Hereditas(Beijing) ›› 2022, Vol. 44 ›› Issue (1): 15-24.doi: 10.16288/j.yczz.21-329
• Orginal Articles • Previous Articles Next Articles
The regulatory effect of protein acetylation modification on autophagy
Jing Liu( ), Cong Yi, Shiming Xu(
), Cong Yi, Shiming Xu( )
)
- School of Medicine, Zhejiang University, Hangzhou 310020, China
-
Received:2021-09-13Revised:2021-12-01Online:2022-01-20Published:2021-12-02 -
Contact:Xu Shiming E-mail:18428302536@163.com;xusm@e-mdic.cn -
Supported by:Supported by the National Natural Science Foundation of China No(31600934)
Cite this article
Jing Liu, Cong Yi, Shiming Xu. The regulatory effect of protein acetylation modification on autophagy[J]. Hereditas(Beijing), 2022, 44(1): 15-24.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
Table 1
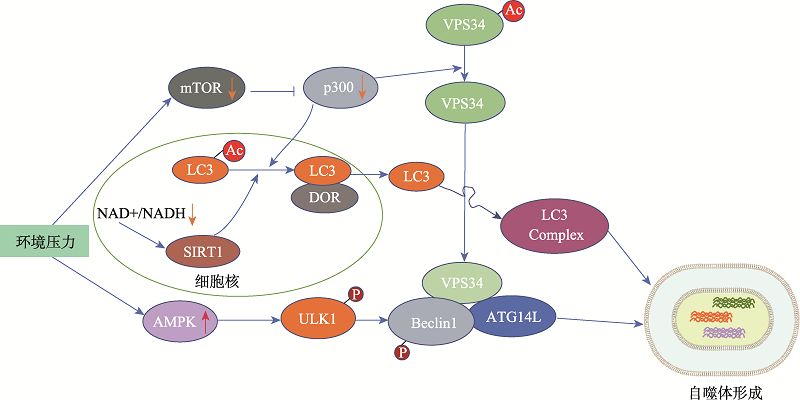
Autophagy-related protein (ATG) acetylation regulates autophagy"
| 蛋白 | 乙酰化位点 | 乙酰化酶/去乙酰化酶 | 对自噬的影响 | 相关文献 |
|---|---|---|---|---|
| ATG12 | — | p300/SIRT1 | ATG12乙酰化抑制自噬发生 | [ |
| ATG9A | K359、K363 | —/SIRT1 | ATG9A的去乙酰不仅影响自噬体膜的形成,也充当内质网(ER)压力诱导自噬的传感器 | [ |
| LC3 | K49、K51 | p300/SIRT1 | 去乙酰化的LC3转位到胞质内,参与自噬复合体的形成 | [ |
| VPS34 | K29、K771 | p300/SIRT1 | p300依赖性乙酰化/去乙酰化是诱导VPS34激酶活化并参与自噬的关键 | [ |
| ULK1 | K162、K606 | TIP60/— | ULK1的乙酰化是生长因子缺乏诱导的自噬所必需的 | [ |
| ATG5 | — | p300/SIRT1 | ATG5乙酰化抑制自噬溶酶体的成熟 | [ |
| ATG7 | — | p300/SIRT1 | ATG7乙酰化抑制自噬 | [ |
| ATG3 | K19、K48 | Esa1/Rpd3 | ATG3的乙酰化通过控制ATG3和ATG8的相互作用以及ATG8的脂化来调节自噬 | [ |
| Pacer | K483、K573 | TIP60/— | Pacer的乙酰化是自噬体成熟和细胞脂质代谢所必需的 | [ |
| STX17 | K219、K223 | CBP/HDAC2 | STX17被募集到自噬体膜后,STX17的去乙酰化是其在自噬溶酶体融合中发挥作用的必不可少的步骤 | [ |
| [1] |
Cecconi F, Levine B. The role of autophagy in mammalian development: cell makeover rather than cell death. Dev Cell, 2008, 15(3):344-357.
doi: 10.1016/j.devcel.2008.08.012 |
| [2] |
Reggiori F, Ungermann C. Autophagosome maturation and fusion. J Mol Biol, 2017, 429(4):486-496.
doi: S0022-2836(17)30016-5 pmid: 28077293 |
| [3] |
Wang LM, Qi H, Tang YC, Shen HM. Post-translational modifications of key machinery in the control of mitophagy. Trends Biochem Sci, 2020, 45(1):58-75.
doi: 10.1016/j.tibs.2019.08.002 |
| [4] |
Hill SM, Wrobel L, Rubinsztein DC. Post-translational modifications of Beclin 1 provide multiple strategies for autophagy regulation. Cell Death Differ, 2019, 26(4):617-629.
doi: 10.1038/s41418-018-0254-9 |
| [5] |
Reiche J, Huber O. Post-translational modifications of tight junction transmembrane proteins and their direct effect on barrier function. Biochim Biophys Acta Biomembr, 2020, 1862(9):183330.
doi: 10.1016/j.bbamem.2020.183330 |
| [6] |
Guerra-Castellano A, Márquez I, Pérez-Mejías G, Díaz-Quintana A, De la Rosa MA, Díaz-Moreno I. Post-translational modifications of cytochrome c in cell life and disease. Int J Mol Sci, 2020, 21(22):8483.
doi: 10.3390/ijms21228483 |
| [7] |
Janke C, Chloë Bulinski J. Post-translational regulation of the microtubule cytoskeleton: mechanisms and functions. Nat Rev Mol Cell Biol, 2011, 12(12):773-786.
doi: 10.1038/nrm3227 |
| [8] |
Mizushima N, Yoshimori T, Ohsumi Y. The role of Atg proteins in autophagosome formation. Annu Rev Cell Dev Biol, 2011, 27:107-132.
doi: 10.1146/annurev-cellbio-092910-154005 pmid: 21801009 |
| [9] |
Allfrey VG, Faulkner R, Mirsky AE. Acetylation and methylation of histones and their possible role in the regulation of RNA synthesis. Proc Natl Acad Sci USA, 1964, 51(5):786-794.
doi: 10.1073/pnas.51.5.786 |
| [10] |
Verdin E, Ott M. 50 years of protein acetylation: from gene regulation to epigenetics, metabolism and beyond. Nat Rev Mol Cell Biol, 2015, 16(4):258-264.
doi: 10.1038/nrm3931 |
| [11] |
Yorimitsu T, Klionsky DJ. Autophagy: molecular machinery for self-eating. Cell Death Differ, 2005, 12(Suppl 2):1542-1552.
doi: 10.1038/sj.cdd.4401765 |
| [12] |
Klionsky DJ, Cuervo AM, Dunn WA, Levine B, van der Klei I, Seglen PO. How shall I eat thee? Autophagy, 2007, 3(5):413-416.
pmid: 17568180 |
| [13] |
Majeski AE, Dice JF. Mechanisms of chaperone-mediated autophagy. Int J Biochem Cell Biol, 2004, 36(12):2435-2444.
doi: 10.1016/j.biocel.2004.02.013 |
| [14] |
Li WW, Li J, Bao JK. Microautophagy: lesser-known self-eating. Cell Mol Life Sci, 2012, 69(7):1125-1136.
doi: 10.1007/s00018-011-0865-5 |
| [15] |
Johansen T, Lamark T. Selective autophagy mediated by autophagic adapter proteins. Autophagy, 2011, 7(3):279-296.
doi: 10.4161/auto.7.3.14487 pmid: 21189453 |
| [16] |
A M, Latario CJ, Pickrell LE, Higgs HN. Lysine acetylation of cytoskeletal proteins: emergence of an actin code. J Cell Biol, 2020, 219(12):e202006151.
doi: 10.1083/jcb.202006151 |
| [17] | Drazic A, Myklebust LM, Ree R, Arnesen T. The world of protein acetylation. Biochim Biophys Acta, 2016, 1864(10):1372-1401. |
| [18] |
Liu YX, Yang H, Liu XC, Gu HH, Li YZ, Sun C. Protein acetylation: a novel modus of obesity regulation. J Mol Med (Berl), 2021, 99(9):1221-1235.
doi: 10.1007/s00109-021-02082-2 |
| [19] | Zhang YJ, Sun ZX, Jia JQ, Du TJ, Zhang NC, Tang Y, Fang Y, Fang D. Overview of histone modification. Adv Exp Med Biol, 2021, 1283:1-16. |
| [20] |
Füllgrabe J, Hajji N, Joseph B. Cracking the death code: apoptosis-related histone modifications. Cell Death Differ, 2010, 17(8):1238-1243.
doi: 10.1038/cdd.2010.58 pmid: 20467440 |
| [21] |
Allis CD, Berger SL, Cote J, Dent S, Jenuwien T, Kouzarides T, Pillus L, Reinberg D, Shi Y, Shiekhattar R, Shilatifard A, Workman J, Zhang Y. New nomenclature for chromatin-modifying enzymes. Cell, 2007, 131(4):633-636.
doi: 10.1016/j.cell.2007.10.039 |
| [22] |
Shao YF, Gao ZH, Marks PA, Jiang XJ. Apoptotic and autophagic cell death induced by histone deacetylase inhibitors. Proc Natl Acad Sci USA, 2004, 101(52):18030-18035.
doi: 10.1073/pnas.0408345102 |
| [23] |
Eisenberg T, Knauer H, Schauer A, Büttner S, Ruckenstuhl C, Carmona-Gutierrez D, Ring J, Schroeder S, Magnes C, Antonacci L, Fussi H, Deszcz L, Hartl R, Schraml E, Criollo A, Megalou E, Weiskopf D, Laun P, Heeren G, Breitenbach M, Grubeck-Loebenstein B, Herker E, Fahrenkrog B, Fröhlich KU, Sinner F, Tavernarakis N, Minois N, Kroemer G, Madeo F. Induction of autophagy by spermidine promotes longevity. Nat Cell Biol, 2009, 11(11):1305-1314.
doi: 10.1038/ncb1975 pmid: 19801973 |
| [24] | Füllgrabe J, Klionsky DJ, Joseph B. The return of the nucleus: transcriptional and epigenetic control of autophagy. Nat Rev Mol Cell Biol, 2014, 15(1):65-74. |
| [25] |
Saidi D, Cheray M, Osman AM, Stratoulias V, Lindberg OR, Shen XL, Blomgren K, Joseph B. Glioma-induced SIRT1-dependent activation of hMOF histone H4 lysine 16 acetyltransferase in microglia promotes a tumor supporting phenotype. Oncoimmunology, 2017, 7(2):e1382790.
doi: 10.1080/2162402X.2017.1382790 |
| [26] |
Füllgrabe J, Lynch-Day MA, Heldring N, Li WB, Struijk RB, Ma Q, Hermanson O, Rosenfeld MG, Klionsky DJ, Joseph B. The histone H4 lysine 16 acetyltransferase hMOF regulates the outcome of autophagy. Nature, 2013, 500(7463):468-471.
doi: 10.1038/nature12313 |
| [27] |
Chen HF, Fan MY, Pfeffer LM, Laribee RN. The histone H3 lysine 56 acetylation pathway is regulated by target of rapamycin (TOR) signaling and functions directly in ribosomal RNA biogenesis. Nucleic Acids Res, 2012, 40(14):6534-6546.
doi: 10.1093/nar/gks345 |
| [28] |
Das C, Lucia MS, Hansen KC, Tyler JK. CBP/p300- mediated acetylation of histone H3 on lysine 56. Nature, 2009, 459(7243):113-117.
doi: 10.1038/nature07861 |
| [29] |
Lee IH, Finkel T. Regulation of autophagy by the p300 acetyltransferase. J Biol Chem, 2009, 284(10):6322-6328.
doi: 10.1074/jbc.M807135200 |
| [30] | Brown AK, Webb AE. Regulation of FoXO factors in mammalian cells. Curr Top Dev Biol, 2018, 127:165-192. |
| [31] |
Bertaggia E, Coletto L, Sandri M. Post-translational modifications control FoxO3 activity during denervation. Am J Physiol Cell Physiol, 2012, 302(3):C587-C596.
doi: 10.1152/ajpcell.00142.2011 |
| [32] |
Mammucari C, Milan G, Romanello V, Masiero E, Rudolf R, Del Piccolo P, Burden SJ, Di Lisi R, Sandri C, Zhao JH, Goldberg AL, Schiaffino S, Sandri M. FoxO3 controls autophagy in skeletal muscle in vivo. Cell Metab, 2007, 6(6):458-471.
doi: 10.1016/j.cmet.2007.11.001 pmid: 18054315 |
| [33] |
Zhao Y, Yang J, Liao WJ, Liu XY, Zhang H, Wang S, Wang DL, Feng JN, Yu L, Zhu WG. Cytosolic FoxO1 is essential for the induction of autophagy and tumour suppressor activity. Nat Cell Biol, 2010, 12(7):665-675.
doi: 10.1038/ncb2069 |
| [34] |
Settembre C, Di Malta C, Polito VA, Garcia Arencibia M, Vetrini F, Erdin S, Erdin SU, Huynh T, Medina D, Colella P, Sardiello M, Rubinsztein DC, Ballabio A. TFEB links autophagy to lysosomal biogenesis. Science, 2011, 332(6036):1429-1433.
doi: 10.1126/science.1204592 pmid: 21617040 |
| [35] |
Napolitano G, Esposito A, Choi H, Matarese M, Benedetti V, Di Malta C, Monfregola J, Medina DL, Lippincott- Schwartz J, Ballabio A. mTOR-dependent phosphorylation controls TFEB nuclear export. Nat Commun, 2018, 9(1):3312.
doi: 10.1038/s41467-018-05862-6 pmid: 30120233 |
| [36] |
Bao JT, Zheng LJ, Zhang Q, Li XY, Zhang XF, Li ZY, Bai X, Zhang Z, Huo W, Zhao XY, Shang SJ, Wang QS, Zhang C, Ji JG. Deacetylation of TFEB promotes fibrillar Aβ degradation by upregulating lysosomal biogenesis in microglia. Protein Cell, 2016, 7(6):417-433.
doi: 10.1007/s13238-016-0269-2 |
| [37] | Zhang JB, Wang JG, Zhou ZH, Park JE, Wang LM, Wu S, Sun X, Lu LQ, Wang TR, Lin QS, Sze SK, Huang DS, Shen HM. Importance of TFEB acetylation in control of its transcriptional activity and lysosomal function in response to histone deacetylase inhibitors. Autophagy, 2018, 14(6):1043-1059. |
| [38] | Wang YS, Huang YW, Liu JQ, Zhang JN, Xu MM, You ZY, Peng C, Gong ZF, Liu W. Acetyltransferase GCN5 regulates autophagy and lysosome biogenesis by targeting TFEB. EMBO Rep, 2020, 21(1):e48335. |
| [39] |
Bánréti A, Sass M, Graba Y. The emerging role of acetylation in the regulation of autophagy. Autophagy, 2013, 9(6):819-829.
doi: 10.4161/auto.23908 pmid: 23466676 |
| [40] |
Pang JQ, Xiong H, Ou YK, Yang HD, Xu YD, Chen SJ, Lai L, Ye YY, Su ZW, Lin HQ, Huang QH, Xu XD, Zheng YQ. SIRT1 protects cochlear hair cell and delays age-related hearing loss via autophagy. Neurobiol Aging, 2019, 80:127-137.
doi: 10.1016/j.neurobiolaging.2019.04.003 |
| [41] |
Pehar M, Jonas MC, Hare TM, Puglielli L. SLC33A1/AT-1 protein regulates the induction of autophagy downstream of IRE1/XBP1 pathway. J Biol Chem, 2012, 287(35):29921-29930.
doi: 10.1074/jbc.M112.363911 |
| [42] |
Huang R, Xu YF, Wan W, Shou X, Qian JL, You ZY, Liu B, Chang CM, Zhou TH, Lippincott-Schwartz J, Liu W. Deacetylation of nuclear LC3 drives autophagy initiation under starvation. Mol Cell, 2015, 57(3):456-466.
doi: 10.1016/j.molcel.2014.12.013 pmid: 25601754 |
| [43] |
Song TT, Su HF, Yin W, Wang LM, Huang R. Acetylation modulates LC3 stability and cargo recognition. FEBS Lett, 2019, 593(4):414-422.
doi: 10.1002/feb2.2019.593.issue-4 |
| [44] |
Su H, Yang F, Wang QT, Shen QH, Huang JT, Peng C, Zhang Y, Wan W, Wong CCL, Sun QM, Wang FD, Zhou TH, Liu W. VPS34 acetylation controls its lipid kinase activity and the initiation of canonical and non-canonical autophagy. Mol Cell, 2017, 67(6): 907-921.e7.
doi: S1097-2765(17)30548-8 pmid: 28844862 |
| [45] |
Lin SY, Li TY, Liu Q, Zhang CX, Li XT, Chen Y, Zhang SM, Lian GL, Liu Q, Ruan K, Wang Z, Zhang CS, Chien KY, Wu JW, Li QX, Han JH, Lin SC. GSK3-TIP60-ULK1 signaling pathway links growth factor deprivation to autophagy. Science, 2012, 336(6080):477-481.
doi: 10.1126/science.1217032 |
| [46] |
Lee IH, Cao L, Mostoslavsky R, Lombard DB, Liu J, Bruns NE, Tsokos M, Alt FW, Finkel T. A role for the NAD-dependent deacetylase Sirt1 in the regulation of autophagy. Proc Natl Acad Sci USA, 2008, 105(9):3374-3379.
doi: 10.1073/pnas.0712145105 |
| [47] |
Sacitharan PK, Bou-Gharios G, Edwards JR. SIRT1 directly activates autophagy in human chondrocytes. Cell Death Discov, 2020, 6:41.
doi: 10.1038/s41420-020-0277-0 pmid: 32528730 |
| [48] |
Sebti S, Prébois C, Pérez-Gracia E, Bauvy C, Desmots F, Pirot N, Gongora C, Bach AS, Hubberstey AV, Palissot V, Berchem G, Codogno P, Linares LK, Liaudet-Coopman E, Pattingre S. BAT3 modulates p300-dependent acetylation of p53 and autophagy-related protein 7 (ATG7) during autophagy. Proc Natl Acad Sci USA, 2014, 111(11):4115-4120.
doi: 10.1073/pnas.1313618111 |
| [49] |
Yi C, Ma MS, Ran LL, Zheng JX, Tong JJ, Zhu J, Ma CY, Sun YF, Zhang SJ, Feng WZ, Zhu LY, Le Y, Gong XQ, Yan XH, Hong B, Jiang FJ, Xie ZP, Miao D, Deng HT, Yu L. Function and molecular mechanism of acetylation in autophagy regulation. Science, 2012, 336(6080):474-477.
doi: 10.1126/science.1216990 |
| [50] |
Li YT, Yi C, Chen CC, Lan H, Pan M, Zhang SJ, Huang YC, Guan CJ, Li YM, Yu L, Liu L. A semisynthetic Atg3 reveals that acetylation promotes Atg3 membrane binding and Atg8 lipidation. Nat Commun, 2017, 8:14846.
doi: 10.1038/ncomms14846 pmid: 28327644 |
| [51] |
Cheng XW, Ma XL, Zhu Q, Song DD, Ding XM, Li L, Jiang X, Wang XY, Tian R, Su H, Shen ZR, Chen S, Liu T, Gong WH, Liu W, Sun QM. Pacer is a mediator of mTORC1 and GSK3-TIP60 signaling in regulation of autophagosome maturation and lipid metabolism. Mol Cell, 2019, 73(4): 788-802.e7.
doi: 10.1016/j.molcel.2018.12.017 |
| [52] |
Shen QH, Shi Y, Liu JQ, Su H, Huang JT, Zhang Y, Peng C, Zhou TH, Sun QM, Wan W, Liu W. Acetylation of STX17 (syntaxin 17) controls autophagosome maturation. Autophagy, 2021, 17(5):1157-1169.
doi: 10.1080/15548627.2020.1752471 |
| [53] |
Fang DM, Xie HZ, Hu T, Shan H, Li M. Binding features and functions of ATG3. Front Cell Dev Biol, 2021, 9:685625.
doi: 10.3389/fcell.2021.685625 |
| [54] |
Nuta GC, Gilad Y, Gershoni M, Sznajderman A, Schlesinger T, Bialik S, Eisenstein M, Pietrokovski S, Kimchi A. A cancer associated somatic mutation in LC3B attenuates its binding to E1-like ATG7 protein and subsequent lipidation. Autophagy, 2019, 15(3):438-452.
doi: 10.1080/15548627.2018.1525476 |
| [55] |
Schaaf MBE, Keulers TG, Vooijs MA, Rouschop KMA. LC3/GABARAP family proteins: autophagy-(un)related functions. FASEB J, 2016, 30(12):3961-3978.
pmid: 27601442 |
| [56] |
Tanida I, Ueno T, Kominami E. LC3 and autophagy. Methods Mol Biol, 2008, 445:77-88.
doi: 10.1007/978-1-59745-157-4_4 pmid: 18425443 |
| [57] |
Tanida I, Ueno T, Kominami E. LC3 conjugation system in mammalian autophagy. Int J Biochem Cell Biol, 2004, 36(12):2503-2518.
doi: 10.1016/j.biocel.2004.05.009 |
| [58] |
Huang R, Liu W. Identifying an essential role of nuclear LC3 for autophagy. Autophagy, 2015, 11(5):852-853.
doi: 10.1080/15548627.2015.1038016 pmid: 25945743 |
| [59] |
Fan Z, Wu J, Chen QN, Lyu AK, Chen JL, Sun Y, Lyu Q, Zhao YX, Guo A, Liao ZY, Yang YF, Zhu SY, Jiang XS, Chen B, Xiao Q. Type 2 diabetes-induced overactivation of p300 contributes to skeletal muscle atrophy by inhibiting autophagic flux. Life Sci, 2020, 258:118243.
doi: 10.1016/j.lfs.2020.118243 |
| [60] |
Huang S, Li Y, Sheng GH, Meng QW, Lv QB. Sirtuin 1 promotes autophagy and proliferation of endometrial cancer cells by reducing acetylation level of LC3. Cell Biol Int, 2021, 45(5):1050-1059.
doi: 10.1002/cbin.v45.5 |
| [61] |
Hill SM, Wrobel L, Rubinsztein DC. Post-translational modifications of Beclin 1 provide multiple strategies for autophagy regulation. Cell Death Differ, 2019, 26(4):617-629.
doi: 10.1038/s41418-018-0254-9 |
| [62] |
Nascimbeni AC, Codogno P, Morel E. Phosphatidylinositol- 3-phosphate in the regulation of autophagy membrane dynamics. FEBS J, 2017, 284(9):1267-1278.
doi: 10.1111/febs.13987 pmid: 27973739 |
| [63] |
Boukhalfa A, Nascimbeni AC, Ramel D, Dupont N, Hirsch E, Gayral S, Laffargue M, Codogno P, Morel E. PI3KC2α-dependent and VPS34-independent generation of PI3P controls primary cilium-mediated autophagy in response to shear stress. Nat Commun, 2020, 11(1):294.
doi: 10.1038/s41467-019-14086-1 pmid: 31941925 |
| [64] |
Russell RC, Tian Y, Yuan HX, Park HW, Chang YY, Kim J, Kim H, Neufeld TP, Dillin A, Guan KL. ULK1 induces autophagy by phosphorylating Beclin-1 and activating VPS34 lipid kinase. Nat Cell Biol, 2013, 15(7):741-750.
doi: 10.1038/ncb2757 pmid: 23685627 |
| [65] |
Munson MJ, Ganley IG. MTOR, PIK3C3, and autophagy: signaling the beginning from the end. Autophagy, 2015, 11(12):2375-2376.
doi: 10.1080/15548627.2015.1106668 |
| [66] |
Wan W, You ZY, Xu YF, Zhou L, Guan ZL, Peng C, Wong CCL, Su H, Zhou TH, Xia HG, Liu W. mTORC1 phosphorylates acetyltransferase p300 to regulate autophagy and lipogenesis. Mol Cell, 2017, 68(2): 323-335.e6.
doi: 10.1016/j.molcel.2017.09.020 |
| [67] |
Holczer M, Hajdú B, Lőrincz T, Szarka A, Bánhegyi G, Kapuy O. Fine-tuning of AMPK-ULK1-mTORC1 regulatory triangle is crucial for autophagy oscillation. Sci Rep, 2020, 10(1):17803.
doi: 10.1038/s41598-020-75030-8 |
| [68] |
Liu PH, Huang GJ, Wei T, Gao J, Huang CL, Sun MW, Zhu LM, Shen WL. Sirtuin 3-induced macrophage autophagy in regulating NLRP3 inflammasome activation. Biochim Biophys Acta Mol Basis Dis, 2018, 1864(3):764-777.
doi: 10.1016/j.bbadis.2017.12.027 |
| [69] |
Cheng XW, Ma XL, Ding XM, Li L, Jiang X, Shen ZR, Chen S, Liu W, Gong WH, Sun QM. Pacer mediates the function of Class III PI3K and HOPS complexes in autophagosome maturation by engaging stx17. Mol Cell, 2017, 65(6): 1029-1043.e5.
doi: 10.1016/j.molcel.2017.02.010 |
| [70] | Chen YY, Chen HY, Lu DR. Molecular mechanisms of SNARE proteins in regulating autophagy. Hereditas (Beijing), 2014, 36(6):547-551. |
| 陈元渊, 陈红岩, 卢大儒. SNARE蛋白调控细胞自噬的分子机制. 遗传, 2014, 36(6):547-551. | |
| [71] |
Cadwell K, Liu J, Brown SL, Miyoshi H, Loh J, Lennerz J, Kishi C, Wumesh KC, Carrero JA, Hunt S, Stone C, Brunt EM, Xavier RJ, Sleckman BP, Li E, Mizushima N, Stappenbeck TS, Virgin HW. A unique role for autophagy and Atg16L1 in Paneth cells in murine and human intestine. Nature, 2008, 456(7219):259-263.
doi: 10.1038/nature07416 |
| [72] | Nakai A, Yamaguchi O, Takeda T, Higuchi Y, Hikoso S, Taniike M, Omiya S, Mizote I, Matsumura Y, Asahi M, Nishida K, Hori M, Mizushima N, Otsu K. The role of autophagy in cardiomyocytes in the basal state and in response to hemodynamic stress. Nat Med, 2007, 13(5):619-624. |
| [73] |
Taneike M, Yamaguchi O, Nakai A, Hikoso S, Takeda T, Mizote I, Oka T, Tamai T, Oyabu J, Murakawa T, Nishida K, Shimizu T, Hori M, Komuro I, Takuji Shirasawa TS, Mizushima N, Otsu K. Inhibition of autophagy in the heart induces age-related cardiomyopathy. Autophagy, 2010, 6(5):600-606.
doi: 10.4161/auto.6.5.11947 pmid: 20431347 |
| [74] |
Hamano T, Gendron TF, Causevic E, Yen SH, Lin WL, Isidoro C, Deture M, Ko LW. Autophagic-lysosomal perturbation enhances tau aggregation in transfectants with induced wild-type tau expression. Eur J Neurosci, 2008, 27(5):1119-1130.
doi: 10.1111/j.1460-9568.2008.06084.x |
| [75] |
Wold MS, Lim J, Lachance V, Deng ZQ, Yue ZY. ULK1-mediated phosphorylation of ATG14 promotes autophagy and is impaired in Huntington's disease models. Mol Neurodegener, 2016, 11(1):76.
doi: 10.1186/s13024-016-0141-0 |
| [76] |
Komatsu M, Waguri S, Ueno T, Iwata J, Murata S, Tanida I, Ezaki J, Mizushima N, Ohsumi Y, Uchiyama Y, Kominami E, Tanaka K, Chiba T. Impairment of starvation- induced and constitutive autophagy in Atg7-deficient mice. J Cell Biol, 2005, 169(3):425-434.
doi: 10.1083/jcb.200412022 |
| [77] |
Feng X, Zhang H, Meng LB, Song HW, Zhou QX, Qu C, Zhao P, Li QH, Zou C, Liu X, Zhang ZY. Hypoxia-induced acetylation of PAK1 enhances autophagy and promotes brain tumorigenesis via phosphorylating ATG5. Autophagy, 2021, 17(3):723-742.
doi: 10.1080/15548627.2020.1731266 |
| [78] |
Richter-Landsberg C, Leyk J. Inclusion body formation, macroautophagy, and the role of HDAC6 in neurodegeneration. Acta Neuropathol, 2013, 126(6):793-807.
doi: 10.1007/s00401-013-1158-x pmid: 23912309 |
| [79] |
Esteves AR, Arduíno DM, Silva DF, Viana SD, Pereira FC, Cardoso SM. Mitochondrial metabolism regulates microtubule acetylome and autophagy trough sirtuin-2: impact for Parkinson's disease. Mol Neurobiol, 2018, 55(2):1440-1462.
doi: 10.1007/s12035-017-0420-y pmid: 28168426 |
| [80] |
Eckschlager T, Plch J, Stiborova M, Hrabeta J. Histone deacetylase inhibitors as anticancer drugs. Int J Mol Sci, 2017, 18(7):1414.
doi: 10.3390/ijms18071414 |
| [81] |
Cao DJ, Wang ZV, Battiprolu PK, Jiang N, Morales CR, Kong YL, Rothermel BA, Gillette TG, Hill JA. Histone deacetylase (HDAC) inhibitors attenuate cardiac hypertrophy by suppressing autophagy. Proc Natl Acad Sci USA, 2011, 108(10):4123-4128.
doi: 10.1073/pnas.1015081108 |
| [82] |
Chiao MT, Cheng WY, Yang YC, Shen CC, Ko JL. Suberoylanilide hydroxamic acid (SAHA) causes tumor growth slowdown and triggers autophagy in glioblastoma stem cells. Autophagy, 2013, 9(10):1509-1526.
doi: 10.4161/auto.25664 |
| [83] |
Beltrao P, Bork P, Krogan NJ, van Noort V. Evolution and functional cross-talk of protein post-translational modifications. Mol Syst Biol, 2013, 9:714.
doi: 10.1002/msb.201304521 pmid: 24366814 |
| [84] |
Grégoire S, Tremblay AM, Xiao L, Yang Q, Ma KW, Nie JY, Mao ZX, Wu ZG, Giguère V, Yang XJ. Control of MEF2 transcriptional activity by coordinated phosphorylation and sumoylation. J Biol Chem, 2006, 281(7):4423-4433.
pmid: 16356933 |
| [85] |
Swaney DL, Beltrao P, Starita L, Guo AL, Rush J, Fields S, Krogan NJ, Villén J. Global analysis of phosphorylation and ubiquitylation cross-talk in protein degradation. Nat Methods, 2013, 10(7):676-682.
doi: 10.1038/nmeth.2519 |
| [86] |
Vu LD, Gevaert K, De Smet I. Protein language: post-translational modifications talking to each other. Trends Plant Sci, 2018, 23(12):1068-1080.
doi: 10.1016/j.tplants.2018.09.004 |
| [1] | Kexue Ma, Rui Li, Fangying Guo, Gege Song, Meng Wu, Guangwen Chen, Dezeng Liu. Functional analysis of autophagy-related gene Atg6 in planarian central nervous system regeneration [J]. Hereditas(Beijing), 2021, 43(8): 792-801. |
| [2] | Junyi Ju,Quan Zhao. Regulation of γ-globin gene expression and its clinical applications [J]. Hereditas(Beijing), 2018, 40(6): 429-444. |
| [3] | Wang Shiming, Song Xiao, Zhao Xueying, Chen Hongyan, Wang Jiucun, Wu Junjie, Gao Zhiqiang, Qian Ji, Bai Chunxue, Li Qiang, Han Baohui, Lu Daru. Association between polymorphisms of autophagy pathway and responses in non-small cell lung cancer patients treated with platinum-based chemotherapy [J]. Hereditas(Beijing), 2017, 39(3): 250-262. |
| [4] | Xiaowei Zeng, Cuicui Liu, Ning Han, Hongwu Bian, Muyuan Zhu. Progress on the autophagic regulators and receptors in plants [J]. HEREDITAS(Beijing), 2016, 38(7): 644-650. |
| [5] | Jianhui Li, Chunfu Wang, Fan Bai, Yan Zhuang, Zhuojun Mao, Yongtao Sun. Establishment and application of a flow cytometry-based method for detecting histone acetylation levels [J]. HEREDITAS(Beijing), 2016, 38(6): 581-587. |
| [6] | Xiaomeng Qiao,Fangyuan Yin,Yunxiao Li,Shuguang Wei,Jianghua Lai. The role of histone acetylation in the basolateral amygdala in morphine-associated memory in rats [J]. HEREDITAS(Beijing), 2015, 37(4): 382-387. |
| [7] | Qiwen Wang, Wei Jin, Cuifang Chang, Cunshuan Xu. Regulation of autophagy on dendritic cells during rat liver regeneration by IPA [J]. HEREDITAS(Beijing), 2015, 37(3): 276-282. |
| [8] | Qiwen Wang,Cuifang Chang,Ningning Gu,Cuiyun Pan,Cunshuan Xu. Effect of autophagy on liver regeneration [J]. HEREDITAS(Beijing), 2015, 37(11): 1116-1124. |
| [9] | Li Chen, Fang Ding, Yong Liu, Fengrui Wu, Biao Ding, Rong Wang, Wenyong Li. Comparative analysis of H3K9 acetylation level in parthenogenetic, and in vitro and in vivo developed mouse embryos [J]. HEREDITAS(Beijing), 2015, 37(1): 77-83. |
| [10] | Yuanyuan Chen, Hongyan Chen, Daru Lu. Molecular mechanisms of SNARE proteins in regulating autophagy [J]. HEREDITAS(Beijing), 2014, 36(6): 547-551. |
| [11] | Yuanchao Sun, Xunsi Qin, Hong Chen, Wei Shen. Epigenetic control of autophagy [J]. HEREDITAS(Beijing), 2014, 36(5): 447-455. |
| [12] | . Molecular mechanisms and functions of autophagy and the ubiq-uitin-proteasome pathway [J]. HEREDITAS, 2012, 34(1): 5-18. |
| [13] | GE Shao-Qin, LI Jian-Zhong, ZHANG Xiao-Jing. Methylation and acetylation of histones during spermatogenesis [J]. HEREDITAS, 2011, 33(9): 939-946. |
| [14] | WANG Shi-Yao, JIN Wei-Na, WU Dan. Mechanisms of juvenile neuronal ceroid lipofuscinosis (JNCL) [J]. HEREDITAS, 2009, 31(8): 779-784. |
| [15] | YU Jian-Ning, WANG Meng, WANG Dan-Qiu, LI Shao-Hua, SHAO Gen-Bao, WU Cai-Feng, LIU Hong-Lin . Chromosome changes of aged oocytes after ovulation [J]. HEREDITAS, 2007, 29(2): 225-225―229. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||