9 |
| [19] |
Hsiao YM, Zheng MH, Hu RM, Yang TC, Tseng YH. Regulation of the pehA gene encoding the major polygalacturonase of Xanthomonas campestris by Clp and RpfF. Microbiology, 2008, 154(Pt 3):705-713.
doi: 10.1099/mic.0.2007/012930-0 |
| [20] |
Hsiao YM, Fang MC, Sun PF, Tseng YH. Clp and RpfF up-regulate transcription of pelA1 gene encoding the major pectate lyase in Xanthomonas campestris pv. campestris. J Agric Food Chem, 2009, 57(14):6207-6215.
doi: 10.1021/jf900701n |
| [21] |
Browning DF, Busby SJ. The regulation of bacterial transcription initiation. Nat Rev Microbiol, 2004, 2(1):57-65.
pmid: 15035009 |
| [22] |
Lee DJ, Minchin SD, Busby SJW. Activating transcription in bacteria. Annu Rev Microbiol, 2012, 66:125-152.
doi: 10.1146/annurev-micro-092611-150012 |
| [1] | Dandan Wu, Mingkun Zhu, Zhongyan Fang, Wei Ma. Progress on molecular composition and genetic mechanism of plant B chromosomes [J]. Hereditas(Beijing), 2022, 44(9): 772-782. |
| [2] | Yuan Zhang, Yuting Zhao, Lenan Zhuang, Jin He. Transcriptional regulation of transcriptional Mediator complexes in cardiovascular development and disease [J]. Hereditas(Beijing), 2022, 44(5): 383-397. |
| [3] | Tianyi Wang, Yingxiang Wang, Chenjiang You. Structural and functional characteristics of plant PHD domain-containing proteins [J]. Hereditas(Beijing), 2021, 43(4): 323-339. |
| [4] | Yuanyuan Hao, Xiangqian Zhao, Fudeng Huang, Chunshou Li. The role of PPR proteins in posttranscriptional regulation of organelle components in plants [J]. Hereditas(Beijing), 2021, 43(11): 1050-1065. |
| [5] | Xiaofen Qiu, Dong’e Tang, Haiyan Yu, Qiuyan Liao, Zhiyang Hu, Jun Zhou, Xin Zhao, Huiyan He, Zhuojian Liang, Chengming Xu, Ming Yang, Yong Dai. Analysis of transcription factors in accessible open chromatin in the 18-trisomy syndrome based on single cell ATAC sequencing technique [J]. Hereditas(Beijing), 2021, 43(1): 74-83. |
| [6] | Xiaomeng Gao, Zhihua Zhang. Three-dimensional structure and function of chromatin regulated by “liquid-liquid phase separation” of biological macromolecules. [J]. Hereditas(Beijing), 2020, 42(1): 45-56. |
| [7] | Yu Zhang, Yuda Fang. Progresses on the structure and function of cohesin [J]. Hereditas(Beijing), 2020, 42(1): 57-72. |
| [8] | Xiaofei Zheng,Haiyan Huang,Qiang Wu. Chromatin architectural protein CTCF regulates gene expression of the UGT1 cluster [J]. Hereditas(Beijing), 2019, 41(6): 509-523. |
| [9] | Chunyou Ning,Mengnan He,Qianzi Tang,Qing Zhu,Mingzhou Li,Diyan Li. Advances in mammalian three-dimensional genome by using Hi-C technology approach [J]. Hereditas(Beijing), 2019, 41(3): 215-233. |
| [10] | Lili Liu, Aiwei Guo, Peifu Wu, Fenfen Chen, Yajin Yang, Qin Zhang. Regulation of VPS28 gene knockdown on the milk fat synthesis in Chinese Holstein dairy [J]. Hereditas(Beijing), 2018, 40(12): 1092-1100. |
| [11] | Min Cheng, Wenjian Zhang, Tianyu Xing, Xiaohong Yan, Yumao Li, Hui Li, Ning Wang. Functional analysis of the upstream regulatory region of chicken miR-17-92 cluster [J]. Hereditas(Beijing), 2016, 38(8): 724-735. |
| [12] | Feng Yang, Fan Yi, Huiqing Cao, Zicai Liang, Quan Du. The emerging landscape of long non-coding RNAs [J]. HEREDITAS(Beijing), 2014, 36(5): 456-468. |
| [13] | Deling Lin, Ying Luo, Yi Song. The post-transcriptional regulation of the DNA damage response [J]. HEREDITAS, 2014, 36(4): 309-318. |
| [14] | Nan Ding, Hongzhu Qu, Xiangdong Fang. The ENCODE project and functional genomics studies [J]. HEREDITAS, 2014, 36(3): 237-247. |
| [15] | ZHANG Jun-Fang ZHU Hua-Bin ZHANG Liu-Guang HAO Hai-Sheng ZHAO Xue-Ming QIN Tong LU Yong-Qiang WANG Dong. Advance on research of gene expression during spermiogenesis at transcription level [J]. HEREDITAS, 2013, 35(5): 587-594. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||