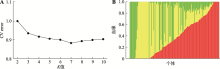
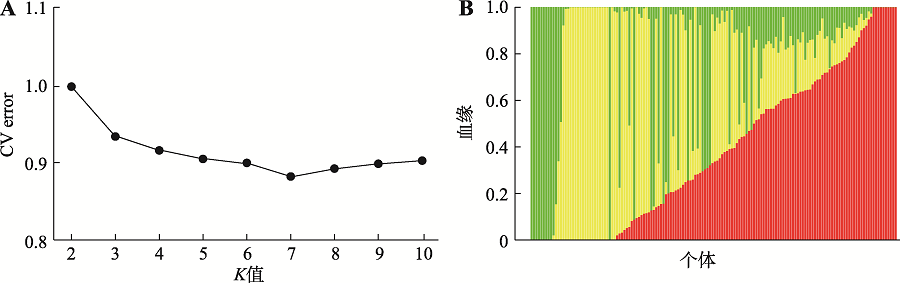
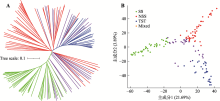
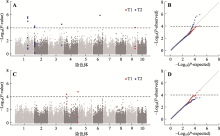
Hereditas(Beijing) ›› 2021, Vol. 43 ›› Issue (12): 1159-1169.doi: 10.16288/j.yczz.21-298
• Research Article • Previous Articles Next Articles
Genome-wide association study and candidate gene prediction of salt tolerance related traits in maize
Tingyu Shan( ), Wen Shi, Yiting Wang, Ziyi Cao, Baohua Wang, Hui Fang(
), Wen Shi, Yiting Wang, Ziyi Cao, Baohua Wang, Hui Fang( )
)
- Ministry of Agricultural Scientific Observing and Experimental Station of Maize in Plain Area of Southern Region, School of Life Sciences, Nantong University, Nantong 226019, China
-
Received:2021-08-13Revised:2021-10-02Online:2021-12-20Published:2021-12-07 -
Contact:Fang Hui E-mail:shan_0822@126.com;fanghui8912@126.com -
Supported by:Supported by the Science and Technology Project of Nantong City, China(MS22020033);Talent Introduction Project of Nantong University No(135420609055)
Cite this article
Tingyu Shan, Wen Shi, Yiting Wang, Ziyi Cao, Baohua Wang, Hui Fang. Genome-wide association study and candidate gene prediction of salt tolerance related traits in maize[J]. Hereditas(Beijing), 2021, 43(12): 1159-1169.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
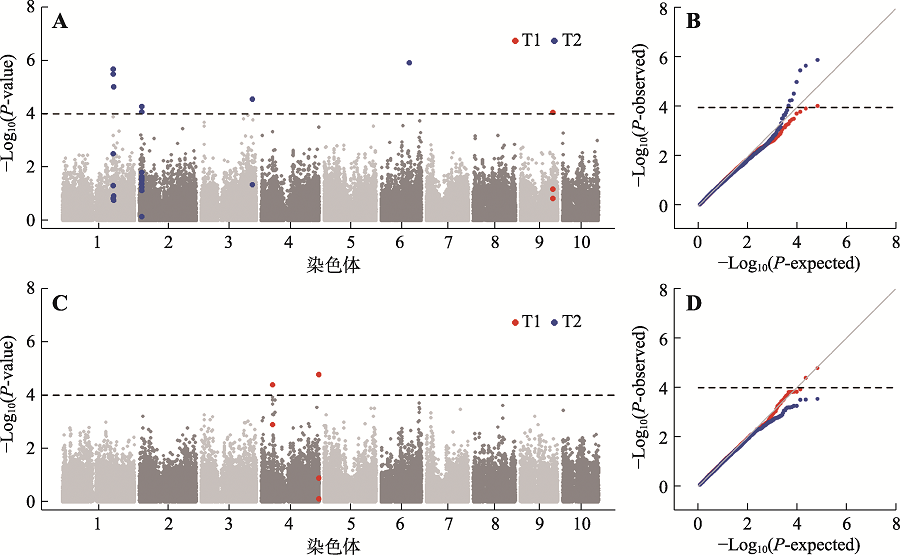
Table 3
The GWAS Results of two maize salt tolerance traits"
| 性状a | 标记 | 染色体 | 物理位置 | P值 | R2 | 候选基因 | 注释 | 分子功能 | 备注 |
|---|---|---|---|---|---|---|---|---|---|
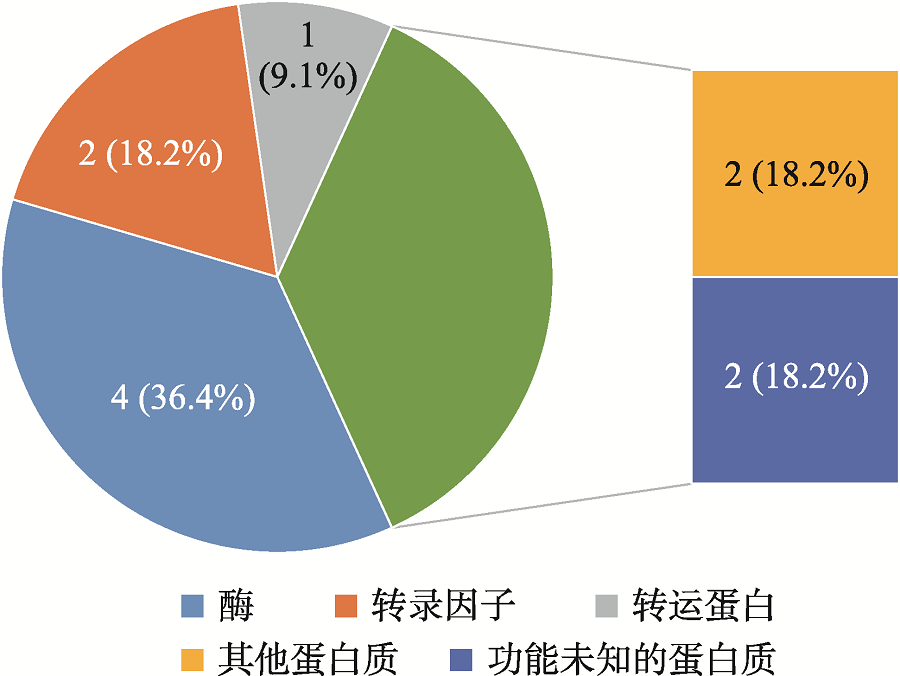
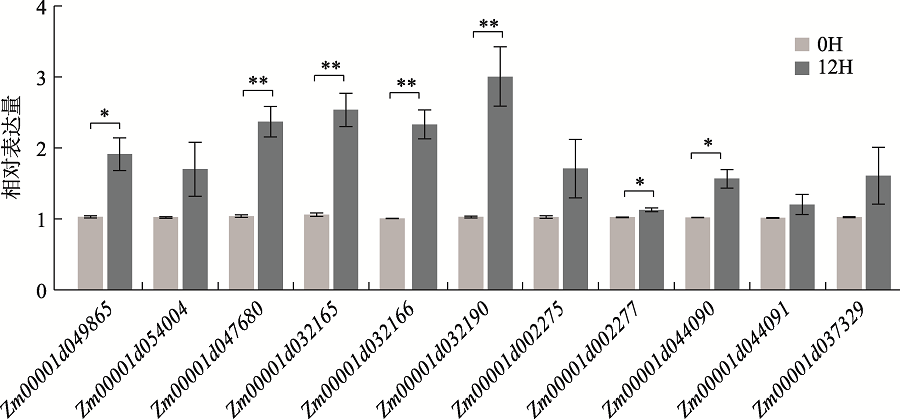
| T1 | PZE-104036105 | 4 | 47241320 | 3.95×10-5 | 0.12 | Zm00001d049865 | CASP-like protein | dehydrogenase | 条件 分析 |
| T1 | SYN16455 | 4 | 239628761 | 1.61×10-5 | 0.13 | Zm00001d054004 | plant-specific domain TIGR01589 family protein | NA | 条件 分析 |
| T1 | SYN38269 | 9 | 136583562 | 8.64×10-5 | 0.12 | Zm00001d047680 | calmodulin-binding protein 60G | DNA-binding transcription factor activity | |
| T2 | SYNGENTA5755 | 1 | 212194098 | 2.02×10-6 | 0.18 | Zm00001d032165 | defective in cullin neddylation protein | ubiquitin-like protein conjugating enzyme binding | |
| Zm00001d032166 | DUF1336 domain- containing protein | defense/immunity protein | |||||||
| T2 | PZE-101169962 | 1 | 213269897 | 9.28×10-6 | 0.15 | Zm00001d032190 | MYB family transcription factor | DNA-binding transcription factor | |
| T2 | SYN1142 | 2 | 9085410 | 5.07×10-5 | 0.12 | Zm00001d002275 | abc transporter c family member 2-like | ATP-binding cassette (ABC) transporter | |
| Zm00001d002277 | Ubiquitin-like domain- containing protein | ATPase activity; chaperone binding | |||||||
| T2 | PHM13742.5 | 3 | 215181142 | 2.77×10-5 | 0.13 | Zm00001d044090 | 5ʹ-deoxynucleotidase | phosphatase activity | |
| Zm00001d044091 | serine threonine-protein kinase ht1-like | non-receptor serine/threonine protein kinase | |||||||
| T2 | PZE-106065133 | 6 | 117419198 | 1.18×10-6 | 0.18 | Zm00001d037329 | uncharacterized protein | NA |
Table 4
Comparison of GWAS results for two statistical models"
| 统计模型 | 标记a | 染色体 | 物理位置 | 关联性状 |
|---|---|---|---|---|
| 一般线性模型 | PZE-101167639 | 1 | 211083097 | 株高变化率 |
| SYNGENTA5755 | 1 | 212194098 | 株高变化率 | |
| PZE-101168916 | 1 | 212192906 | 株高变化率 | |
| PZE-101169962 | 1 | 213269897 | 株高变化率 | |
| PHM13742.5 | 3 | 215181142 | 株高变化率 | |
| PZE-106065133 | 6 | 117419198 | 株高变化率 | |
| SYN4789 | 6 | 150688349 | 鲜重变化率 | |
| 混合线性模型 | SYNGENTA5755 | 1 | 212194098 | 株高变化率 |
| PZE-101169962 | 1 | 213269897 | 株高变化率 | |
| SYN1142 | 2 | 9085410 | 株高变化率 | |
| PHM13742.5 | 3 | 215181142 | 株高变化率 | |
| PZE-106065133 | 6 | 117419198 | 株高变化率 | |
| PZE-104036105 | 4 | 47241320 | 枯萎度 | |
| SYN16455 | 4 | 239628761 | 枯萎度 | |
| SYN38269 | 9 | 136583562 | 枯萎度 |
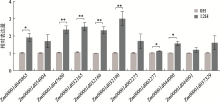
Supplementary Table 1
Primers sequences of qRT-PCR"
| Gene_ID | Primer-F | Primer-R |
|---|---|---|
| Zm00001d049865 | AATCACCGTCACCTTCAA | GCAGTAGAGGCGGTAGAA |
| Zm00001d049867 | AGGGCTACTTCAACGACC | GCAGCATTCCTCCAGCAC |
| Zm00001d054004 | TATGGGATGCCTGCTATG | CACGGTCTCATTGCCTAA |
| Zm00001d047680 | CTGGAAGCCGTGAGCAAC | CCTTGAACTCGGCTATGC |
| Zm00001d032165 | ATGGGTTCCTGTTTCTCC | TGTTTCAATGCCTTCTGG |
| Zm00001d032166 | TTCAGTTGCCTTACATCC | ACCATCCAATAACCCTCA |
| Zm00001d050434 | CACCTCTTCCGCCAAACA | AGCATCGTCAGAACCTCC |
| Zm00001d032190 | TGCCTGTCCTCCACTTCC | CCAACGTCGGCTTCCTCT |
| Zm00001d002275 | AACCCTTGGCTGTTACGA | AGCCGATGAGAACGAAAG |
| Zm00001d002277 | GCCGAGTACCTGGACGCA | TCTCCCATTTGGTCGTCA |
| Zm00001d044090 | TGGCTCTGATTGCTGGTG | TCCTGCTTTCCGCATCCC |
| Zm00001d044091 | AGTTGTGGTCCAAATAGC | TCTTTACCGTTCTCGTCT |
| Zm00001d037329 | ATGGGCAACAGCATCGGC | GCGTGAGCATGAGCGACT |
| ACTIN | GCATCCATGAGACCACCTACAAC | GATGGACCCTCCTATCCAGACAC |
Supplementary Table 2
Clustering of 150 inbred lines"
| NSS | SS | TST | Mixed | NSS | SS | TST | Mixed |
|---|---|---|---|---|---|---|---|
| PHR47 | 764 | 83IBI3 | PHG71 | PHZ51 | CR14 | LH164 | ND203 |
| PHK05 | 78002A | L135 | PHK29 | OH40B | 2369 | PHR62 | PHW03 |
| PHGG7 | B14 | NS501 | PHM81 | MBSJ | PHK35 | 912 | T226 |
| P737M20 | B14A | PHG29 | HB8229 | LH160 | A635 | 11430 | CO109 |
| LH93 | LH132 | PHG72 | 4676A | PHBA6 | PHW17 | PHG83 | |
| PHV78 | LH196 | PHH93 | PHVA9 | LH39 | LH143 | ||
| MO22 | LH197 | PHJ90 | PHW30 | PHR58 | PHP85 | ||
| L317 | LH202 | PHK42 | PHV37 | E8501 | 87916W | ||
| PHP60 | LH204 | PHN11 | FR19 | PHK76 | LH145 | ||
| PHR55 | LH205 | PHP02 | B47 | MBST | FAPW | ||
| PHR32 | LH220HT | PHP55 | PHR63 | LH128 | 6F629 | ||
| T8 | LH222 | Q381 | RS710 | 78551S | PHJ70 | ||
| PHM57 | LH74 | IBB15 | LH82 | 78371A | PHG39 | ||
| PHG84 | NS701 | PHM10 | L139 | C103 | PHN29 | ||
| PA91 | PB80 | PHG50 | NC250 | LH213 | F42 | ||
| MBPM | PF010 | PHN82 | PHT22 | LH214 | PHT55 | ||
| PHJ40 | PHG86 | PHR25 | LH163 | LH216 | PHW51 | ||
| PHV53 | PHT10 | NQ508 | OH7B | LH52 | PHHH9 | ||
| PHT60 | W8304 | IBO14 | PHG35 | LH54 | B37 | ||
| PHN73 | PHW52 | IB02 | PHR36 | LH57 | H93 | ||
| PHG47 | W8555 | 904 | POP-N-EAT | LH61 | PHV07 | ||
| PHWG5 | LH146HT | PHJ75 | 779 | SG17 | PHN66 | ||
| VA35 | LH149 | PHKE6 | PHW20 | WIL903 | |||
| LH162 | LP5 | PHN47 | W22 |
Supplementary Table 3
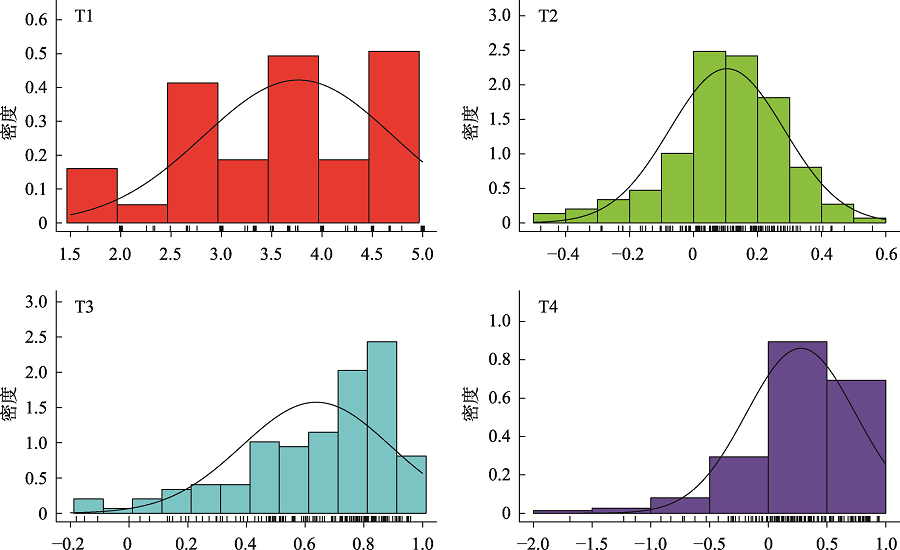
Phenotypic results of salt stress-related traits"
| 家系名称 | T1 | T2 | T3 | T4 |
|---|---|---|---|---|
| 764 | 4 | 0.106685807 | 0.859905959 | 0.775090325 |
| 779 | 3.666666667 | 0.053892216 | 0.441441441 | 0.034965035 |
| 904 | 3.333333333 | 0.120395688 | 0.885410628 | 0.525030525 |
| 912 | 3.75 | -0.022568998 | 0.640350877 | 0.20970266 |
| 2369 | 4 | 0.188785703 | 0.785634119 | 0.527058824 |
| 11430 | 3.333333333 | 0.138089758 | 0.487804878 | 0.385481852 |
| 4676A | 2 | 0.019224102 | 0.688906798 | 0.130612245 |
| 6F629 | 5 | 0.065575558 | 0.703703704 | -0.715789474 |
| 78002A | 4.5 | 0.161691542 | 0.5 | -0.015384615 |
| 78371A | 2.666666667 | 0.087169299 | 0.450910931 | -0.247563353 |
| 78551S | 5 | 0.332941696 | 0.834848485 | 0.55 |
| 83IBI3 | 3.5 | 0.23180624 | 0.75049891 | -0.30625 |
| 87916W | 5 | 0.139334378 | 0.783488028 | -0.195845697 |
| A635 | 5 | 0.182547395 | 0.720695971 | 0.268096515 |
| B14 | 4.666666667 | -0.421686747 | 0.363636364 | -1.133757962 |
| B14A | 4.5 | 0.301216743 | 0.831049398 | 0.715843489 |
| B37 | 3 | 0.136442918 | 0.8255563 | 0.142857143 |
| B47 | 5 | 0.309312793 | 0.871871872 | 0.490701001 |
| C103 | 4 | 0.558739226 | 0.910714286 | 0.464174455 |
| CO109 | 2 | -0.076696165 | 0 | 0.07826087 |
| CR14 | 4.333333333 | 0.014563107 | 0.52173913 | 0.536516854 |
| E8501 | 3 | 0.216734631 | 0.827926421 | 0.843537415 |
| F42 | 2.666666667 | 0.134222222 | 0.5 | 0.093406593 |
| FAPW | 4 | 0.286407767 | 0.636363636 | 0.203597122 |
| FR19 | 5 | 0.310602547 | 0.875838947 | 0.338830585 |
| H93 | 5 | 0.053082505 | 0.692640693 | -0.528767123 |
| HB8229 | 1.666666667 | -0.166666667 | 0.272727273 | 0.273139746 |
| IB02 | 5 | 0.382641225 | 0.558823529 | 0.389112903 |
| IBB15 | 2.666666667 | 0.125596897 | 0.792174797 | 0.340588988 |
| IBO14 | 5 | 0.310928226 | 0.829650504 | 0.083916084 |
| L135 | 5 | -0.015375964 | 0.298611111 | -0.00538358 |
| L139 | 2 | 0.094623146 | 0.728282828 | 0.07106599 |
| L317 | 4.666666667 | 0.432276531 | 0.873738254 | 0.348484848 |
| LH128 | 3 | -0.477574144 | 0.741428571 | 0.484827586 |
| LH132 | 4 | 0.180683002 | 0.755934783 | 0.773415133 |
| LH143 | 3.333333333 | -0.285917994 | 0.468148148 | 0.063334983 |
| LH145 | 3.666666667 | -0.128747795 | 0.296296296 | -0.339694656 |
| LH146HT | 5 | 0.031914894 | 0.6 | 0.158371041 |
| LH149 | 5 | -0.81443299 | 0.25 | -0.047619048 |
| LH160 | 3 | 0.143435352 | 0.774744974 | 0.776025237 |
| LH162 | 4 | -0.198924699 | 0.192639805 | 0.679364669 |
| LH163 | 3 | -0.160241915 | 0.631410256 | 0.074198988 |
| LH164 | 4 | 0.366197183 | 0.95 | 0.053254438 |
| LH196 | 2 | 0.150510204 | 0.5 | 0.247654784 |
| LH197 | 3 | 0.071724138 | 0.613636364 | -0.625 |
| LH202 | 2.666666667 | -0.289041096 | 0.037037037 | 0.263665595 |
| LH204 | 4.25 | 0.250292071 | 0.841452991 | 0.285356696 |
| LH205 | 5 | -0.149584076 | 0.919309524 | 0.807927812 |
| LH213 | 4 | 0.155 | 0.069767442 | 0.605 |
| LH214 | 4 | 0.146307858 | 0.75870936 | 0.356514788 |
| LH216 | 3 | 0.08213168 | 0.594179894 | 0.272829763 |
| LH220HT | 2.75 | 0.320799608 | 0.79386883 | 0.482022472 |
| LH222 | 3.75 | 0.250100281 | 0.704426129 | -0.047038328 |
| LH39 | 4 | 0.232224104 | 0.806684905 | 0.45472973 |
| LH52 | 3 | 0.191662197 | 0.897622655 | 0.853852481 |
| LH54 | 3.75 | 0.20716033 | 0.803333221 | 0.615072464 |
| LH57 | 5 | -0.013888889 | 0.476190476 | 0.363538296 |
| LH61 | 4.8 | 0.22531233 | 0.718266254 | -0.297577855 |
| LH74 | 4 | 0.091427566 | 0.807241965 | 0.617501166 |
| LH82 | 3 | 0.075389408 | 0.691056911 | 0.345581802 |
| LH93 | 3 | 0.3253386 | 0.874730053 | 0.645687646 |
| LP5 | 5 | 0.392215722 | 0.767102397 | -0.051948052 |
| MBPM | 4.5 | 0.429179416 | 0.906234289 | 0.627483444 |
| MBSJ | 3 | 0.070597856 | 0.176470588 | 0.210869565 |
| MBST | 2 | 0.031718062 | 0.496296296 | 0.191637631 |
| Mo22 | 3.75 | -0.011951503 | 0.79111927 | 0.626065341 |
| NC250 | 5 | 0.068195561 | 0.81547619 | -0.145363409 |
| ND203 | 3.5 | 0.034778671 | 0.526195286 | -0.279346211 |
| NQ508 | 4 | -0.221556886 | -1.058141026 | 0.169768754 |
| NS501 | 4 | 0.182549203 | 0.558760684 | -0.131880734 |
| NS701 | 3 | -0.392568659 | 0.148148148 | 0.355339806 |
| Oh40B | 5 | -0.009803922 | 0.428571429 | 0.081451061 |
| OH7B | 3.5 | 0.177205024 | 0.63962704 | 0.224550898 |
| P737M20 | 2.333333333 | 0.141843972 | 0.333333333 | 0.210884354 |
| Pa91 | 2 | 0.034689864 | 0.808045815 | 0.723135146 |
| PB80 | 4.333333333 | 0.161861037 | 0.693253923 | 0.165021157 |
| PF010 | 3.333333333 | 0.309720548 | 0.860636507 | 0.557239057 |
| PHBA6 | 3 | -0.038989439 | 0.388678052 | -0.830548926 |
| PHG29 | 3.333333333 | 0.065765251 | 0.871539081 | 0.74143057 |
| PHG35 | 4.666666667 | 0.293120097 | 0.923564127 | 0.862402512 |
| PHG39 | 5 | 0.114242424 | -0.545079365 | 0.283173077 |
| PHG47 | 3 | 0.133514678 | 0.92 | 0.830970776 |
| PHG50 | 2 | 0.2125746 | 0.869800202 | 0.8170164 |
| PHG71 | 4 | 0.281727747 | 0.875491259 | 0.774668813 |
| PHG72 | 5 | 0.40382879 | 0.951678788 | 0.847798341 |
| PHG83 | 2.5 | -0.325203252 | -0.151515152 | -1.230769231 |
| PHG84 | 5 | 0.470915549 | 0.924074074 | 0.836134454 |
| PHG86 | 3 | 0.063305513 | 0.799387904 | 0.739177883 |
| PHGG7 | 3 | 0.064839051 | 0.880388466 | 0.801129332 |
| PHH93 | 4 | 0.075818696 | 0.73994832 | 0.938946046 |
| PHHH9 | 3.666666667 | 0.186025829 | 0.841795255 | 0.851485149 |
| PHJ40 | 4.666666667 | 0.016144349 | 0.515789474 | 0.016645327 |
| PHJ70 | 4 | 0.182412791 | 0.658227848 | 0.370417193 |
| PHJ75 | 2.666666667 | 0.202660478 | 0.771780405 | 0.710249474 |
| PHJ90 | 2 | 0.105318062 | 0.785738301 | 0.74165577 |
| PHK05 | 3.666666667 | -0.064676617 | -0.179487179 | -0.730200174 |
| PHK29 | 3 | 0.125299248 | 0.778988805 | 0.772440526 |
| PHK35 | 5 | 0.252361057 | 0.929857143 | 0.849577532 |
| PHK42 | 2 | -0.086715867 | 0.130434783 | 0.305232558 |
| PHK76 | 4.5 | 0.202912215 | 0.310810811 | 0.05 |
| PHKE6 | 3.333333333 | -0.041915065 | 0.533501604 | 0.124 |
| PHM10 | 4.333333333 | 0.025943396 | 0.481481481 | 0.06446281 |
| PHM57 | 2.666666667 | 0.38240269 | 0.851551971 | 0.860177467 |
| PHM81 | 3.666666667 | -0.221052632 | -0.107142857 | 0.312684366 |
| PHN11 | 4 | 0.2244816 | 0.719628647 | 0.196078431 |
| PHN29 | 5 | 0.263157895 | 0.555555556 | 0.944954128 |
| PHN47 | 4.25 | 0.024830793 | 0.739251768 | 0.164233577 |
| PHN66 | 4 | 0.139673354 | 0.742966277 | 0.021935484 |
| PHN73 | 4 | 0.020422018 | 0.811269556 | 0.743728068 |
| PHN82 | 4.5 | 0.049443442 | 0.802449965 | 0.597969543 |
| PHP02 | 4 | 0.035364483 | 0.602286325 | 0.484480432 |
| PHP55 | 5 | 0.261301942 | 0.785714286 | 0.31686747 |
| PHP60 | 4.5 | 0.009009009 | 0.44 | 0.050179211 |
| PHP85 | 5 | 0.237179487 | 0.403508772 | 0.212627669 |
| PHR25 | 3 | 0.098412698 | 0.229166667 | 0.261989343 |
| PHR32 | 4 | 0.114851636 | 0.912760137 | 0.488294314 |
| PHR36 | 4 | 0.27199986 | 0.838235294 | 0.115 |
| PHR47 | 5 | 0.281074811 | 0.5875 | 0.427966102 |
| PHR55 | 4 | -0.072343744 | 0.607460317 | -1.69001387 |
| PHR58 | 4 | -0.234189084 | 0.837209302 | 0.417682927 |
| PHR62 | 4.333333333 | -0.015039405 | 0.723585859 | 0.092957746 |
| PHR63 | 3 | -0.025642716 | 0.879562948 | 0.76727909 |
| PHT10 | 3 | 0.036237933 | 0.866034337 | 0.807460176 |
| PHT22 | 3.333333333 | 0.208751022 | 0.742037037 | 0.746268657 |
| PHT55 | 5 | 0.210235665 | 0.959651515 | 0.928606153 |
| PHT60 | 3.666666667 | 0.273846154 | 0.49122807 | -0.107317073 |
| PHV07 | 3 | 0.059171598 | 0.203389831 | -0.100628931 |
| PHV37 | 2 | 0.063039724 | 0.479166667 | -0.169934641 |
| PHV53 | 5 | 0.071045068 | 0.946530604 | 0.866765496 |
| PHV78 | 5 | -0.101263146 | 0.565121136 | -0.078014184 |
| PHVA9 | 3.25 | 0.190359348 | 0.806177156 | 0.805031447 |
| PHW03 | 2.333333333 | 0.00662121 | 0.318347271 | -0.342763874 |
| PHW17 | 5 | -0.104384214 | 0.815278788 | 0.69679773 |
| PHW20 | 4 | 0.069364162 | 0.5125 | 0.129181084 |
| PHW30 | 5 | -0.324631794 | 0.698446637 | 0.521840614 |
| PHW51 | 4 | 0.132500019 | 0.764473304 | 0.645142637 |
| PHW52 | 5 | 0.012268182 | 0.137931034 | 0.176470588 |
| PHWG5 | 3 | -0.082323159 | 0.665372199 | -0.05483871 |
| PHZ51 | 3 | 0.139534884 | 0.5625 | 0.023224044 |
| POP-N-EAT | 2.25 | -0.03435569 | 0.650768766 | 0.71042471 |
| Q381 | 4.333333333 | 0.093844601 | 0.378787879 | 0.236961451 |
| RS710 | 3.25 | 0.246142459 | 0.841076856 | 0.850202429 |
| SG17 | 3.666666667 | 0.279799616 | 0.861968465 | 0.529527559 |
| T226 | 5 | 0.195014617 | 0.735802728 | -0.318181818 |
| T8 | 2.666666667 | 0.122820737 | 0.689078642 | 0.212806026 |
| Va35 | 2 | 0.178753558 | 0.898576185 | 0.783600802 |
| W22 | 4.333333333 | 0.261864034 | 0.068783069 | -0.426829268 |
| W8304 | 4.666666667 | 0.114802673 | 0.757346491 | -0.010791367 |
| W8555 | 3.333333333 | 0.197577587 | 0.522999223 | -0.91321499 |
| WIL903 | 3.333333333 | 0.243623993 | 0.626071044 | 0.033678756 |
Supplementary Table 4
All genes within the 50 Kb range before and after the significant SNP locus"
| 性状 | 显著SNP | 染色体 | start | end | 候选基因_V2 | 候选基因_V4 | 注释 |
|---|---|---|---|---|---|---|---|
| T1 | PZE-104036105 | 4 | 47199131 | 47199937 | GRMZM2G325580 | Zm00001d049865 | CASP-like protein |
| T1 | SYN16455 | 4 | 239623748 | 239627091 | GRMZM2G074423 | Zm00001d054004 | plant-specific domain tigr01589 family protein |
| T1 | SYN16455 | 4 | 239627275 | 239630488 | GRMZM2G074414 | Zm00001d054005 | biotinyl-lipoyl domain- containing protein |
| T1 | SYN16455 | 4 | 239630746 | 239640417 | GRMZM2G074278 | Zm00001d054005 | ap-3 complex subunit mu-1-like |
| T1 | SYN16455 | 4 | 239640750 | 239641632 | GRMZM2G074252 | Zm00001d054006 | hypothetical protein SORBIDRAFT_04g001625 |
| T1 | SYN16455 | 4 | 239643193 | 239651050 | GRMZM2G074015 | Zm00001d054008 | mevalonate kinase |
| T1 | SYN38269 | 9 | 136552200 | 136555676 | GRMZM2G006429 | Zm00001d047679 | probable protein phosphatase 2c 5-like |
| T1 | SYN38269 | 9 | 136558046 | 136561365 | GRMZM2G006714 | Zm00001d047680 | calmodulin-binding protein |
| T1 | SYN38269 | 9 | 136572037 | 136574076 | GRMZM2G306482 | Zm00001d047681 | pentatricopeptide repeat- containing protein at5g50990-like |
| T1 | SYN38269 | 9 | 136576621 | 136577373 | GRMZM2G006830 | Zm00001d047681 | protein cobra |
| T1 | SYN38269 | 9 | 136583501 | 136589780 | GRMZM2G006884 | Zm00001d047682 | --NA-- |
| T1 | SYN38269 | 9 | 136615573 | 136620806 | GRMZM5G830983 | Zm00001d047683 | udp-glucose 4-epimerase |
| T2 | SYNGENTA5755 | 1 | 212166415 | 212168249 | GRMZM2G067235 | Zm00001d032164 | hypothetical protein |
| T2 | SYNGENTA5755 | 1 | 212191637 | 212196339 | GRMZM2G067067 | Zm00001d032165 | Defective in cullin neddylation protein |
| T2 | SYNGENTA5755 | 1 | 212287664 | 212305641 | GRMZM2G160927 | Zm00001d032166 | DUF1336 domain-containing protein |
| T2 | SYNGENTA5755 | 1 | 212332408 | 212358537 | GRMZM2G005955 | Zm00001d032170 | lipid binding |
| T2 | PZE-101169962 | 1 | 213230607 | 213233044 | GRMZM2G117193 | Zm00001d032190 | MYB family transcription factor |
| T2 | PZE-101169962 | 1 | 213309345 | 213314586 | GRMZM2G031677 | Zm00001d032192 | ap-3 complex subunit mu-1-like |
| T2 | SYN1142 | 2 | 9085071 | 9088878 | GRMZM2G084046 | Zm00001d002274 | dna binding |
| T2 | SYN1142 | 2 | 9114721 | 9126810 | GRMZM2G084181 | Zm00001d002275 | abc transporter c family member 2-like |
| T2 | SYN1142 | 2 | 9127513 | 9128809 | GRMZM2G084325 | Zm00001d002276 | 5a2 protein precursor |
| T2 | SYN1142 | 2 | 9128588 | 9133414 | GRMZM2G383122 | Zm00001d002277 | Ubiquitin-like domain- containing protein |
| T2 | PHM13742.5 | 3 | 215169901 | 215182117 | GRMZM2G088436 | Zm00001d044089 | rna polymerase sigma factor-like |
| T2 | PHM13742.5 | 3 | 215207099 | 215211653 | GRMZM2G088397 | Zm00001d044090 | 5'-deoxynucleotidase |
| T2 | PHM13742.5 | 3 | 215213789 | 215219139 | GRMZM2G088299 | Zm00001d044091 | serine threonine-protein kinase ht1-like |
| T2 | PZE-106065133 | 6 | 117370662 | 117372555 | GRMZM2G012717 | Zm00001d037327 | zinc finger |
| T2 | PZE-106065133 | 6 | 117452361 | 117454400 | GRMZM2G168304 | Zm00001d037328 | 3-ketoacyl-CoA synthase |
| T2 | PZE-106065133 | 6 | 117595563 | 117599517 | GRMZM2G382711 | Zm00001d037329 | uncharacterized protein |
| [1] | Deinlein U, Stephan AB, Horie T, Luo W, Xu GH, Schroeder JI. Plant salt-tolerance mechanisms. Trends Plant Sci, 2014,19(6):371-379. |
| [2] | Hu T, Zhang GX, Zheng FC, Cao Y. Research progress in plant salt stress response. Mol Plant Breed, 2018,16(9):3006-3015. |
| 胡涛, 张鸽香, 郑福超, 曹钰. 植物盐胁迫响应的研究进展. 分子植物育种, 2018,16(9):3006-3015. | |
| [3] | Munns R, Tester M. Mechanisms of salinity tolerance. Annu Rev Plant Biol, 2008,59:651-681. |
| [4] | Liang WJ, Ma XL, Wan P, Liu LY. Plant salt-tolerance mechanism: a review. Biochem Bioph Res Co, 2018,495(1):286-291. |
| [5] | Luo X, Wang BC, Gao S, Zhang F, Terzaghi W, Dai MQ. Genome-wide association study dissects the genetic bases of salt tolerance in maize seedlings. J Integr Plant Biol, 2019,61(6):658-674. |
| [6] | Prasad SR, Bagali PG, Hittalmani S, Shashidhar HE. Molecular mapping of quantitative trait loci associated with seedling tolerance to salt stress in rice (Oryza sativa L.). Curr Sci India, 2000,78(2):162-164. |
| [7] | Hamwieh A, Tuyen DD, Cong H, Benitez ER, Takahashi R, Xu DH. Identification and validation of a major QTL for salt tolerance in soybean. Euphytica, 2011,179(3):451-459. |
| [8] | Ren ZH, Zheng ZM, Chinnusamy V, Zhu JH, Cui XP, Iida K, Zhu JK. RAS1, a quantitative trait locus for salt tolerance and ABA sensitivity in Arabidopsis. Proc Natl Acad Sci USA, 2010,107(12):5669-5674. |
| [9] | Luo MJ, Zhao YX, Zhang RY, Xing JF, Duan MX, Li JN, Wang NS, Wang WG, Zhang SS, Chen ZH, Zhang HS, Shi Z, Song W, Zhao JR. Mapping of a major QTL for salt tolerance of mature field-grown maize plants based on SNP markers. BMC Plant Biol, 2017,17(1):140. |
| [10] | Luo MJ, Zhang YX, Chen K, Kong MS, Song W, Lu BS, Shi YX, Zhao YX, Zhao JR. Mapping of quantitative trait loci for seedling salt tolerance in maize. Mol Breeding, 2019,39(5):1-12. |
| [11] | Ren ZH, Gao JP, Li LG, Cai XL. Huang W, Chao DY, Zhu MZ, Wang ZY, Luan S, Liu HX. A rice quantitative trait locus for salt tolerance encodes a sodium transporter. Nat Genet, 2005,37(10):1141-1146. |
| [12] | Cui D, Wu D, Somarathna Y, Xu CY, Li S, Li P, Zhang H, Chen HB, Li Z. QTL mapping for salt tolerance based on SNP markers at the seedling stage in maize (Zea mays L.). Euphytica, 2015,203(2):273-283. |
| [13] | Zhang M, Cao YB, Wang ZP, Wang ZQ, Shi JP, Liang XY, Song WB, Chen QJ, Lai JS, Jiang CF. A retrotransposon in an HKT1 family sodium transporter causes variation of leaf Na + exclusion and salt tolerance in maize . New Phytol, 2018,217(3):1161-1176. |
| [14] | Zhang M, Liang XY, Wang LM, Cao YB, Song WB, Shi JP, Lai JS, Jiang CF. A HAK family Na + transporter confers natural variation of salt tolerance in maize . Nat Plants, 2019,5(12):1297-1308. |
| [15] | Cao YB, Zhang M, Liang XY, Li FR, Shi YL, Yang XH, Jiang CF. Natural variation of an EF-hand Ca 2+-binding- protein coding gene confers saline-alkaline tolerance in maize . Nat Commun, 2020,11(1):186. |
| [16] | Xie YH, Feng Y, Chen Q, Zhao FK, Zhou SJ, Ding Y, Song XL, Li P, Wang BH. Genome-wide association analysis of salt tolerance QTLs with SNP markers in maize (Zea mays L.). Genes Genomics, 2019,41(10):1135-1145. |
| [17] | Ganal MW, Durstewitz G, Polley A, Bérard A, Buckler ES, Charcosset A, Clarke JD, Graner E, Hansen M, Joets J, LePaslier M, McMullen MD, Montalent P, Rose M, Schön C, Sun Q, Walter H, Martin OC, Falque M. A large maize (Zea mays L.) SNP genotyping array: development and germplasm genotyping, and genetic mapping to compare with the B73 reference genome. PLoS One, 2011,6(12):e28334. |
| [18] | Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, Maller J, Sklar P, de Bakker PIW, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet, 2007,81(3):559-575. |
| [19] | Alexander D, Lange K. Enhancements to the ADMIXTURE algorithm for individual ancestry estimation. BMC Bioinformatics, 2011,12(1):1-6. |
| [20] | Bradbury PJ, Zhang ZW, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES. TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics, 2007,23(19):2633-2635. |
| [21] | Kumar S, Tamura K, Nei M. MEGA: molecular evolutionary genetics analysis software for microcomputers. Bioinformatics, 1994,10(2):189-191. |
| [22] | Yu JM, Pressoir G, Briggs WH, Bi IV, Yamasaki M, Doebley JF, McMullen MD, Gaut BS, Nielsen DM, Holland JB, Kresovich S, Buckle ES. A unified mixed- model method for association mapping that accounts for multiple levels of relatedness. Nat Genet, 2006,38(2):203-208. |
| [23] | Yu SB, Li JX, Xu CG, Tan YF, Gao YJ, Li XH, Zhang Q, Maroof MAF. Importance of epistasis as the genetic basis of heterosis in an elite rice hybrid. Proc Natl Acad Sci USA, 1997,94(17):9226-9231. |
| [24] | Wen WW, Liu HJ, Zhou Y, Jin M, Yang N, Li D, Luo J, Xiao YJ, Pan QC, Tohge T, Fernie AR, Yan JB. Combining quantitative genetics approaches with regulatory network analysis to dissect the complex metabolism of the maize kernel. Plant Physiol, 2016,170(1):136-146. |
| [25] | Li H, Peng ZY, Yang XH, Wang WD, Fu JJ, Wang JH, Han YJ, Chai YC, Guo TT, Yang N, Liu J, Warburton ML, Cheng YB, Hao XM, Zhang P, Zhao JY, Liu YJ, Wang GY, Li JS, Yan JB. Genome-wide association study dissects the genetic architecture of oil biosynthesis in maize kernels. Nat Genet, 2013,45(1):43-50. |
| [26] | Hoopes GM, Hamilton JP, Wood JC, Esteban E, Pasha A, Vaillancourt B, Provart NJ, Buell CR. An updated gene atlas for maize reveals organ-specific and stress-induced genes. Plant J, 2019,97(6):1154-1167. |
| [27] | Yang XH, Gao SB, Xu ST, Zhang ZX, Prasanna BM, Li L, Li JS, Yan JB. Characterization of a global germplasm collection and its potential utilization for analysis of complex quantitative traits in maize. Mol Breeding, 2011,28(4):511-526. |
| [28] | Yang XH, Yan JB, Zheng YP, Yu JM, Li JS. Reviews of association analysis for quantitative traits in plants. Acta Agron Sin, 2007,33(4):523-530. |
| 杨小红, 严建兵, 郑艳萍, 余建明, 李建生. 植物数量性状关联分析研究进展. 作物学报, 2007,33(4):523-530. | |
| [29] | Meng YY, Wang ZY, Wang YQ, Wang CN, Zhu BT, Liu H, Ji WK, Wen JQ, Chu CC, Tadege M, Niu LF, Lin H. The MYB activator WHITE PETAL1 associates with MtTT8 and MtWD40-1 to regulate carotenoid-derived flower pigmentation in medicago truncatula. Plant Cell, 2019,31(11):2751-2767. |
| [30] | Wang J, Zhou L, Shi H, Chern M, Yu H, Yi H, He M, Yin JJ, Zhu XB, Li Y, Li WT, Liu JL, Wang JC, Chen XQ, Qing H, Wang YP, Liu GF, Wang WM, Li P, Wu XJ, Zhu LH, Zhou JM, Ronald PC, Li SG, Li JY, Chen WX. A single transcription factor promotes both yield and immunity in rice. Science, 2018,361(6406):1026-1028. |
| [31] | Ambawat S, Sharma P, Yadav NR, Yadav RC. MYB transcription factor genes as regulators for plant responses: an overview. Physiol Mol Biol Plants, 2013,19(3):307-321. |
| [32] | Qin YX, Wang MC, Tian YC, He WX, Han L, Xia GM. Over-expression of TaMYB33 encoding a novel wheat MYB transcription factor increases salt and drought tolerance in Arabidopsis. Mol Biol Rep, 2012,39(6):7183-7192. |
| [33] | He YN, Li W, Lv J, Jia YB, Wang MC, Xia GM. Ectopic expression of a wheat MYB transcription factor gene, TaMYB73, improves salinity stress tolerance in Arabidopsis thaliana. J Exp Bot, 2012,63(3):1511-1522. |
| [34] | Chen YH, Cao YY, Wang LJ, Li LM, Yang J, Zou MX. Identification of MYB transcription factor genes and their expression during abiotic stresses in maize. Biol Plantarum, 2018,62(2):222-230. |
| [35] | Wu JD, Jiang YL, Liang YN, Chen L, Chen WJ, Cheng BJ. Expression of the maize MYB transcription factor ZmMYB3R enhances drought and salt stress tolerance in transgenic plants. Plant Physiol Bioch, 2019,137:179-188. |
| [36] | Dong W, Liu XJ, Li DL, Gao TX, Song YG. Transcriptional profiling reveals that a MYB transcription factor MsMYB4 contributes to the salinity stress response of alfalfa. PLoS One, 2018,13(9):e0204033. |
| [37] | Guo HY, Wang YC, Wang LQ, Hu P, Wang YM, Jia YY, Zhang CR, Zhang Y, Zhang YM, Wang C, Yang CP. Expression of the MYB transcription factor gene Bpl MYB46 affects abiotic stress tolerance and secondary cell wall deposition in betula platyphylla. Plant Biotechnol J, 2017,15(1):107-121. |
| [38] | Yang N, Lu YL, Yang XH, Huang J, Zhou Y, Ali F, Wen WW, Liu J, Li JS, Yan JB. Genome wide association studies using a new nonparametric model reveal the genetic architecture of 17 agronomic traits in an enlarged maize association panel. PLoS Genet, 2014,10(9):e1004573. |
| [39] | Li N, Lin B, Wang H, Li XM, Yang FF, Ding XH, Yan JB, Chu ZH. Natural variation in ZmFBL41 confers banded leaf and sheath blight resistance in maize. Nat Genet, 2019,51(10):1540-1548. |
| [40] | Wang XL, Wang HW, Liu SX, Ferjani A, Li JS, Yan JB, Yang XH, Qin F. Genetic variation in ZmVPP1 contributes to drought tolerance in maize seedlings. Nat Genet, 2016,48(10):1233-1241. |
| [41] | Mao HD, Wang HW, Liu SX, Li ZG, Yang XH, Yan JB, Li JS, Tran LP, Qin F. A transposable element in a NAC gene is associated with drought tolerance in maize seedlings. Nat Commun, 2015,6:8326. |
| [42] | Luo MJ, Zhang YX, Li JN, Zhang PP, Chen K, Song W, Wang XQ, Yang JX, Lu XD, Lu YX, Zhao JR. Molecular dissection of maize seedling salt tolerance using a genome-wide association analysis method. Plant Biotechnol J, 2021,19(10):1937-1951. |
| [1] | Yu Chen, Xiaoyun Chen, Cheng Peng, Junfeng Xu, Jie Shen, Yueying Li, Xiaofu Wang. Establishment of a field visual detection method for genetically modified maize ‘Shuangkang’12-5 by fluorescence RPA [J]. Hereditas(Beijing), 2021, 43(8): 802-812. |
| [2] | Yidan Sun, Zizhao Tian, Wei Zhou, Mo Li, Cong Huai, Lin He, Shengying Qin. Genome-wide association study on liver function tests in Chinese [J]. Hereditas(Beijing), 2021, 43(3): 249-260. |
| [3] | Guoqing Qian. Advances in genome-wide association study of chronic obstructive pulmonary disease [J]. Hereditas(Beijing), 2020, 42(9): 832-846. |
| [4] | Wang Dezhou,Mo Xiaoting,Zhang Xia,Xu Miaoyun,Zhao Jun,Wang Lei. Isolation and functional characterization of a stress-responsive transcription factor ZmC2H2-1 in Zea mays [J]. Hereditas(Beijing), 2018, 40(9): 767-778. |
| [5] | Zhuofan Zhao, Ling Huang, Yongming Liu, Peng Zhang, Gui Wei, Moju Cao. Genetics of fertility restoration in the isocytoplasm allonuclear C-group of cytoplasmic male sterility in maize [J]. Hereditas(Beijing), 2018, 40(5): 402-414. |
| [6] | Chao Yang, Ruifu Yang, Yujun Cui. Bacterial genome-wide association study: methodologies and applications [J]. Hereditas(Beijing), 2018, 40(1): 57-65. |
| [7] | Yuyan Wang,Zixing Wang,Yaoda Hu,Lei Wang,Ning Li,Biao Zhang,Wei Han,Jingmei Jiang. Current status of pathway analysis in genome-wide association study [J]. Hereditas(Beijing), 2017, 39(8): 707-716. |
| [8] | Zhaohua Yin, Yan Li, Chunqing Zhang, Cuicui Yang, Chenglai Wu, Xiangpan Liu, Mingming Wang. Cloning and promoter activity analyses of the promoter of 2’-deoxymugineic acid (DMA) secretion channel gene YS3 in maize [J]. HEREDITAS(Beijing), 2016, 38(6): 560-568. |
| [9] | Kaixu Chen, Weilan Wang, Fuchun Zhang, Xiufen Zheng. Progress in genetic research of human height [J]. HEREDITAS(Beijing), 2015, 37(8): 741-755. |
| [10] | Li Wang, Yanmei Xu, Zhujun Cheng, Zhaoping Xiong, Libin Deng. The progress of genetics of cholesterol metabolism disorder [J]. HEREDITAS(Beijing), 2014, 36(9): 857-863. |
| [11] | Xiongwei Zhao, Haijian Lin, Zhiming Zhang, Yaou Shen, Guangtang Pan. Mapping of QTLs controlling Pb2+ content in maize kernels under Pb2+stress [J]. HEREDITAS(Beijing), 2014, 36(8): 821-826. |
| [12] | Jiao Li, Yuqi Guo, Weiling Cui, Aihua Xu, Zengyuan Tian. Response of maize serine/arginine-rich protein gene family in seedlings to drought stress [J]. HEREDITAS(Beijing), 2014, 36(7): 697-706. |
| [13] | Xuehuan Gao, Fengling Fu, Wei Mu, Shufeng Zhou, Suzhi Zhang, Wanchen Li. Cloning and functional validation of promoter of mo-molybdopterin cofactor sulfurase gene in maize [J]. HEREDITAS(Beijing), 2014, 36(6): 584-591. |
| [14] | Jiapeng Zhou, Zhiyong Pei, Yubao Chen, Runsheng Chen. Strategies of genome-wide association study based on high-throughput sequencing [J]. HEREDITAS(Beijing), 2014, 36(11): 1099-1111. |
| [15] | Yanhua Zhang, Hongyang Yi, Ming Fang, Tingzhao Rong, Moju Cao. Cytological observation and DNA methylation analysis of two new cytoplasmic male sterile lines of maize during microsporogenesis [J]. HEREDITAS(Beijing), 2014, 36(10): 1021-1026. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||