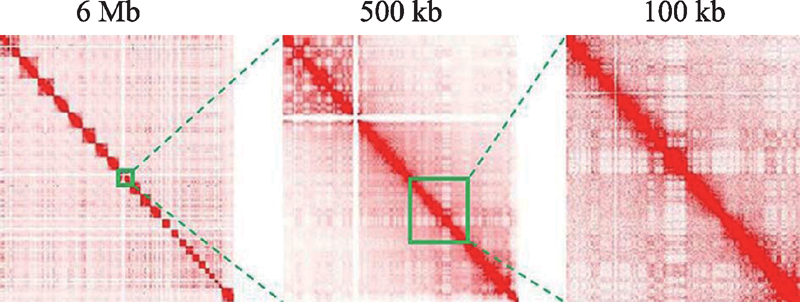
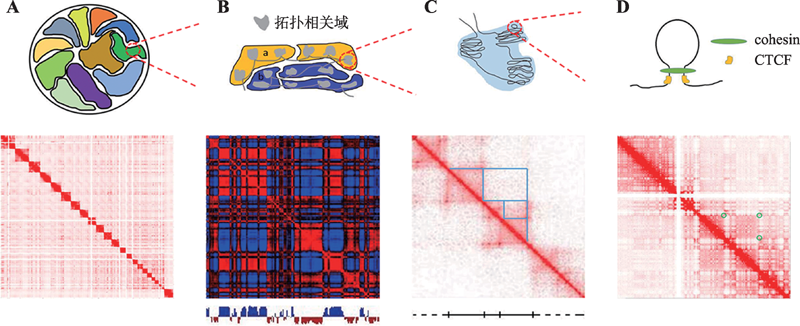
Hereditas(Beijing) ›› 2020, Vol. 42 ›› Issue (1): 87-99.doi: 10.16288/j.yczz.19-163
• Special Section: 3D Genome • Previous Articles Next Articles
Current status and future perspectives in bioinformatical analysis of Hi-C data
Hongqiang Lyu, Lele Hao, Erhu Liu, Zhifang Wu, Jiuqiang Han, Yuan Liu
- School of Electronic and Information Engineering, Xi’an Jiaotong University, Xi’an 710049, China
-
Received:2019-07-23Revised:2019-11-26Online:2020-01-20Published:2019-12-05 -
Supported by:Supported by the National Natural Science Foundation of China No(61602367)
Cite this article
Hongqiang Lyu, Lele Hao, Erhu Liu, Zhifang Wu, Jiuqiang Han, Yuan Liu. Current status and future perspectives in bioinformatical analysis of Hi-C data[J]. Hereditas(Beijing), 2020, 42(1): 87-99.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
Table 1
Normalization methods of Hi-C data"
| 方法 | 分类 | 特点 | 实现语言 | 典型程序 |
|---|---|---|---|---|
| SCN | 隐式,单样本 | 行列归一化的矩阵平衡 | MATLAB | SCN_sumV2.m |
| HiCNorm | 显式,单样本 | 泊松回归估计系统偏差 | R | HiCNorm.R/HiTC |
| ICE | 隐式,单样本 | 迭代修正的矩阵平衡 | R,C,Python | HiTC/Hi-Corrector |
| KR | 隐式,单样本 | 内外迭代的快速矩阵平衡 | MATLAB | BNEWT.m |
| caICB | 显式,单样本 | 移除拷贝数偏差的改进ICE | R,perl | HiCapp |
| HiCcompare | 隐式,跨样本 | 双样本,局部加权线性回归 | R | HiCcompare |
| MultiHiCcompare | 隐式,跨样本 | 多样本,局部加权线性回归 | R | multiHiCcompare |
| Binless | 隐式,跨样本 | 配对末端序列片段的统计显著性分析 | R | Binless |
Table 2
Methods for identification of TADs"
| 方法 | 分类 | 特点 | 实现语言 | 典型程序 |
|---|---|---|---|---|
| DI | 边界点,非差异 | 隐马尔科夫模型 | R,Python | HiTC/TADtool |
| HiCseg | 边界点,非差异 | 二维分割矩阵 | R | HiCseg |
| TopDom | 边界点,非差异 | 钻石形滑窗法 | R | TopDom.R |
| TADtree | 层级式,非差异 | 交互频率经验分布 | Python | TADtree |
| TADbit | 边界点,非差异 | 基于BIC惩罚的概率模型 | Python | TADbit |
| HiTAD | 层级式,非差异 | 隐马尔科夫模型 | Python | TADLib |
| IC-Finder | 层级式,非差异 | 层次聚类 | MATLAB | IC-Finder.m |
| GMAP | 层级式,非差异 | 高斯混合模型 | R | GMAP |
| 3DNetMod | 层级式,非差异 | 基于图理论 | Python | 3DNetMod |
| deDoc | 层级式,非差异 | 基于图结构墒理论 | R | deDoc |
| HiCDB | 边界点,差异性 | 局部相对隔绝指数和多尺度聚类 | R,MATLAB | RHiCDB/HiCDB.m |
Table 4
Visual software tools for Hi-C data"
| 方法 | 交互 | 网址 |
|---|---|---|
| WashU Epigenome Browser | 浏览器 | http://epigenomegateway.wustl.edu/ |
| HiCPlotter | Python软件工具 | https://github.com/kcakdemir/HiCPlotter |
| 3Disease Browser | 浏览器 | http://3dgb.cbi.pku.edu.cn/disease/ |
| Juicebox | 浏览器,Java软件工具 | http://aidenlab.org/juicebox |
| HiC-3Dviewer | 浏览器 | http://bioinfo.au.tsinghua.edu.cn/member/nadhir/HiC3DViewer/ |
| Delta | Java软件工具 | http://delta.big.ac.cn |
| GITAR | Python软件工具 | http://genomegitar.org |
| 3D Genome browser | 浏览器 | http://3dgenome.org |
| Galaxy HiCExplorer | 浏览器 | https://hicexplorer.usegalaxy.eu |
| [1] |
Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, Sandstrom R, Bernstein B, Bender MA, Groudine M, Gnirke A, Stamatoyannopoulos J, Mirny LA, Lander ES, Dekker J . Comprehensive mapping of long- range interactions reveals folding principles of the human genome. Science, 2009,326(5950):289-293.
doi: 10.1126/science.1181369 pmid: 19815776 |
| [2] |
Schmitt AD, Hu M, Ren B . Genome-wide mapping and analysis of chromosome architecture. Nat Rev Mol Cell Biol, 2016,17(12):743-755.
doi: 10.1038/nrm.2016.104 pmid: 27580841 |
| [3] |
Li GL, Ruan YJ, Gu RS, Du SM . Emergence of 3D genomics. Chin Sci Bull, 2014,59(13):1165-1172.
doi: 10.1360/N972014-00163 |
|
李国亮, 阮一骏, 谷瑞升, 杜生明 . 起航三维基因组学研究. 科学通报, 2014,59(13):1165-1172.
doi: 10.1360/N972014-00163 |
|
| [4] |
Dekker J, Rippe K, Dekker M, Kleckner N . Capturing chromosome conformation. Science, 2002,295(5558):1306-1311.
doi: 10.1126/science.1067799 pmid: 11847345 |
| [5] |
Zhao ZH, Tavoosidana G, Sjölinder M, Göndör A, Mariano P, Wang S, Kanduri C, Lezcano M, Sandhu KS, Singh U, Pant V, Tiwari V, Kurukuti S, Ohlsson R . Circular chromosome conformation capture (4C) uncovers extensive networks of epigenetically regulated intra- and interchromosomal interactions. Nat Genet, 2006,38(11):1341-1347.
doi: 10.1038/ng1891 pmid: 17033624 |
| [6] |
Dostie J, Richmond TA, Arnaout RA, Selzer RR, Lee WL, Honan TA, Rubio ED, Krumm A, Lamb J, Nusbaum C, Green RD, Dekker J . Chromosome conformation capture carbon copy (5C): a massively parallel solution for mapping interactions between genomic elements. Genome Res, 2006,16(10):1299-1309.
doi: 10.1101/gr.5571506 pmid: 16954542 |
| [7] |
Zhang XY, He C, Ye BY, Xie DJ, Shi ML, Zhang Y, Shen WL, Li P, Zhao ZH . Optimization and quality control of genome-wide Hi-C library preparation. Hereditas(Beijing), 2017,39(9):847-855.
doi: 10.16288/j.yczz.17-152 pmid: 28936982 |
|
张香媛, 何超, 叶丙雨, 谢德健, 师明磊, 张彦, 沈文龙, 李平, 赵志虎 . 全基因组染色质相互作用Hi-C文库制备的优化及其质量控制. 遗传, 2017,39(9):847-855.
doi: 10.16288/j.yczz.17-152 pmid: 28936982 |
|
| [8] |
de Wit E, de Laat W . A decade of 3C technologies: insights into nuclear organization. Genes Dev, 2012,26(1):11-24.
doi: 10.1101/gad.179804.111 pmid: 22215806 |
| [9] |
Schoenfelder S, Furlan-Magaril M, Mifsud B, Tavares- Cadete F, Sugar R, Javierre BM, Nagano T, Katsman Y, Sakthidevi M, Wingett SW, Dimitrova E, Dimond A, Edelman LB, Elderkin S, Tabbada K, Darbo E, Andrews S, Herman B, Higgs A, LeProust E, Osborne CS, Mitchell JA, Luscombe NM, Fraser P,. The pluripotent regulatory circuitry connecting promoters to their long-range interacting elements. Genome Res, 2015,25(4):582-597.
doi: 10.1101/gr.185272.114 pmid: 25752748 |
| [10] |
Takashi Nagano, Yaniv Lubling, Tim J. Stevens, Stefan Schoenfelder, Eitan Yaffe, Wendy Dean, Ernest D,. Laue, Amos Tanay, Peter Fraser. Single-cell Hi-C reveals cell-to- cell variability in chromosome structure. Nature, 2013,502(7469):59-64.
doi: 10.1038/nature12593 |
| [11] |
Liang ZY, Li GP, Wang ZJ, Djekidel MN, Li YJ, Qian MP, Zhang MQ, Chen Y . BL-Hi-C is an efficient and sensitive approach for capturing structural and regulatory chromatin interactions. Nat Commun, 2017,8(1):1622.
doi: 10.1038/s41467-017-01754-3 pmid: 29158486 |
| [12] |
Lin D, Hong P, Zhang SH, Xu WZ, Jamal M, Yan KJ, Lei YY, Li L, Ruan YJ, Fu Z, Li GL, Cao G . Digestion- ligation-only Hi-C is an efficient and cost-effective method for chromosome conformation capture. Nat Genet, 2018,50(5):754-763.
doi: 10.1038/s41588-018-0111-2 pmid: 29700467 |
| [13] |
Barrett T, Edgar R . Gene expression omnibus: microarray data storage, submission, retrieval, and analysis. Method Enzymol, 2006,411:352-369.
doi: 10.1016/S0076-6879(06)11019-8 pmid: 16939800 |
| [14] |
Qu HZ, Fang XD . A brief review on the human encyclopedia of DNA elements (encode) project. Genomics Proteomics Bioinformatics, 2013,11(3):135-141.
doi: 10.1016/j.gpb.2013.05.001 pmid: 23722115 |
| [15] | Moore D, Dines J, Doss MM, Vepa J, Cheng O, Hain T . Juicer: A weighted finite-state transducer speech decoder. International Workshop on Machine Learning for Multimodal Interaction, 2006,4299:285-296. |
| [16] |
de Wit E, de Laat W . A decade of 3C technologies: insights into nuclear organization. Genes Dev, 2012,26(1):11-24.
doi: 10.1101/gad.179804.111 pmid: 22215806 |
| [17] |
Shavit Y, Merelli I, Milanesi L, Lio’ P . How computer science can help in understanding the 3D genome architecture. Brief Bioinform, 2016,17(5):733-744.
doi: 10.1093/bib/bbv085 pmid: 26433013 |
| [18] |
Schmitt AD, Hu M, Ren B . Genome-wide mapping and analysis of chromosome architecture. Nat Rev Mol Cell Biol, 2016,17(12):743-755.
doi: 10.1038/nrm.2016.104 pmid: 27580841 |
| [19] |
Eagen KP . Principles of chromosome architecture revealed by Hi-C. Trends Biochem Sci, 2018,43(6):469-478.
doi: 10.1016/j.tibs.2018.03.006 pmid: 29685368 |
| [20] | Zhang XL, Fang H, Wang XW . The progress of methods for analysing 3D genome data. Prog Biochem Biophys, 2018,45(11):1093-1105. |
| 张祥林, 方欢, 汪小我 . 三维基因组数据分析方法进展. 生物化学与生物物理进展, 2018,45(11):1093-1105. | |
| [21] |
Yaffe E, Tanay A . Probabilistic modeling of Hi-C contact maps eliminates systematic biases to characterize global chromosomal architecture. Nat Genet, 2011,43(11):1059-1065.
doi: 10.1038/ng.947 pmid: 22001755 |
| [22] |
Cournac A, Marie-Nelly H, Marbouty M, Koszul R, Mozziconacci J . Normalization of a chromosomal contact map. BMC Genomics, 2012,13(1):436.
doi: 10.1093/nar/gkx644 pmid: 28973466 |
| [23] |
Hu M, Deng K, Selvaraj S, Qin ZH, Ren B, Liu JS . HiCNorm: removing biases in Hi-C data via poisson regression. Bioinformatics, 2012,28(23):3131-3133.
doi: 10.1093/bioinformatics/bts570 |
| [24] |
Imakaev M, Fudenberg F, McCord RP, Naumova N, Goloborodko A, Lajoie BR, Dekker J, Mirny LA,. Iterative correction of Hi-C data reveals hallmarks of chromosome organization. Nat Methods, 2012,9(10):999-1003.
doi: 10.1038/NMETH.2148 |
| [25] |
Knight PA, Ruiz D . A fast algorithm for matrix balancing. IMA J Numer Anal, 2013,33(3):1029-1047.
doi: 10.1093/imanum/drs019 |
| [26] |
Wu HJ, Michor F . A computational strategy to adjust for copy number in tumor Hi-C data. Bioinformatics, 2016,32(24):3695-3701.
doi: 10.1093/bioinformatics/btw540 pmid: 27531101 |
| [27] |
Stansfield JC, Cresswell KG, Vladimirov VI, Dozmorov MG . HiCcompare: an R-package for joint normalization and comparison of Hi-C datasets. BMC Bioinformatics, 2018,19(1):279.
doi: 10.1186/s12859-018-2288-x pmid: 30064362 |
| [28] |
Stansfield JC, Cresswell KG, Dozmorov MG,. multiHiCcompare: joint normalization and comparative analysis of complex Hi-C experiments. Bioinformatics, 2019,35(17):2916-2923.
doi: 10.1093/bioinformatics/btz048 pmid: 30668639 |
| [29] |
Spill YG, Castillo D, Vidal E, Marti-Renom MA . Binless normalization of Hi-C data provides significant interaction and difference detection independent of resolution. Nat Commun, 2019,10(1):1938.
doi: 10.1038/s41467-019-09907-2 pmid: 31028255 |
| [30] | Ning CY, He MN, Tang QZ, Zhu Q, Li MZ, Li DY . Advances in mammalian three-dimensional genome by using Hi-C technology approach. Hereditas(Beijing), 2019,41(3):215-233. |
| 宁椿游, 何梦楠, 唐茜子, 朱庆, 李明洲, 李地艳 . 基于Hi-C技术哺乳动物三维基因组研究进展. 遗传, 2019,41(3):215-233. | |
| [31] |
Fortin JP, Hansen KD . Reconstructing A/B compartments as revealed by Hi-C using long-range correlations in epigenetic data. Genome Biol, 2015,16(1):180.
doi: 10.1186/s13059-015-0741-y pmid: 26316348 |
| [32] |
Dong PF, Tu XY, Chu PY, Lu P, Zhu N, Grierson D, Du BJ, Li PH, Zhong SL . 3D chromatin architecture of large plant genomes determined by local A/B compartments. Mol Plant, 2017,10(12):1497-1509.
doi: 10.1016/j.molp.2017.11.005 pmid: 29175436 |
| [33] |
Miura H, Poonperm R, Takahashi S, Hiratani I . Practical analysis of Hi-C data: generating A/B compartment profiles. Methods Mol Biol, 2018: 221-245.
doi: 10.1007/978-1-0716-0247-8_19 pmid: 31939184 |
| [34] |
Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B . Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature, 2012,485(7398):376-380.
doi: 10.1038/nature11082 |
| [35] |
Phillips-Cremins JE, Sauria MEG, Sanyal A, Gerasimova TI, Lajoie BR, Bell JSK, Ong CT, Hookway TA, Guo CY, Sun YH, Bland NJ, Wagstaff W, Dalton S, McDevitt TC, Sen R, Dekker J, Taylor J, Corces VG,. Architectural protein subclasses shape 3D organization of genomes during lineage commitment. Cell, 2013,153(6):1281-1295.
doi: 10.1016/j.cell.2013.04.053 |
| [36] |
Pope BD, Ryba T, Dileep V, Yue F, Wu WS, Denas O, Vera DL, Wang YL, Hansen RS, Canfield TK, Thurman RE, Cheng Y, Gülsoy G, Dennis JH, Snyder MP, Stamatoyannopoulos JA, Taylor J, Hardison RC, Kahveci T, Ren B, Gilbert DM . Topologically associating domains are stable units of replication-timing regulation. Nature, 2014,515(7527):402-405.
doi: 10.1038/nature13986 |
| [37] |
Narendra V, Bulajić M, Dekker J, Mazzoni EO, Reinberg D . Corrigendum: CTCF-mediated topological boundaries during development foster appropriate gene regulation. Genes Dev, 2016,30(24):2657-2662.
doi: 10.1101/gad.288324.116 pmid: 28087711 |
| [38] |
Lévy-Leduc C, Delattre M, Mary-Huard T, Robin S . Two- dimensional segmentation for analyzing Hi-C data. Bioinformatics, 2014,30(17):i386-i392.
doi: 10.1093/bioinformatics/btu443 |
| [39] |
Shin HJ, Shi Y, Dai C, Tjong H, Gong K, Alber F, Zhou XJ . TopDom: an efficient and deterministic method for identifying topological domains in genomes. Nucleic Acids Res, 2015,44(7):e70.
doi: 10.1093/nar/gkv1505 pmid: 26704975 |
| [40] |
Weinreb C, Raphael BJ . Identification of hierarchical chromatin domains. Bioinformatics, 2016,32(11):1601-1609.
doi: 10.1093/bioinformatics/btv485 pmid: 26315910 |
| [41] |
Serra F, Baù D, Goodstadt M, Castillo D, Filion GJ, Marti-Renom MA . Automatic analysis and 3D-modelling of Hi-C data using TADbit reveals structural features of the fly chromatin colors. PLoS Comput Biol, 2017,13(7):e1005665.
doi: 10.1371/journal.pcbi.1005665 pmid: 28723903 |
| [42] |
Wang XT, Cui W, Peng C . HiTAD: detecting the structural and functional hierarchies of topologically associating domains from chromatin interactions. Nucleic Acids Res, 2017,45(19):e163.
doi: 10.1093/nar/gkx735 pmid: 28977529 |
| [43] |
Haddad N, Vaillant C, Jost D . IC-Finder: inferring robustly the hierarchical organization of chromatin folding. Nucleic Acids Res, 2017,45(10):e81.
doi: 10.1093/nar/gkx036 pmid: 28130423 |
| [44] |
Yu WB, He B, Tan K . Identifying topologically associating domains and subdomains by gaussian mixture model and proportion test. Nat Commun, 2017,8(1):535.
doi: 10.1038/s41467-017-00478-8 pmid: 28912419 |
| [45] |
Norton HK, Emerson DJ, Huang H, Kim J, Titus KR, Gu S, Bassett DS, Phillips-Cremins JE . Detecting hierarchical genome folding with network modularity. Nat Methods, 2018,15(2):119-122.
doi: 10.1038/nmeth.4560 pmid: 29334377 |
| [46] |
Li AS, Yin XC, Xu BX, Wang DY, Han JM, Wei Y, Deng Y, Xiong Y, Zhang ZH . Decoding topologically associating domains with ultra-low resolution Hi-C data by graph structural entropy. Nat Commun, 2018,9(1):3265.
doi: 10.1038/s41467-018-05691-7 pmid: 30111883 |
| [47] |
Chen FL, Li GP, Zhang MQ, Chen Y . HiCDB: a sensitive and robust method for detecting contact domain boundaries. Nucleic Acids Res, 2018,46(21):11239-11250.
doi: 10.1093/nar/gky789 pmid: 30184171 |
| [48] |
Sexton T, Bantignies F, Cavalli G . Genomic interactions: chromatin loops and gene meeting points in transcriptional regulation. Semin Cell Dev Biol, 2009,20(7):849-855.
doi: 10.1016/j.semcdb.2009.06.004 |
| [49] |
Lu YL, Zhou YP, Tian WD . Combining Hi-C data with phylogenetic correlation to predict the target genes of distal regulatory elements in human genome. Nucleic Acids Res, 2013,41(22):10391-10402.
doi: 10.1093/nar/gkt785 pmid: 24003029 |
| [50] |
Ay F, Bailey TL, Noble WS . Statistical confidence estimation for Hi-C data reveals regulatory chromatin contacts. Genome Res, 2014,24(6):999-1011.
doi: 10.1101/gr.160374.113 |
| [51] |
Rao SSP, Huntley MH, Durand NC, Stamenova EK, Bochkov ID, Robinson JT, Sanborn AL, Machol I, Omer AD, Lander ES, Aiden EL . A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell, 2014,159(7):1665-1680.
doi: 10.1016/j.cell.2014.11.021 |
| [52] |
Hwang YC, Lin CF, Valladares O, Malamon J, Kuksa PP, Zheng Q, Gregory BD, Wang LS . HIPPIE: a high-throughput identification pipeline for promoter interacting enhancer elements. Bioinformatics, 2014,31(8):1290-1292.
doi: 10.1093/bioinformatics/btu801 pmid: 25480377 |
| [53] |
Lun ATL, Smyth GK,. diffHic: a bioconductor package to detect differential genomic interactions in Hi-C data. BMC Bioinformatics, 2015,16(1):258.
doi: 10.1186/s12870-016-0945-7 pmid: 27905870 |
| [54] |
Zhang H, Li FF, Jia Y, Xu BX, Zhang YQ, Li XL, Zhang ZH . Characteristic arrangement of nucleosomes is predictive of chromatin interactions at kilobase resolution. Nucleic Acids Res, 2017,45(22):12739-12751.
doi: 10.1093/nar/gkx885 pmid: 29036650 |
| [55] |
Djekidel MN, Chen Y, Zhang MQ . FIND: difFerential chromatin interactions detection using a spatial poisson process. Genome Res, 2018,28(3):412-422.
doi: 10.1101/gr.212241.116 pmid: 29440282 |
| [56] |
Manduchi E, Chesi A, Hall MA, Grant SFA, Moore JH . Leveraging putative enhancer-promoter interactions to investigate two-way epistasis in type 2 diabetes GWAS. Pac Symp Biocomput, 2018,23:548-558.
pmid: 29218913 |
| [57] |
Zhou X, Lowdon RF, Li DF, Lawson HA, Madden PAF, Costello JF, Wang T . Exploring long-range genome interactions using the WashU epigenome browser. Nat Methods, 2013,10(5):375-376.
doi: 10.1038/nmeth.2440 pmid: 23629413 |
| [58] |
Akdemir KC, Chin L . HiCPlotter integrates genomic data with interaction matrices. Genome Biol, 2015,16(1):198.
doi: 10.1186/s13059-015-0767-1 pmid: 26392354 |
| [59] |
Li RF, Liu YY, Li TT, Li C . 3Disease Browser: a web server for integrating 3D genome and disease-associated chromosome rearrangement data. Sci Rep, 2016,6:34651.
doi: 10.1038/srep34651 pmid: 27734896 |
| [60] |
Durand NC, Robinson JT, Shamim MS, Machol I, Mesirov JP, Lander ES, Aiden EL . Juicebox provides a visualization system for Hi-C contact maps with unlimited zoom. Cell Syst, 2016,3(1):99-101.
doi: 10.1016/j.cels.2015.07.012 pmid: 27467250 |
| [61] |
Djekidel MN, Wang MJ, Zhang MQ, Gao JT . HiC- 3DViewer: a new tool to visualize Hi-C data in 3D space. Quant Biol, 2017,5(2):183-190.
doi: 10.1007/s40484-017-0091-8 |
| [62] |
Tang BX, Li FF, Li J, Zhao WM, Zhang ZH . Delta: a new web-based 3D genome visualization and analysis platform. Bioinformatics, 2017,34(8):1409-1410.
doi: 10.1093/bioinformatics/btx805 pmid: 29253110 |
| [63] |
Calandrelli R, Wu QY, Guan JH, Zhong S . GITAR: an open source tool for analysis and visualization of Hi-C data. Genomics, Proteomics & Bioinformatics, 2018,16(5):365-372.
doi: 10.1016/j.cbd.2020.100654 pmid: 31954363 |
| [64] |
Wang YL, Song F, Zhang B, Zhang LJ, Xu J, Kuang D, Li DF, Choudhary MNK, Li Y, Hu M, Hardison R, Wang T, Yue F . The 3D Genome Browser: a web-based browser for visualizing 3D genome organization and long-range chromatin interactions. Genome Biol, 2018,19(1):151.
doi: 10.1186/s13059-018-1519-9 pmid: 30286773 |
| [65] |
Wolff J, Bhardwaj V, Nothjunge S, Richard G, Renschler G, Gilsbach R, Manke T, Backofen R, Ramírez F, Grüning BA . Galaxy HiCExplorer: a web server for reproducible Hi-C data analysis, quality control and visualization. Nucleic Acids Res, 2018,46(W1):W11-W16.
doi: 10.1093/nar/gky504 pmid: 29901812 |
| [66] |
Rousseau M, Fraser J, Ferraiuolo MA, Dostie J, Blanchette M . Three-dimensional modeling of chromatin structure from interaction frequency data using markov chain monte carlo sampling. BMC Bioinformatics, 2011,12(1):414.
doi: 10.1186/1471-2105-12-414 pmid: 22026390 |
| [67] | Zhang ZZ, Li GL, Toh KC, Sung WK . Inference of spatial organizations of chromosomes using semi-definite embedding approach and Hi-C data. Annual International Conference on Research in Computational Molecular Biology, 2013: 317-332. |
| [68] |
Peng C, Fu LY, Dong PF, Deng ZL, Li JX, Wang XT, Zhang HY . The sequencing bias relaxed characteristics of Hi-C derived data and implications for chromatin 3D modeling. Nucleic Acids Res, 2013,41(19):e183.
doi: 10.1093/nar/gkt745 pmid: 23965308 |
| [69] |
Trieu T, Cheng JL . MOGEN: a tool for reconstructing 3D models of genomes from chromosomal conformation capturing data. Bioinformatics, 2015,32(9):1286-1292.
doi: 10.1093/bioinformatics/btv754 pmid: 26722115 |
| [70] |
Paulsen J, Sekelja M, Oldenburg AR, Barateau A, Briand N, Delbarre E, Shah A, Sørensen AL, Vigouroux C, Buendia B, Collas P . Chrom3D: three-dimensional genome modeling from Hi-C and nuclear lamin-genome contacts. Genome Biol, 2017,18(1):21.
doi: 10.1186/s13059-016-1146-2 pmid: 28137286 |
| [71] |
Segal MR, Bengtsson HL . Improved accuracy assessment for 3D genome reconstructions. BMC Bioinformatics, 2018,19(1):196.
doi: 10.1186/s12859-018-2214-2 pmid: 29848293 |
| [72] |
Zhu GX, Deng WX, Hu HL, Ma R, Zhang S, Yang JL, Peng J, Kaplan T, Zeng JY . Reconstructing spatial organizations of chromosomes through manifold learning. Nucleic Acids Res, 2018,46(8):e50.
doi: 10.1093/nar/gky065 pmid: 29408992 |
| [73] |
Fraser J, Ferrai C, Chiariello AM, Schueler M, Rito T, Laudanno G, Barbieri M, Moore BL, Kraemer DCA, Aitken S, Xie SQ, Morris KJ, Itoh M, Kawaji H, Jaeger I, Hayashizaki Y, Carninci P, Forrest ARR, Semple CA, Dostie J, Pombo A, Nicodemi N . Hierarchical folding and reorganization of chromosomes are linked to transcriptional changes in cellular differentiation. Mol Syst Biol, 2015,11(12):852.
doi: 10.15252/msb.20156492 pmid: 26700852 |
| [74] |
Liu T, Wang Z. scHiCNorm: a software package to eliminate systematic biases in single-cell Hi-C data. Bioinformatics, 2017,34(6):1046-1047.
doi: 10.1093/bioinformatics/btx747 pmid: 29186290 |
| [75] |
Liu ST, Stroncek DF, Zhao YD, Chen V, Shi RY, Chen JG, Ren JQ, Liu H, Bae HJ, Highfill SL, Jin P . Single cell sequencing reveals gene expression signatures associated with bone marrow stromal cell subpopulations and time in culture. . Transl Med, 2019,17(1):23.
doi: 10.1186/s12967-018-1766-2 pmid: 30635013 |
| [76] |
Wang Q, Sun Q, Czajkowsky DM, Shao ZF . Sub-kb Hi-C in D. melanogaster reveals conserved characteristics of TADs between insect and mammalian cells. Nat Commun, 2018,9(1):188.
doi: 10.1038/s41467-017-02526-9 pmid: 29335463 |
| [1] | Shunze Wang, Feng Jiang, Dongli Zhu, Tie-Lin Yang, Yan Guo. Application of Hi-C technology in three-dimensional genomics research and disease pathogenesis analysis [J]. Hereditas(Beijing), 2023, 45(4): 279-294. |
| [2] | Shanshan Wang, Wanyi Zhao, Huixiao Wu, Meng Shu, Jiaxin Yuan, Li Fang, Chao Xu. Research on the variants of FGFR1 and CEP290 genes in idiopathic hypogonadotropin hypogonadism [J]. Hereditas(Beijing), 2022, 44(10): 937-949. |
| [3] | Hong Xiang, Xiaohu Yang, Liangxia Ai, Yanping Pan, Yong Hu. Bioinformatics analysis of differentially expressed genes on alopecia [J]. Hereditas(Beijing), 2020, 42(2): 172-182. |
| [4] | Qianli Dong, Jinbin Wang, Xiaochong Li, Lei Gong. Progresses in the plant 3D chromatin architecture [J]. Hereditas(Beijing), 2020, 42(1): 73-86. |
| [5] | Chao He,Wenlong Shen,Ping Li,Yan Zhang,Jing Zeng,Zuoming Yin,Zhihu Zhao. Bioinformatics analysis of Alu components at the level of genome 3D structure [J]. Hereditas(Beijing), 2019, 41(3): 254-261. |
| [6] | Chunyou Ning,Mengnan He,Qianzi Tang,Qing Zhu,Mingzhou Li,Diyan Li. Advances in mammalian three-dimensional genome by using Hi-C technology approach [J]. Hereditas(Beijing), 2019, 41(3): 215-233. |
| [7] | Yuansheng Zhang,Lin Xia,Jian Sang,Man Li,Lin Liu,Mengwei Li,Guangyi Niu,Jiabao Cao,Xufei Teng,Qing Zhou,Zhang Zhang. The BIG Data Center’s database resources [J]. Hereditas(Beijing), 2018, 40(11): 1039-1043. |
| [8] | Xiangyuan Zhang,Chao He,Bingyu Ye,Dejian Xie,Minglei Shi,Yan Zhang,Wenlong Shen,Ping Li,Zhihu Zhao. Optimization and quality control of genome-wide Hi-C library preparation [J]. Hereditas(Beijing), 2017, 39(9): 847-855. |
| [9] | Yajun Liu,Feng Zhang,Hongde Liu,Xiao Sun. The application of next-generation sequencing techniques in studying transcriptional regulation in embryonic stem cells [J]. Hereditas(Beijing), 2017, 39(8): 717-725. |
| [10] | Xiaohua Xiang, Xinru Wu, Jiangtao Chao, Minglei Yang, Fan Yang, Guo Chen, Guanshan Liu, Yuanying Wang. Genome-wide identification and expression analysis of the WRKY gene family in common tobacco (Nicotiana tabacum L.) [J]. Hereditas(Beijing), 2016, 38(9): 840-856. |
| [11] | Xiaoxu Li, Cheng Liu, Wei Li, Zenglin Zhang, Xiaoming Gao, Hui Zhou, Yongfeng Guo. Genome-wide identification, phylogenetic analysis and expression profiling of the WOX family genes in Solanum lycopersicum [J]. HEREDITAS(Beijing), 2016, 38(5): 444-460. |
| [12] | Xue Zhou, Yilan Du, Ping Jin, Fei Ma. Bioinformatic analysis of cancer-related microRNAs and their target genes [J]. HEREDITAS(Beijing), 2015, 37(9): 855-864. |
| [13] | Xiang Fang, Ningqiu Li, Xiaozhe Fu, Kaibin Li, Qiang Lin, Lihui Liu, Cunbin Shi, Shuqin Wu. Construction and application of bioinformatic analysis platform for aquatic pathogen based on the MilkyWay-2 supercomputer [J]. HEREDITAS(Beijing), 2015, 37(7): 702-710. |
| [14] | Hongmei Qiu, Wenyuan Hao, Shuqin Gao, Xiaoping Ma, Yuhong Zheng, Fanfan Meng, Xuhong Fan, Yang Wang, Yueqiang Wang, Shuming Wang. Gene mining of sulfur-containing amino acid metabolic enzymes in soybean [J]. HEREDITAS(Beijing), 2014, 36(9): 934-942. |
| [15] | Yang Shi, Xiao Xu, Haoyang Li, Qian Xu, Jichen Xu. Bioinformatics analysis of the expansin gene family in rice [J]. HEREDITAS(Beijing), 2014, 36(8): 809-820. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||