Hereditas(Beijing) ›› 2023, Vol. 45 ›› Issue (4): 324-340.doi: 10.16288/j.yczz.22-385
• Research Article • Previous Articles Next Articles
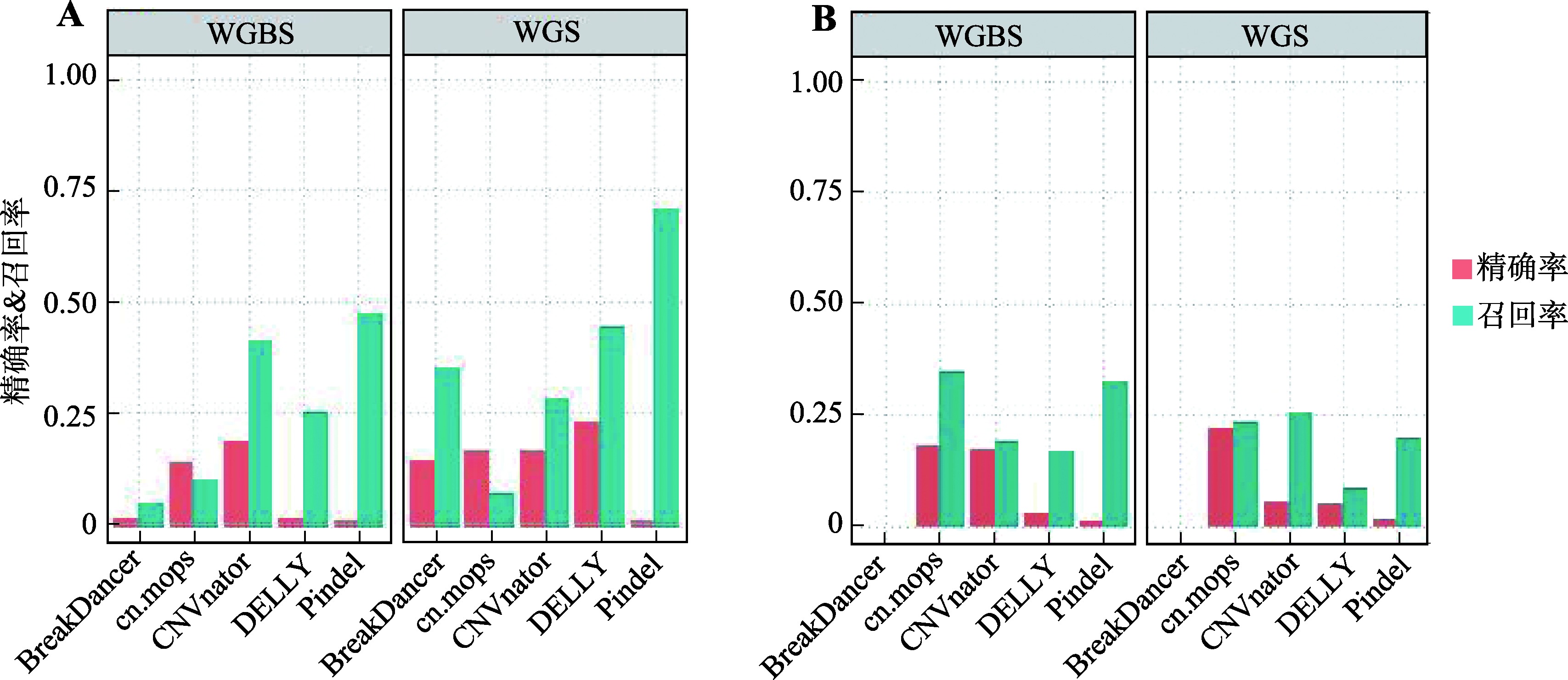
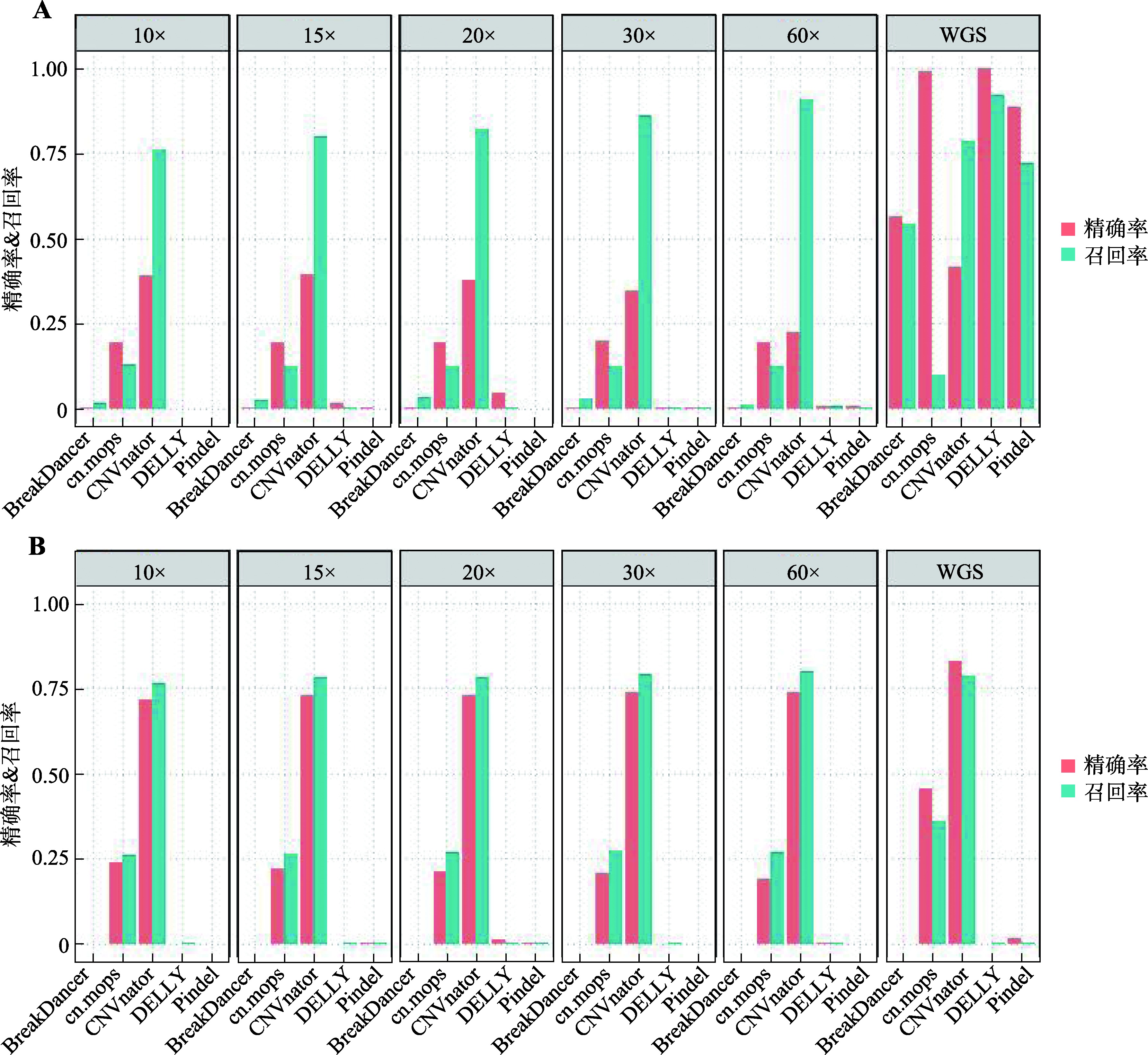
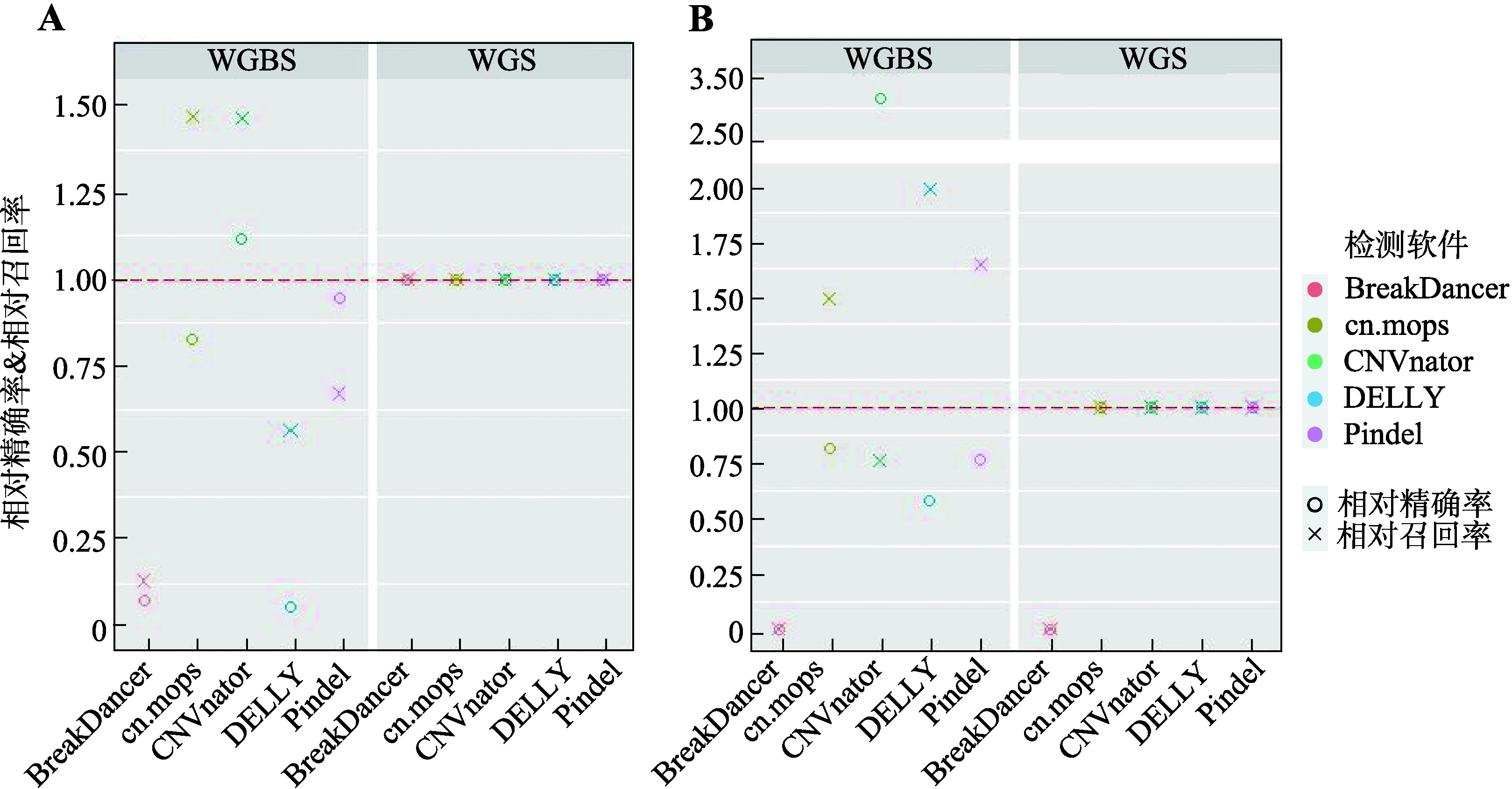
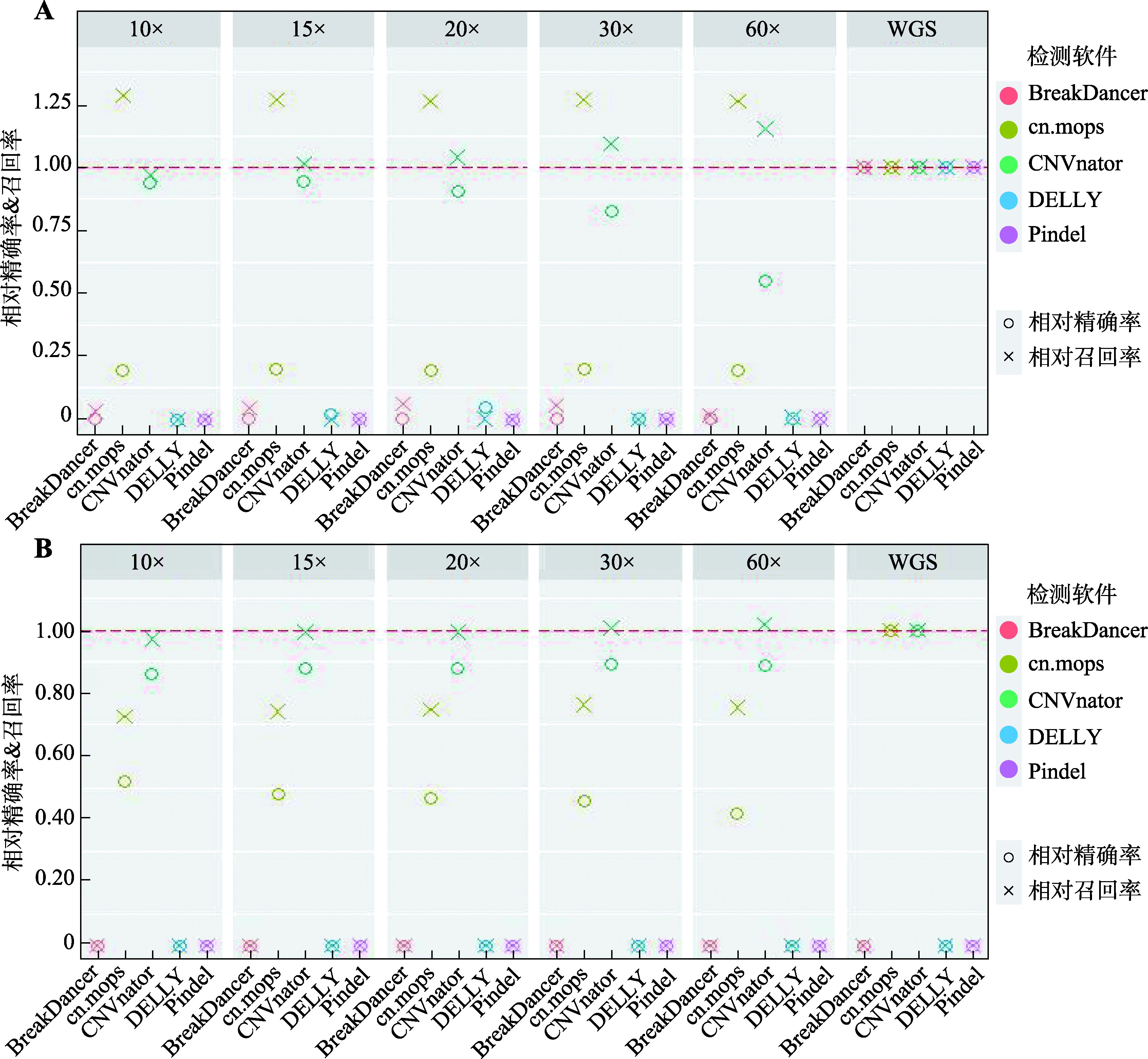
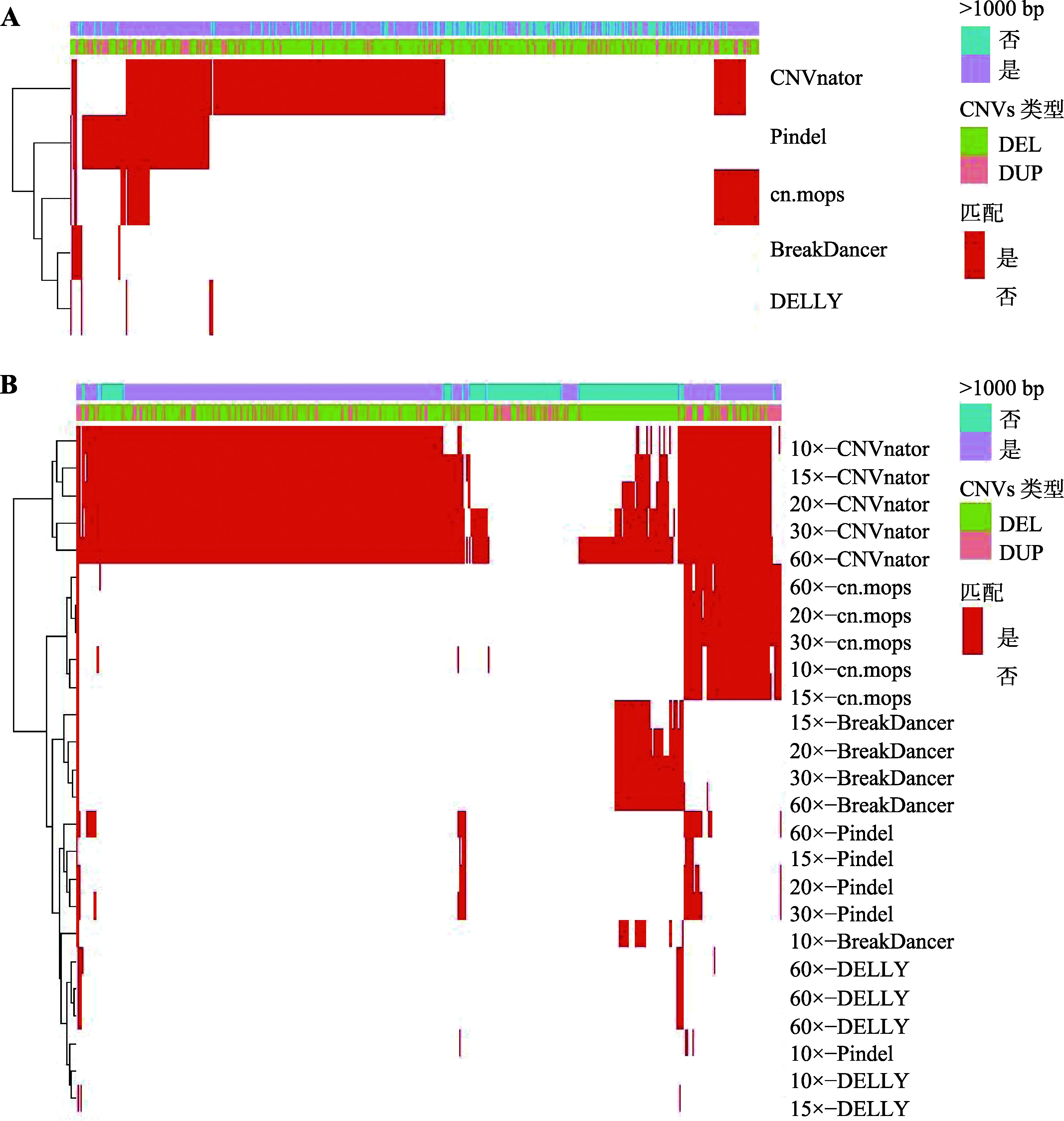
Study on detection of CNVs using human whole genome bisulfite sequencing data
Dantong Xu( ), Yifei Wang, Jiali Cai, Wentao Gong, Xiangchun Pan, Yuhan Tian, Qingpeng Shen, Jiaqi Li, Xiaolong Yuan(
), Yifei Wang, Jiali Cai, Wentao Gong, Xiangchun Pan, Yuhan Tian, Qingpeng Shen, Jiaqi Li, Xiaolong Yuan( )
)
- College of Animal Science, South China Agricultural University/Guangdong Provincial Key Laboratory of Agro-animal Genomics and Molecular Breeding/National Engineering Research Center for Breeding Swine Industry, Guangzhou 510642, China
-
Received:2023-01-18Revised:2023-03-02Online:2023-04-20Published:2023-03-06 -
Contact:Yuan Xiaolong E-mail:xdt2020@163.com;yxl@scau.edu.cn -
Supported by:Earmarked Fund for China Agriculture Research System(CARS-35);Earmarked Fund for Guangdong Provincial Science and Technology Project(210713156902656)
Cite this article
Dantong Xu, Yifei Wang, Jiali Cai, Wentao Gong, Xiangchun Pan, Yuhan Tian, Qingpeng Shen, Jiaqi Li, Xiaolong Yuan. Study on detection of CNVs using human whole genome bisulfite sequencing data[J]. Hereditas(Beijing), 2023, 45(4): 324-340.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
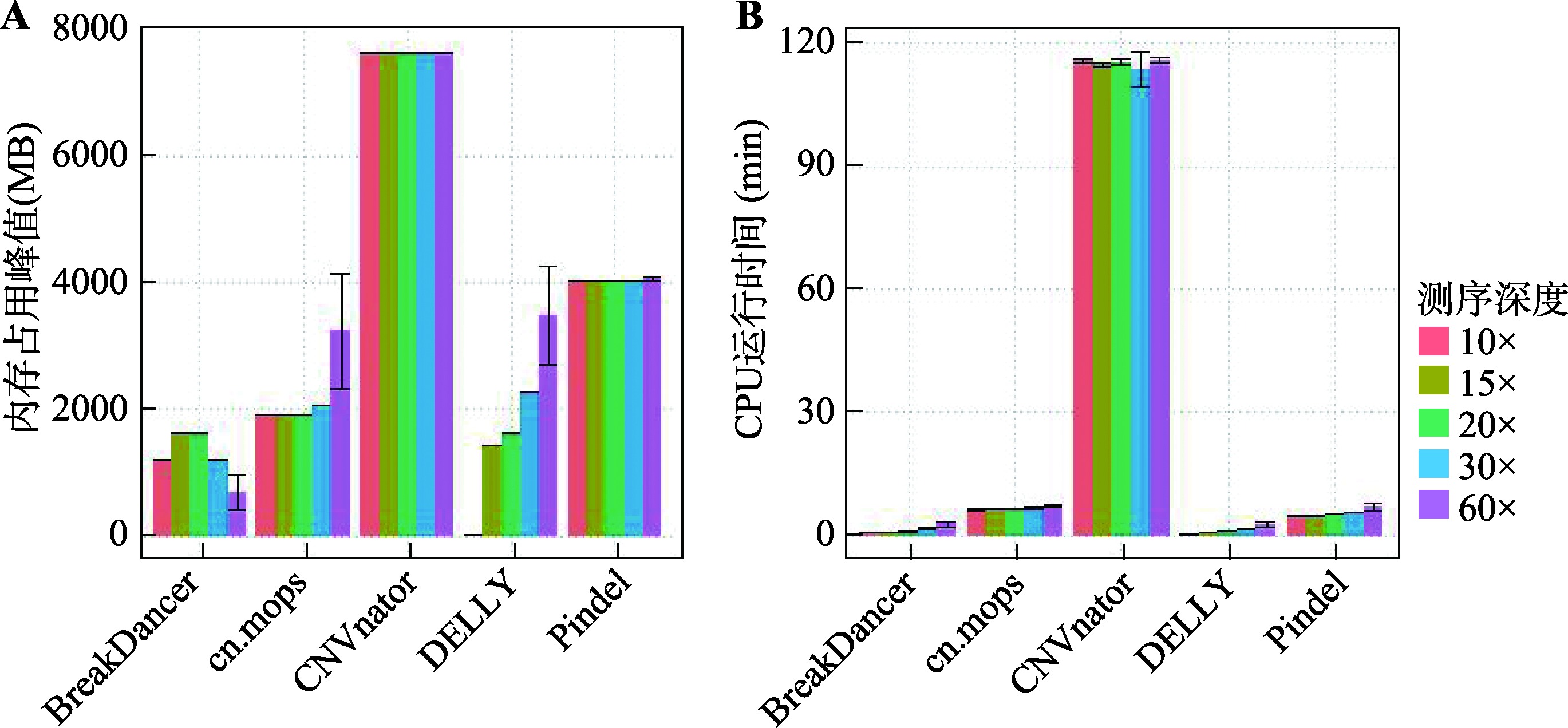
Table 1
Source and number of real data"
| BAM文件编号 | 数据类型 | 测序数据来源(来自NCBI) | 样本个体名称 |
|---|---|---|---|
| 1 | WGS | SRR622457 | NA12878 |
| 2 | WGS | ERR3239334 | NA12878 |
| 3 | WGS | ERR194147 | NA12878 |
| 4 | WGBS | SRR10532133、SRR20318446、SRR20318448、SRR20318450 | NA12878 |
| 5 | WGBS | SRR10532131、SRR10532135、SRR6006942、SRR6006945 | NA12878 |
| 6 | WGBS | SRR10532128、SRR6006943、SRR6006944、SRR6006947 | NA12878 |
Table 2
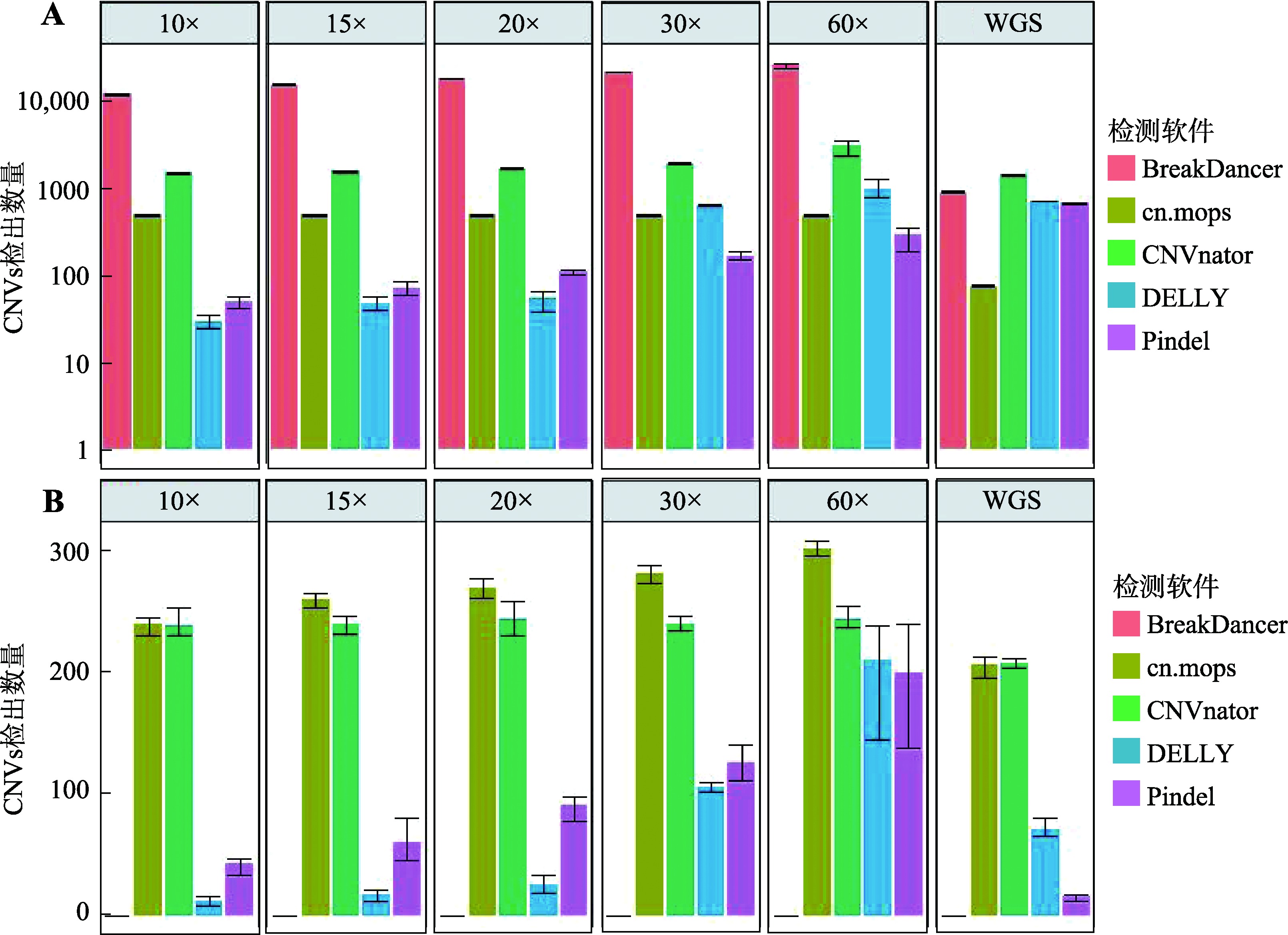
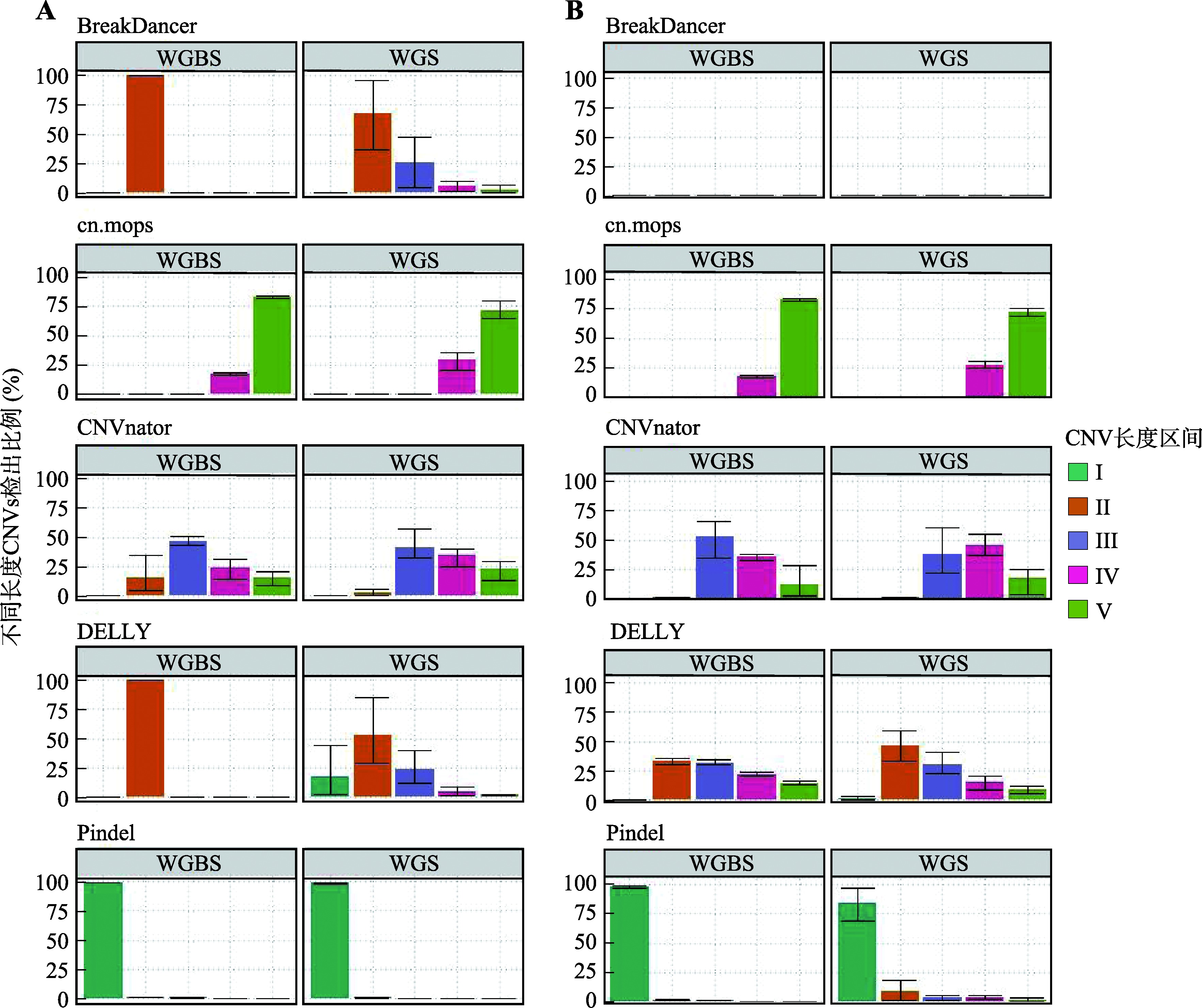
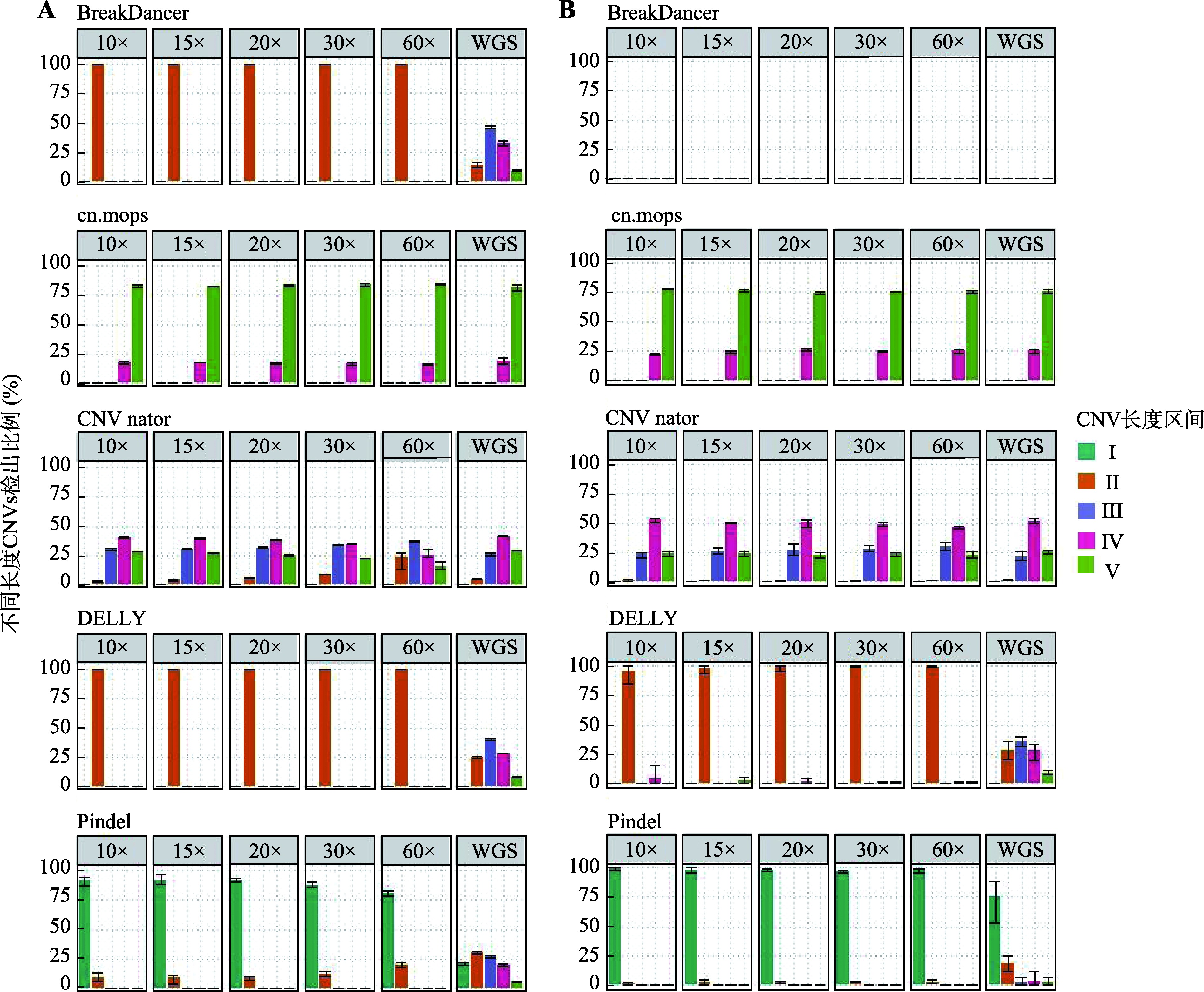
Descriptive statistics of CNVs number detected in real data by five software"
| 软件 | 数据类型 | CNVs类型 | 检出数量均值 | 标准差 | 最小值 | 最大值 |
|---|---|---|---|---|---|---|
| BreakDancer | WGBS | DEL | 23,049.67 | 19,583.18 | 723.00 | 37,318.00 |
| WGBS | DUP | 0 | 0 | 0 | 0 | |
| WGS | DEL | 4,335.67 | 5,361.69 | 992.00 | 10,520.00 | |
| WGS | DUP | 0 | 0 | 0 | 0 | |
| cn.mops | WGBS | DEL | 424.67 | 19.86 | 409.00 | 447.00 |
| WGBS | DUP | 392.00 | 12.77 | 378.00 | 403.00 | |
| WGS | DEL | 201.67 | 37.07 | 160.00 | 231.00 | |
| WGS | DUP | 157.00 | 18.33 | 137.00 | 173.00 | |
| CNVnator | WGBS | DEL | 3,066.00 | 1,506.88 | 1,954.00 | 4,781.00 |
| WGBS | DUP | 985.33 | 291.57 | 815.00 | 1,322.00 | |
| WGS | DEL | 2,066.33 | 1,124.41 | 1,324.00 | 3,360.00 | |
| WGS | DUP | 2,998.67 | 1,861.44 | 963.00 | 4,614.00 | |
| DELLY | WGBS | DEL | 42,788.00 | 32,305.60 | 19,843.00 | 79,732.00 |
| WGBS | DUP | 1,215.33 | 353.48 | 873.00 | 1,579.00 | |
| WGS | DEL | 3,021.67 | 1,719.65 | 1,036.00 | 4,022.00 | |
| WGS | DUP | 461.00 | 384.04 | 182.00 | 899.00 | |
| Pindel | WGBS | DEL | 458,175.33 | 234,456.02 | 291,944.00 | 726,345.00 |
| WGBS | DUP | 21,022.67 | 13,394.01 | 12,997.00 | 36,485.00 | |
| WGS | DEL | 577,734.00 | 201,609.58 | 414,822.00 | 803,208.00 | |
| WGS | DUP | 8,879.33 | 4,080.03 | 4,790.00 | 12,950.00 |
| [1] |
Zarrei M, MacDonald JR, Merico D, Scherer SW. A copy number variation map of the human genome. Nat Rev Genet, 2015, 16(3): 172-183.
doi: 10.1038/nrg3871 pmid: 25645873 |
| [2] |
Hollox EJ, Zuccherato LW, Tucci S. Genome structural variation in human evolution. Trends Genet, 2022, 38(1): 45-58.
doi: 10.1016/j.tig.2021.06.015 |
| [3] |
Stranger BE, Forrest MS, Dunning M, Ingle CE, Beazley C, Thorne N, Redon R, Bird CP, de Grassi A, Lee C, Tyler-Smith C, Carter N, Scherer SW, Tavaré S, Deloukas P, Hurles ME, Dermitzakis ET. Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science, 2007, 315(5813): 848-853.
doi: 10.1126/science.1136678 pmid: 17289997 |
| [4] |
Sudmant PH, Rausch T, Gardner EJ, Handsaker RE, Abyzov A, Huddleston J, Zhang Y, Ye K, Jun G, Fritz MHY, Konkel MK, Malhotra A, Stütz AM, Shi XH, Casale FP, Chen JM, Hormozdiari F, Dayama G, Chen K, Malig M, Chaisson MJP, Walter K, Meiers S, Kashin S, Garrison E, Auton A, Lam HYK, Mu XJ, Alkan C, Antaki D, Bae T, Cerveira E, Chines P, Chong ZC, Clarke L, Dal E, Ding L, Emery S, Fan X, Gujral M, Kahveci F, Kidd JM, Kong Y, Lameijer EW, McCarthy S, Flicek P, Gibbs RA, Marth G, Mason CE, Menelaou A, Muzny DM, Nelson BJ, Noor A, Parrish NF, Pendleton M, Quitadamo A, Raeder B, Schadt EE, Romanovitch M, Schlattl A, Sebra R, Shabalin AA, Untergasser A, Walker JA, Wang M, Yu FL, Zhang CS, Zhang J, Zheng-Bradley X, Zhou WD, Zichner T, Sebat J, Batzer MA, McCarroll SA; 1000 Genomes Project Consortium; Mills RE, Gerstein MB, Bashir A, Stegle O, Devine SE, Lee C, Eichler EE, Korbel JO. An integrated map of structural variation in 2,504 human genomes. Nature, 2015, 526(7571): 75-81.
doi: 10.1038/nature15394 |
| [5] |
Monks S, Niarchou M, Davies AR, Walters JTR, Williams N, Owen MJ, van den Bree MBM, Murphy KC. Further evidence for high rates of schizophrenia in 22q11.2 deletion syndrome. Schizophr Res, 2014, 153(1-3): 231-236.
doi: 10.1016/j.schres.2014.01.020 pmid: 24534796 |
| [6] |
Rose AM, Krishan A, Chakarova CF, Moya L, Chambers SK, Hollands M, Illingworth JC, Williams SMG, McCabe HE, Shah AZ, Palmer CNA, Chakravarti A, Berg JN, Batra J, Bhattacharya SS. MSR1 repeats modulate gene expression and affect risk of breast and prostate cancer. Ann Oncol, 2018, 29(5): 1292-1303.
doi: S0923-7534(19)34547-8 pmid: 29509840 |
| [7] |
Liu B, Yang L, Huang BF, Cheng M, Wang H, Li YY, Huang DS, Zheng J, Li QC, Zhang X, Ji WD, Zhou YF, Lu JC. A functional copy-number variation in MAPKAPK2 predicts risk and prognosis of lung cancer. Am J Hum Genet, 2012, 91(2): 384-390.
doi: 10.1016/j.ajhg.2012.07.003 pmid: 22883146 |
| [8] |
Wheeler E, Huang N, Bochukova EG, Keogh JM, Lindsay S, Garg S, Henning E, Blackburn H, Loos RJF, Wareham NJ, O'Rahilly S, Hurles ME, Barroso I, Farooqi IS. Genome-wide SNP and CNV analysis identifies common and low-frequency variants associated with severe early- onset obesity. Nat Genet, 2013, 45(5): 513-517.
doi: 10.1038/ng.2607 |
| [9] |
Shi XH, Radhakrishnan S, Wen J, Chen JY, Chen JJ, Lam BA, Mills RE, Stranger BE, Lee C, Setlur SR. Association of CNVs with methylation variation. NPJ Genom Med, 2020, 5: 41.
doi: 10.1038/s41525-020-00145-w pmid: 34556651 |
| [10] |
Sun W, Bunn P, Jin C, Little P, Zhabotynsky V, Perou CM, Hayes DN, Chen MJ, Lin DY. The association between copy number aberration, DNA methylation and gene expression in tumor samples. Nucleic Acids Res, 2018, 46(6): 3009-3018.
doi: 10.1093/nar/gky131 pmid: 29529299 |
| [11] |
Li J, Harris RA, Cheung SW, Coarfa C, Jeong M, Goodell MA, White LD, Patel A, Kang SH, Shaw C, Chinault AC, Gambin T, Gambin A, Lupski JR, Milosavljevic A. Genomic hypomethylation in the human germline associates with selective structural mutability in the human genome. PLoS Genet, 2012, 8(5): e1002692.
doi: 10.1371/journal.pgen.1002692 |
| [12] |
Schenkel LC, Aref-Eshghi E, Rooney K, Kerkhof J, Levy MA, McConkey H, Rogers RC, Phelan K, Sarasua SM, Jain L, Pauly R, Boccuto L, DuPont B, Cappuccio G, Brunetti-Pierri N, Schwartz CE, Sadikovic B. DNA methylation epi-signature is associated with two molecularly and phenotypically distinct clinical subtypes of Phelan-McDermid syndrome. Clin Epigenetics, 2021, 13(1): 2.
doi: 10.1186/s13148-020-00990-7 pmid: 33407854 |
| [13] |
Frommer M, McDonald LE, Millar DS, Collis CM, Watt F, Grigg GW, Molloy PL, Paul CL. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc Natl Acad Sci USA, 1992, 89(5): 1827-1831.
doi: 10.1073/pnas.89.5.1827 pmid: 1542678 |
| [14] |
Cho S, Kim HS, Zeiger MA, Umbricht CB, Cope LM. Measuring DNA copy number variation using high-density methylation microarrays. J Comput Biol, 2019, 26(4): 295-304.
doi: 10.1089/cmb.2018.0143 pmid: 30789293 |
| [15] |
Teng HJ, Xue MY, Liang JL, Wang XX, Wang L, Wei WQ, Li C, Zhang Z, Li QL, Ran X, Shi XH, Cai WS, Wang WH, Gao HJ, Sun ZS. Inter-and intratumor DNA methylation heterogeneity associated with lymph node metastasis and prognosis of esophageal squamous cell carcinoma. Theranostics, 2020, 10(7): 3035-3048.
doi: 10.7150/thno.42559 |
| [16] |
Chen K, Wallis JW, McLellan MD, Larson DE, Kalicki JM, Pohl CS, McGrath SD, Wendl MC, Zhang QY, Locke DP, Shi XQ, Fulton RS, Ley TJ, Wilson RK, Ding L, Mardis ER. BreakDancer: an algorithm for high-resolution mapping of genomic structural variation. Nat Methods, 2009, 6(9): 677-681.
doi: 10.1038/nmeth.1363 pmid: 19668202 |
| [17] |
Klambauer G, Schwarzbauer K, Mayr A, Clevert DA, Mitterecker A, Bodenhofer U, Hochreiter S. cn. MOPS: mixture of Poissons for discovering copy number variations in next-generation sequencing data with a low false discovery rate. Nucleic Acids Res, 2012, 40(9): e69.
doi: 10.1093/nar/gks003 |
| [18] |
Abyzov A, Urban AE, Snyder M, Gerstein M. CNVnator: an approach to discover, genotype, and characterize typical and atypical CNVs from family and population genome sequencing. Genome Res, 2011, 21(6): 974-984.
doi: 10.1101/gr.114876.110 pmid: 21324876 |
| [19] |
Rausch T, Zichner T, Schlattl A, Stütz AM, Benes V, Korbel JO. DELLY: structural variant discovery by integrated paired-end and split-read analysis. Bioinformatics, 2012, 28(18): i333-i339.
doi: 10.1093/bioinformatics/bts378 |
| [20] |
Ye K, Schulz MH, Long Q, Apweiler R, Ning Z. Pindel: a pattern growth approach to detect break points of large deletions and medium sized insertions from paired-end short reads. Bioinformatics, 2009, 25(21): 2865-2871.
doi: 10.1093/bioinformatics/btp394 pmid: 19561018 |
| [21] |
1000 Genomes Project Consortium, Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, Handsaker RE, Kang HM, Marth GT, McVean GA. An integrated map of genetic variation from 1,092 human genomes. Nature, 2012, 491(7422): 56-65.
doi: 10.1038/nature11632 |
| [22] | 1000 Genomes Project Consortium, Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, Korbel JO, Marchini JL, McCarthy S, McVean GA, Abecasis GR. A global reference for human genetic variation. Nature, 2015, 526(7571): 68-74. |
| [23] | MacDonald JR, Ziman R, Yuen RK, Feuk L, Scherer SW. The database of genomic variants: a curated collection of structural variation in the human genome. Nucleic Acids Res, 2014, 42(Database issue): D986-D992. |
| [24] |
Kosugi S, Momozawa Y, Liu XX, Terao C, Kubo M, Kamatani Y. Comprehensive evaluation of structural variation detection algorithms for whole genome sequencing. Genome Biol, 2019, 20(1): 117.
doi: 10.1186/s13059-019-1720-5 pmid: 31159850 |
| [25] |
Gabrielaite M, Torp MH, Rasmussen MS, Andreu-Sánchez S, Vieira FG, Pedersen CB, Kinalis S, Madsen MB, Kodama M, Demircan GS, Simonyan A, Yde CW, Olsen LR, Marvig RL, Østrup O, Rossing M, Nielsen FC, Winther O, Bagger FO. A comparison of tools for copy-number variation detection in germline whole exome and whole genome sequencing data. Cancers (Basel), 2021, 13(24): 6283.
doi: 10.3390/cancers13246283 |
| [26] |
Cock PJ, Antao T, Chang JT, Chapman BA, Cox CJ, Dalke A, Friedberg I, Hamelryck T, Kauff F, Wilczynski B, de Hoon MJL. Biopython: freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics, 2009, 25(11): 1422-1423.
doi: 10.1093/bioinformatics/btp163 pmid: 19304878 |
| [27] |
Xi YX, Li W. BSMAP: whole genome bisulfite sequence mapping program. BMC Bioinformatics, 2009, 10: 232.
doi: 10.1186/1471-2105-10-232 pmid: 19635165 |
| [28] |
Danecek P, Bonfield JK, Liddle J, Marshall J, Ohan V, Pollard MO, Whitwham A, Keane T, McCarthy SA, Davies RM, Li H. Twelve years of SAMtools and BCFtools. Gigascience, 2021, 10(2): giab008.
doi: 10.1093/gigascience/giab008 |
| [29] |
Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics, 2010, 26(6): 841-842.
doi: 10.1093/bioinformatics/btq033 pmid: 20110278 |
| [30] |
Nunn A, Otto C, Stadler PF, Langenberger D. Comprehensive benchmarking of software for mapping whole genome bisulfite data: from read alignment to DNA methylation analysis. Brief Bioinform, 2021, 22(5): bbab021.
doi: 10.1093/bib/bbab021 |
| [31] |
Chatterjee A, Rodger EJ, Morison IM, Eccles MR, Stockwell PA. Tools and strategies for analysis of genome-wide and gene-specific DNA methylation patterns. Methods Mol Biol, 2017, 1537: 249-277.
pmid: 27924599 |
| [32] |
Ji LX, Sasaki T, Sun XX, Ma P, Lewis ZA, Schmitz RJ. Methylated DNA is over-represented in whole-genome bisulfite sequencing data. Front Genet, 2014, 5: 341.
doi: 10.3389/fgene.2014.00341 pmid: 25374580 |
| [33] | Gong WT, Pan XC, Xu DT, Ji GY, Wang YF, Tian YH, Cai JL, Li JQ, Zhang Z, Yuan XL. Benchmarking DNA methylation analysis of 14 alignment algorithms for whole genome bisulfite sequencing in mammals. Comput Struct Biotechnol J, 2022, 20: 4704-4716. |
| [34] |
Haraksingh RR, Abyzov A, Urban AE. Comprehensive performance comparison of high-resolution array platforms for genome-wide Copy Number Variation (CNV) analysis in humans. BMC Genomics, 2017, 18(1): 321.
doi: 10.1186/s12864-017-3658-x pmid: 28438122 |
| [35] |
Pirooznia M, Goes FS, Zandi PP. Whole-genome CNV analysis: advances in computational approaches. Front Genet, 2015, 6: 138.
doi: 10.3389/fgene.2015.00138 pmid: 25918519 |
| [36] |
Trost B, Walker S, Wang ZZ, Thiruvahindrapuram B, MacDonald JR, Sung WWL, Pereira SL, Whitney J, Chan AJS, Pellecchia G, Reuter MS, Lok S, Yuen RKC, Marshall CR, Merico D, Scherer SW. A comprehensive workflow for read depth-based identification of copy- number variation from whole-genome sequence data. Am J Hum Genet. 2018, 102(1): 142-155.
doi: 10.1016/j.ajhg.2017.12.007 |
| [37] |
Malnic B, Godfrey PA, Buck LB. The human olfactory receptor gene family. Proc Natl Acad Sci USA, 2004, 101(8): 2584-2589.
doi: 10.1073/pnas.0307882100 pmid: 14983052 |
| [38] |
Gaudet P, Livstone MS, Lewis SE, Thomas PD. Phylogenetic-based propagation of functional annotations within the Gene Ontology consortium. Brief Bioinform, 2011, 12(5): 449-462.
doi: 10.1093/bib/bbr042 pmid: 21873635 |
| [39] |
Matsunaga T, Endo S, Maeda S, Ishikura S, Tajima K, Tanaka N, Nakamura KT, Imamura Y, Hara A. Characterization of human DHRS4: an inducible short-chain dehydrogenase/reductase enzyme with 3beta-hydroxysteroid dehydrogenase activity. Arch Biochem Biophys, 2008, 477(2): 339-347.
doi: 10.1016/j.abb.2008.06.002 pmid: 18571493 |
| [40] |
Su ZJ, Zhang QX, Liu GF, Song XH, Li Q, Wang RJ, Chen HB, Xu XY, Sui XX, Huang DY. Bioinformatic analysis of the human DHRS4 gene cluster and a proposed mechanism for its transcriptional regulation. BMC Mol Biol, 2010, 11: 43.
doi: 10.1186/1471-2199-11-43 |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||