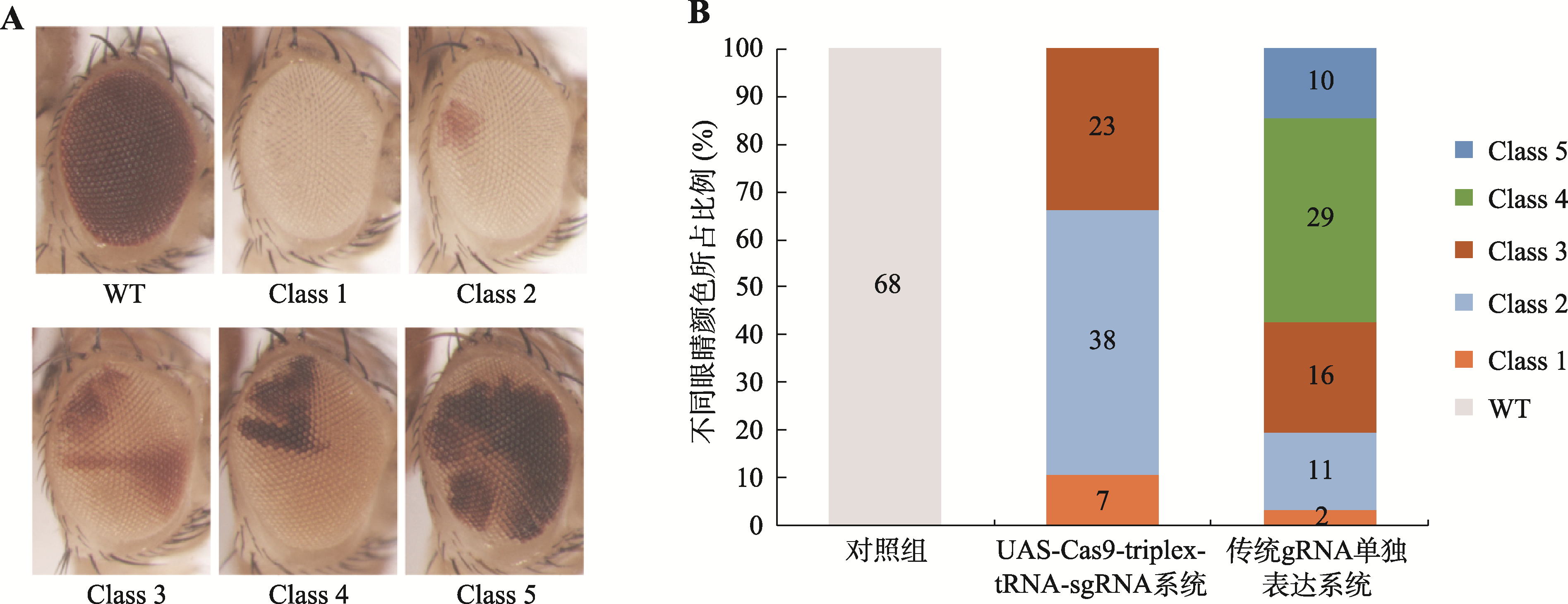
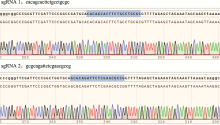
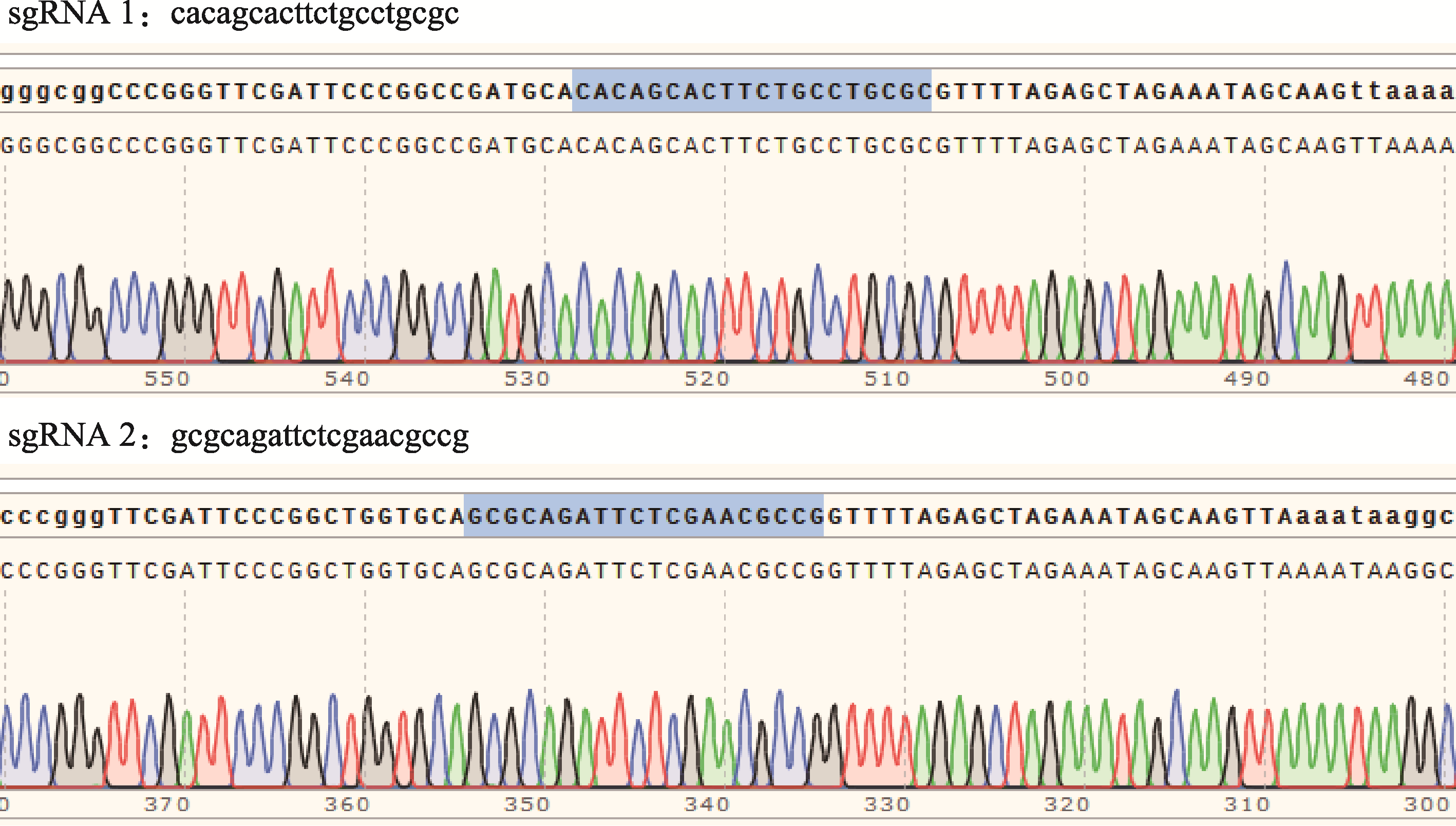
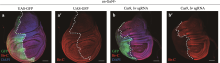
Hereditas(Beijing) ›› 2023, Vol. 45 ›› Issue (7): 593-601.doi: 10.16288/j.yczz.23-099
• Research Article • Previous Articles Next Articles
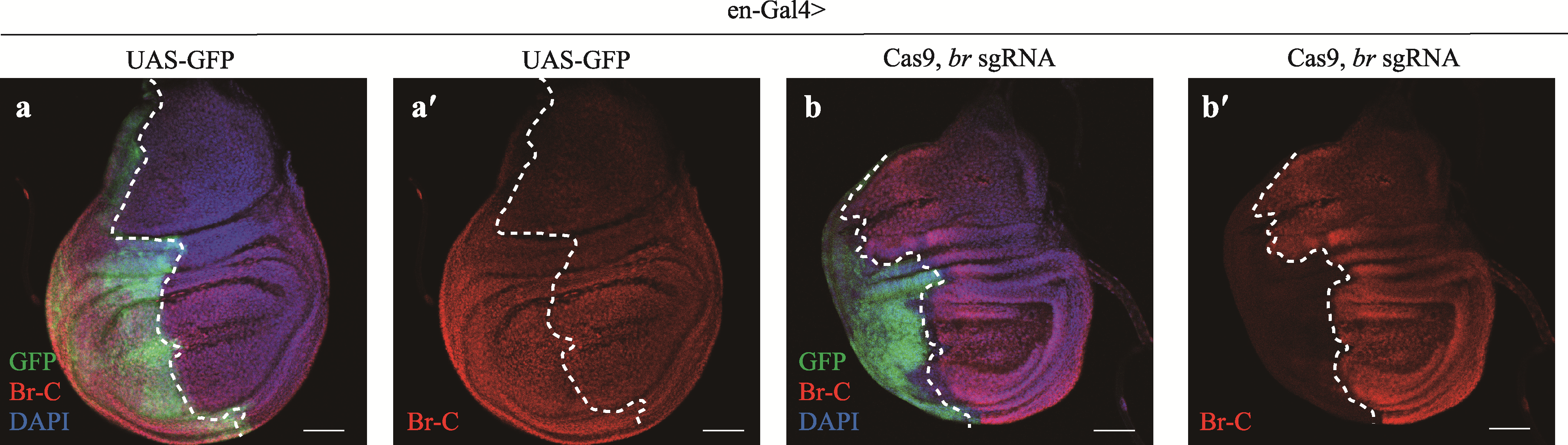
Conditional editing of the Drosophila melanogaster genome using single transcripts expressing Cas9 and sgRNA
Bingzheng Wang1( ), Chao Zhang2, Jiali Zhang2, Jin Sun1(
), Chao Zhang2, Jiali Zhang2, Jin Sun1( )
)
- 1. School of Laboratory Animal & Shandong Laboratory Animal Center, Shandong First Medical University & Shandong Academy of Medical Sciences, Jinan 250024, China
2. School of Clinical and Basic Medical Sciences, Shandong First Medical University, Jinan 250024, China
-
Received:2023-04-15Revised:2023-06-09Online:2023-07-20Published:2023-07-03 -
Contact:Jin Sun E-mail:370756420@qq.com;sunjin@sdfmu.edu.cn -
Supported by:National Natural Science Foundation of China(31801079)
Cite this article
Bingzheng Wang, Chao Zhang, Jiali Zhang, Jin Sun. Conditional editing of the Drosophila melanogaster genome using single transcripts expressing Cas9 and sgRNA[J]. Hereditas(Beijing), 2023, 45(7): 593-601.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
Table 1
Primer sequences used in this study"
| 引物名称 | 引物序列(5′→3′) |
|---|---|
| cas9-F | AGACGCATACCAAAGTCTAGAATGGCCCCAAAGAAGAAGC |
| cas9-R | cctagGTGCTAGATTATTTCTTTTTCTTAGC |
| Triplex-F | AAGAAATAATCTAGCACctaggaggatccactcgAGGTTTAGAG |
| tRNA-sgRNA-R | CCTACCTGATGCCAACAATTctagTTTGCTCTAGTCCTAGtgg |
| White-F | tgcaCAGGAGCTATTAATTCGCGG |
| White-R | aaacCCGCGAATTAATAGCTCCTG |
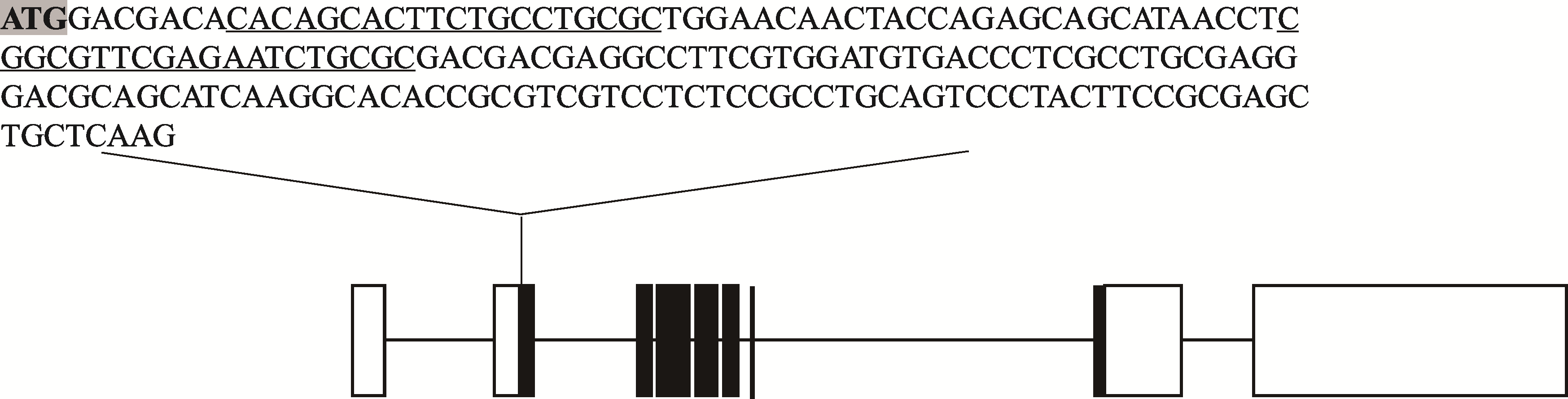
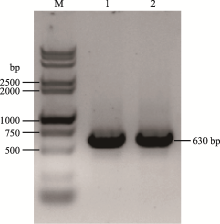
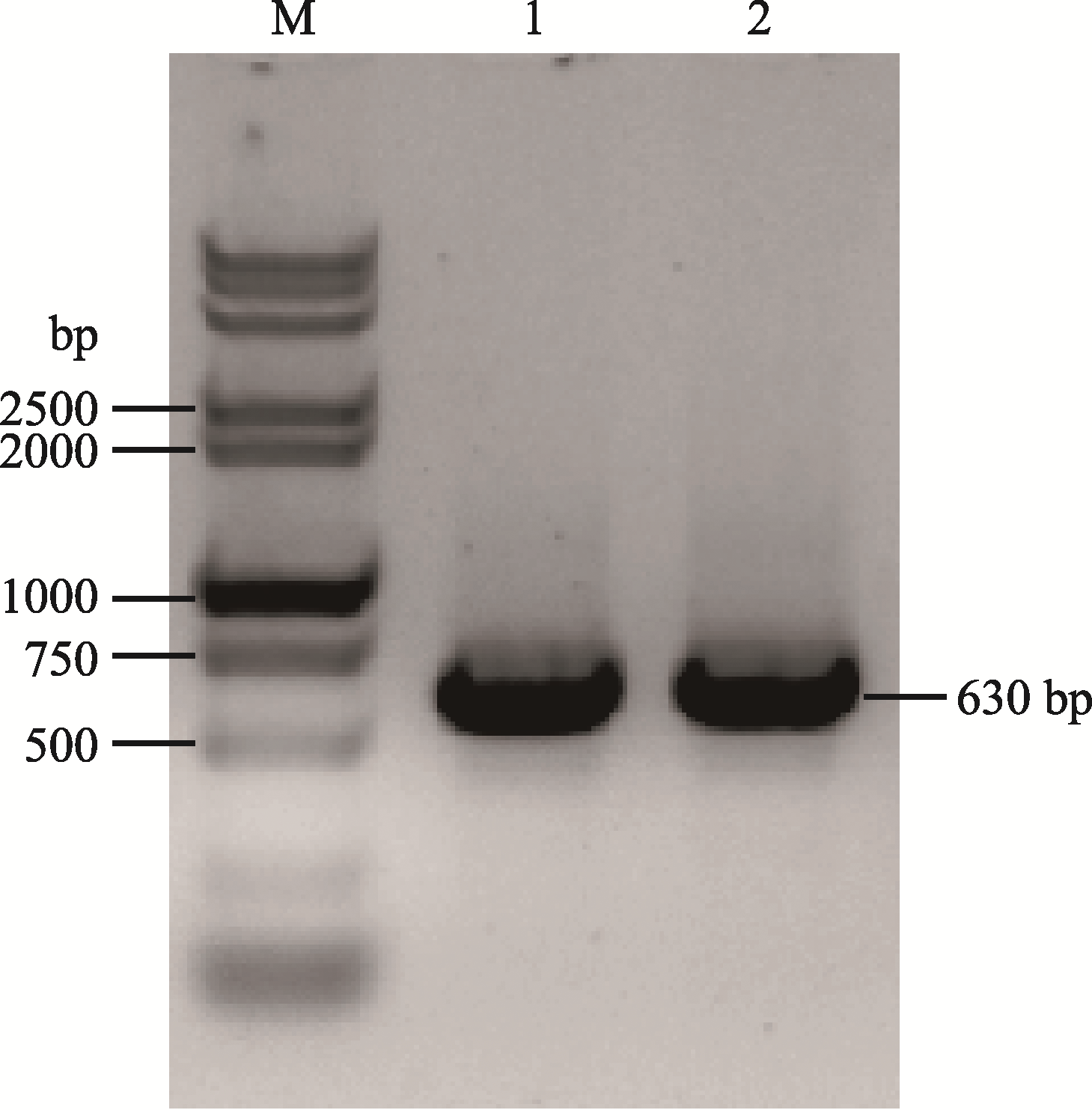
| br-sg-F | CCCGGGTTCGATTCCCGGCCGATGCACACAGCACTTCTGCCTGCGCGTTTTAGAGCTAGAAATAGCAAG |
| br-sg-R | TAACTTGCTATTTCTAGCTCTAAAACCGGCGTTCGAGAATCTGCGCTGCACCAGCCGGGAATCGAA |
| Dmt-Fs | AAGCATCGGTGGTTCAGTGG |
| ftz | GATTGCAAAGAACTAGAGCCG |
| [1] |
Carroll D. Genome editing: past, present, and future. Yale J Biol Med, 2017, 90(4): 653-659.
pmid: 29259529 |
| [2] |
Tyagi S, Kumar R, Das A, Won SY, Shukla P. CRISPR-Cas9 system: a genome-editing tool with endless possibilities. J Biotechnol, 2020, 319: 36-53.
doi: S0168-1656(20)30128-0 pmid: 32446977 |
| [3] |
Cong L, Ran FA, Cox D, Lin SL, Barretto R, Habib N, Hsu PD, Wu XB, Jiang WY, Marraffini LA, Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science, 2013, 339(6121): 819-823.
doi: 10.1126/science.1231143 pmid: 23287718 |
| [4] |
Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science, 2012, 337(6096): 816-821.
doi: 10.1126/science.1225829 pmid: 22745249 |
| [5] |
Mali P, Yang LH, Esvelt KM, Aach J, Guell M, Dicarlo JE, Norville JE, Church GM.RNA-guided human genome engineering via Cas9. Science, 2013, 339(6121): 823-826.
doi: 10.1126/science.1232033 pmid: 23287722 |
| [6] |
Gratz SJ, Ukken FP, Rubinstein CD, Thiede G, Donohue LK, Cummings AM, O'connor-Giles KM.Highly specific and efficient CRISPR/Cas9-catalyzed homology-directed repair in Drosophila. Genetics, 2014, 196(4): 961-971.
doi: 10.1534/genetics.113.160713 pmid: 24478335 |
| [7] |
Ren YX, Xiao RD, Lou XM, Fang XD. Research advance and application in the gene therapy of gene editing technologies. Hereditas(Beijing), 2019, 41(1): 18-27.
doi: 10.16288/j.yczz.18-142 pmid: 30686782 |
|
任云晓, 肖茹丹, 娄晓敏, 方向东. 基因编辑技术及其在基因治疗中的应用. 遗传, 2019, 41(1): 18-27.
doi: 10.16288/j.yczz.18-142 pmid: 30686782 |
|
| [8] |
Wilusz JE, Jnbaptiste CK, Lu LY, Kuhn CD, Joshua-Tor L, Sharp PA. A triple helix stabilizes the 3' ends of long noncoding RNAs that lack poly(A) tails. Genes Dev, 2012, 26(21): 2392-2407.
doi: 10.1101/gad.204438.112 |
| [9] |
Campa CC, Weisbach NR, Santinha AJ, Incarnato D, Platt RJ. Multiplexed genome engineering by Cas12a and CRISPR arrays encoded on single transcripts. Nat Methods, 2019, 16(9): 887-893.
doi: 10.1038/s41592-019-0508-6 pmid: 31406383 |
| [10] |
Ren XJ, Sun J, Housden BE, Hu YH, Roesel C, Lin SL, Liu LP, Yang ZH, Mao DC, Sun LZ, Wu QJ, Ji JY, Xi JZ, Mohr SE, Xu J, Perrimon N, Ni JQ.Optimized gene editing technology for Drosophila melanogaster using germ line-specific Cas9. Proc Natl Acad Sci USA, 2013, 110(47): 19012-19017.
doi: 10.1073/pnas.1318481110 |
| [11] |
Qiao HH, Wang F, Xu RG, Sun J, Zhu RB, Mao DC, Ren XJ, Wang X, Jia Y, Peng P, Shen D, Liu LP, Chang ZJ, Wang GR, Li S, Ji JY, Liu QF, Ni JQ. An efficient and multiple target transgenic RNAi technique with low toxicity in Drosophila. Nat Commun, 2018, 9(1): 4160.
doi: 10.1038/s41467-018-06537-y |
| [12] |
Gurumurthy CB, O'brien AR, Quadros RM, Adams J, Alcaide P, Ayabe S, Ballard J, Batra SK, Beauchamp MC, Becker KA, Bernas G, Brough D, Carrillo-Salinas F, Chan W, Chen HY, Dawson R, Demambro V, D'hont J, Dibb KM, Eudy JD, Gan L, Gao J, Gonzales A, Guntur AR, Guo HP, Harms DW, Harrington A, Hentges KE, Humphreys N, Imai S, Ishii H, Iwama M, Jonasch E, Karolak M, Keavney B, Khin NC, Konno M, Kotani Y, Kunihiro Y, Lakshmanan I, Larochelle C, Lawrence CB, Li L, Lindner V, Liu XD, Lopez-Castejon G, Loudon A, Lowe J, Jerome-Majewska LA, Matsusaka T, Miura H, Miyasaka Y, Morpurgo B, Motyl K, Nabeshima YI, Nakade K, Nakashiba T, Nakashima K, Obata Y, Ogiwara S, Ouellet M, Oxburgh L, Piltz S, Pinz I, Ponnusamy MP, Ray D, Redder RJ, Rosen CJ, Ross N, Ruhe MT, Ryzhova L, Salvador AM, Alam SS, Sedlacek R, Sharma K, Smith C, Staes K, Starrs L, Sugiyama F, Takahashi S, Tanaka T, Trafford AW, Uno Y, Vanhoutte L, Vanrockeghem F, Willis BJ, Wright CS, Yamauchi Y, Yi X, Yoshimi K, Zhang XS, Zhang Y, Ohtsuka M, Das S, Garry DJ, Hochepied T, Thomas P, Parker-Thornburg J, Adamson AD, Yoshiki A, Schmouth JF, Golovko A, Thompson WR, Lloyd KCK, Wood JA, Cowan M, Mashimo T, Mizuno S, Zhu H, Kasparek P, Liaw L, Miano JM, Burgio G. Reproducibility of CRISPR-Cas9 methods for generation of conditional mouse alleles: a multi-center evaluation. Genome Biol, 2019, 20(1): 171.
doi: 10.1186/s13059-019-1776-2 pmid: 31446895 |
| [13] |
Jin M, Eblimit A, Pulikkathara M, Corr S, Chen R, Mardon G. Conditional knockout of retinal determination genes in differentiating cells in Drosophila. FEBS J, 2016, 283(15): 2754-2766.
doi: 10.1111/febs.13772 pmid: 27257739 |
| [14] |
Hamilton DL, Abremski K. Site-specific recombination by the bacteriophage P1 lox-Cre system. Cre-mediated synapsis of two lox sites. J Mol Biol, 1984, 178(2): 481-486.
pmid: 6333513 |
| [15] |
Andrews BJ, Proteau GA, Beatty LG, Sadowski PD. The FLP recombinase of the 2 micron circle DNA of yeast: interaction with its target sequences. Cell, 1985, 40(4): 795-803.
pmid: 3879971 |
| [16] |
Golic KG, Lindquist S. The FLP recombinase of yeast catalyzes site-specific recombination in the Drosophila genome. Cell, 1989, 59(3): 499-509.
pmid: 2509077 |
| [17] |
Germani F, Bergantinos C, Johnston LA. Mosaic analysis in Drosophila. Genetics, 2018, 208(2): 473-490.
doi: 10.1534/genetics.117.300256 pmid: 29378809 |
| [18] |
Siegal ML, Hartl DL. Transgene coplacement and high efficiency site-specific recombination with the Cre/loxP system in Drosophila. Genetics, 1996, 144(2): 715-726.
doi: 10.1093/genetics/144.2.715 pmid: 8889532 |
| [19] |
Heidmann D, Lehner CF. Reduction of Cre recombinase toxicity in proliferating Drosophila cells by estrogen- dependent activity regulation. Dev Genes Evol, 2001, 211(8-9): 458-465.
pmid: 11685583 |
| [20] |
Theodosiou NA, Xu T. Use of FLP/FRT system to study Drosophila development. Methods, 1998, 14(4): 355-365.
pmid: 9608507 |
| [21] |
Nakazawa N, Taniguchi K, Okumura T, Maeda R, Matsuno K. A novel Cre/loxP system for mosaic gene expression in the Drosophila embryo. Dev Dyn, 2012, 241(5): 965-974.
doi: 10.1002/dvdy.23784 |
| [22] |
Charpentier E, Doudna JA. Biotechnology: rewriting a genome. Nature, 2013, 495(7439): 50-51.
doi: 10.1038/495050a |
| [23] |
Ding D, Chen KY, Chen YD, Li H, Xie KB. Engineering introns to express RNA guides for Cas9- and Cpf1- mediated multiplex genome editing. Mol Plant, 2018, 11(4): 542-552.
doi: S1674-2052(18)30055-8 pmid: 29462720 |
| [1] | Meizhen Liu, Liren Wang, Yongmei Li, Xueyun Ma, Honghui Han, Dali Li. Generation of genetically modified rat models via the CRISPR/Cas9 technology [J]. Hereditas(Beijing), 2023, 45(1): 78-87. |
| [2] | Xiaojun Zhang, Kun Xu, Juncen Shen, Lu Mu, Hongrun Qian, Jieyu Cui, Baoxia Ma, Zhilong Chen, Zhiying Zhang, Zehui Wei. A CRISPR/Cas9-Gal4BD donor adapting system for enhancing homology-directed repair [J]. Hereditas(Beijing), 2022, 44(8): 708-719. |
| [3] | Chong Zhang, Zixuan Wei, Min Wang, Yaosheng Chen, Zuyong He. Editing MC1R in human melanoma cells by CRISPR/Cas9 and functional analysis [J]. Hereditas(Beijing), 2022, 44(7): 581-590. |
| [4] | Yao Liu, Xianhui Zhou, Shuhong Huang, Xiaolong Wang. Prime editing: a search and replace tool with versatile base changes [J]. Hereditas(Beijing), 2022, 44(11): 993-1008. |
| [5] | Yuting Han, Bowen Xu, Yutong Li, Xinyi Lu, Xizhi Dong, Yuhao Qiu, Qinyun Che, Ruibao Zhu, Li Zheng, Xiaochen Li, Xu Si, Jianquan Ni. The cutting edge of gene regulation approaches in model organism Drosophila [J]. Hereditas(Beijing), 2022, 44(1): 3-14. |
| [6] | Guangwu Yang, Yuan Tian. The F-box gene Ppa promotes lipid storage in Drosophila [J]. Hereditas(Beijing), 2021, 43(6): 615-622. |
| [7] | Dingwei Peng, Ruiqiang Li, Wu Zeng, Min Wang, Xuan Shi, Jianhua Zeng, Xiaohong Liu, Yaoshen Chen, Zuyong He. Editing the cystine knot motif of MSTN enhances muscle development of Liang Guang Small Spotted pigs [J]. Hereditas(Beijing), 2021, 43(3): 261-270. |
| [8] | Na Wang, Zhilian Jia, Qiang Wu. RFX5 regulates gene expression of the Pcdhα cluster [J]. Hereditas(Beijing), 2020, 42(8): 760-774. |
| [9] | Guoling Li, Shanxin Yang, Zhenfang Wu, Xianwei Zhang. Recent developments in enhancing the efficiency of CRISPR/Cas9- mediated knock-in in animals [J]. Hereditas(Beijing), 2020, 42(7): 641-656. |
| [10] | Yingnan Chen, Jing Lu. Application of CRISPR/Cas9 mediated gene editing in trees [J]. Hereditas(Beijing), 2020, 42(7): 657-668. |
| [11] | Siyuan Liu, Guoqiang Yi, Zhonglin Tang, Bin Chen. Progress on genome-wide CRISPR/Cas9 screening for functional genes and regulatory elements [J]. Hereditas(Beijing), 2020, 42(5): 435-443. |
| [12] | Liwen Bao, Yiye Zhou, Fanyi Zeng. Advances in gene therapy for β-thalassemia and hemophilia based on the CRISPR/Cas9 technology [J]. Hereditas(Beijing), 2020, 42(10): 949-964. |
| [13] | Minting Lin, Lulu Lai, Miao Zhao, Biwei Lin, Xiangping Yao. Construction of a striatum-specific Slc20a2 gene knockout mice model by CRISPR/Cas9 AAV system [J]. Hereditas(Beijing), 2020, 42(10): 1017-1027. |
| [14] | Peifeng Liu, Qiang Wu. Probing 3D genome by CRISPR/Cas9 [J]. Hereditas(Beijing), 2020, 42(1): 18-31. |
| [15] | Jue Wang, Juan Huang, Rui Xu. Seamless genome editing in Drosophila by combining CRISPR/Cas9 and piggyBac technologies [J]. Hereditas(Beijing), 2019, 41(5): 422-429. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||