Hereditas(Beijing) ›› 2020, Vol. 42 ›› Issue (9): 898-915.doi: 10.16288/j.yczz.20-190
• Research Article • Previous Articles Next Articles
Transcriptome heterogeneity of porcine ear fibroblast and its potential influence on embryo development in nuclear transplantation
Jun Zhou1, Chengcheng Zhao1, Xiao Wu1, Junsong Shi2, Rong Zhou2, Zhenfang Wu1, Zicong Li1( )
)
- 1. National Engineering Research Center for Swine Breeding Industry, College of Animal Science, South China Agricultural University, Guangzhou 510642, China
2. Guangdong Wenshi breeding pig Technology Co., Ltd,Xinxing 527400, China
-
Received:2020-06-22Revised:2020-08-11Online:2020-09-20Published:2020-08-31 -
Contact:Li Zicong E-mail:lizicongcong@163.com -
Supported by:Supported by Guangdong Provincial Promotion Project on Preservation and Utilization of Local Breed of Livestock and Poultry (2018)
Cite this article
Jun Zhou, Chengcheng Zhao, Xiao Wu, Junsong Shi, Rong Zhou, Zhenfang Wu, Zicong Li. Transcriptome heterogeneity of porcine ear fibroblast and its potential influence on embryo development in nuclear transplantation[J]. Hereditas(Beijing), 2020, 42(9): 898-915.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
Table 1
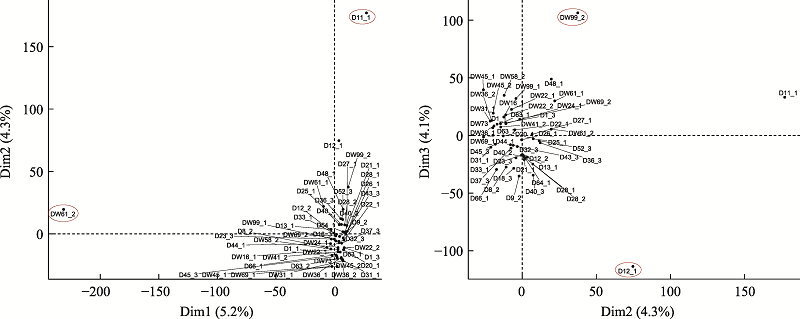
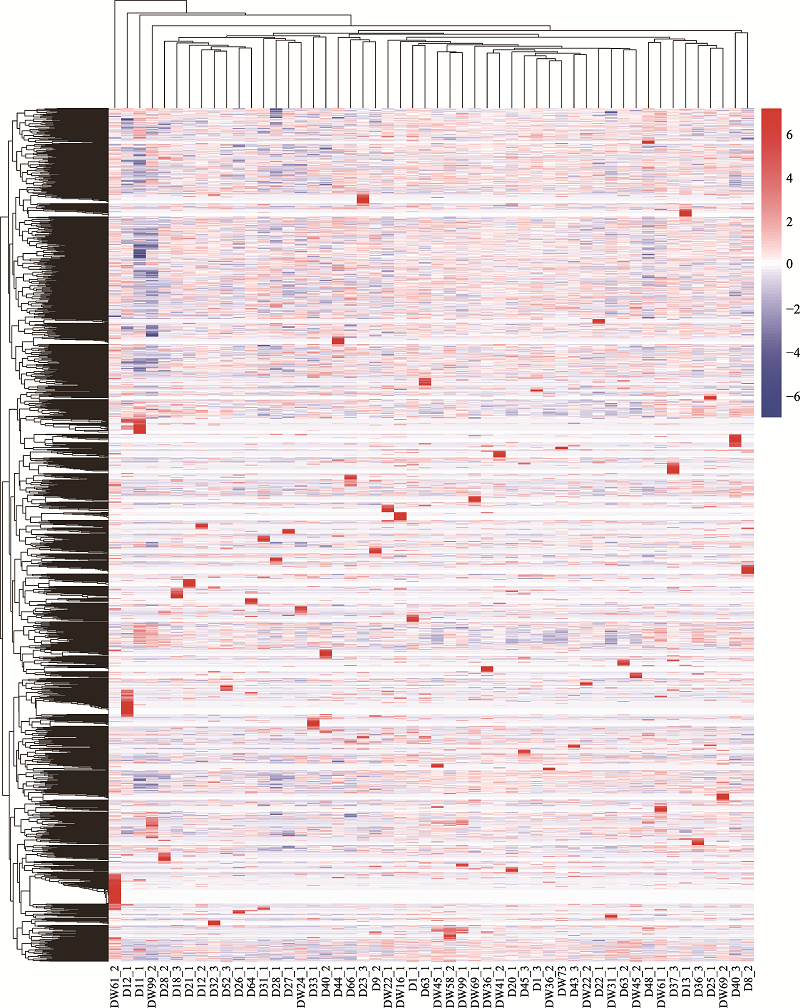
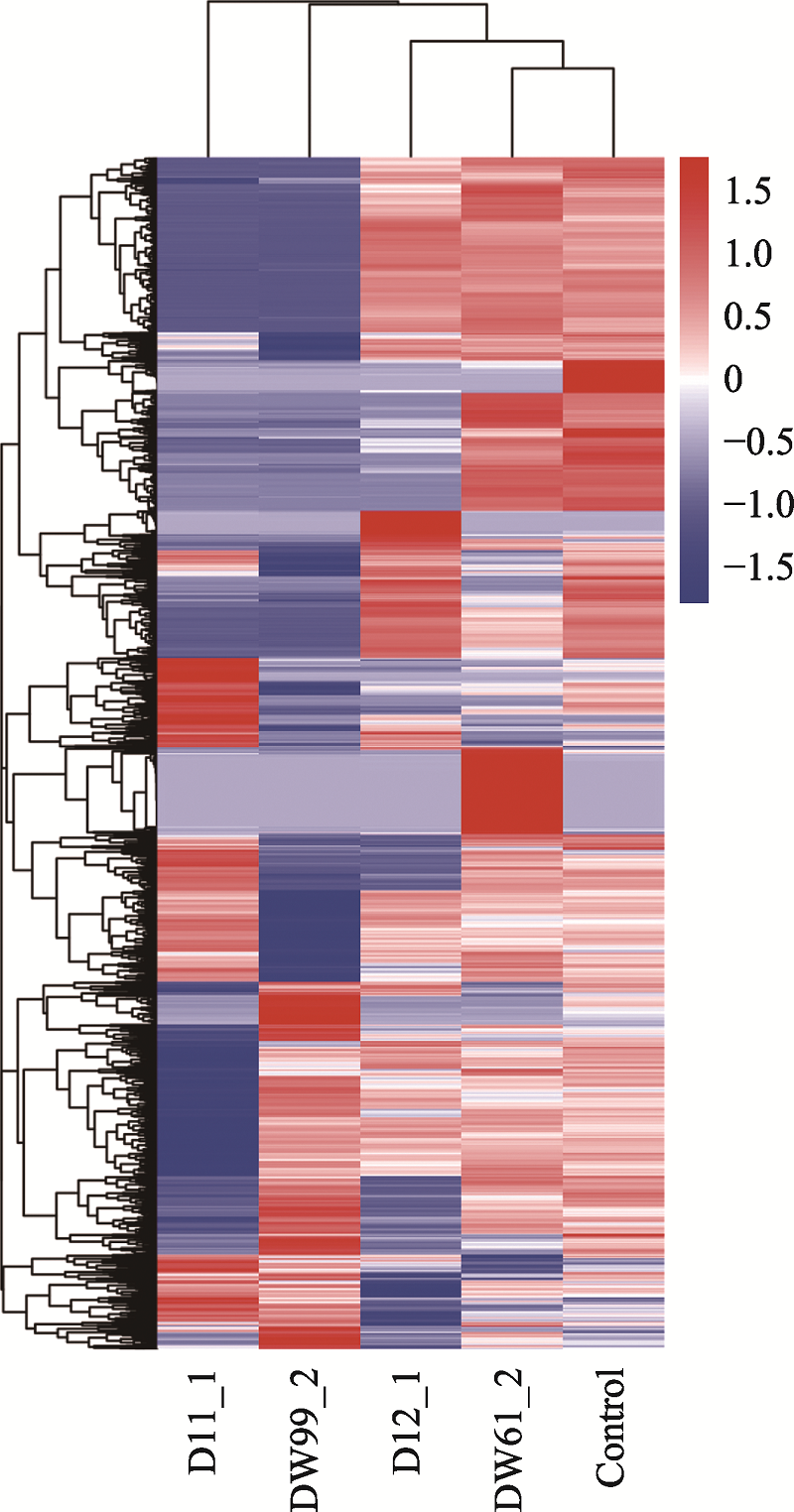
The single cell samples which were successfully amplified and sequenced"
| 序号 | 样品编号 | 序号 | 样品编号 | 序号 | 样品编号 |
|---|---|---|---|---|---|
| 1 | D1_1 | 19 | D31_1 | 37 | DW22_2 |
| 2 | D1_3 | 20 | D32_3 | 38 | DW24_1 |
| 3 | D8_2 | 21 | D33_1 | 39 | DW31_1 |
| 4 | D9_2 | 22 | D36_3 | 40 | DW36_1 |
| 5 | D11_1 | 23 | D37_3 | 41 | DW36_2 |
| 6 | D12_1 | 24 | D40_2 | 42 | DW41_2 |
| 7 | D12_2 | 25 | D40_3 | 43 | DW45_1 |
| 8 | D13_1 | 26 | D43_3 | 44 | DW45_2 |
| 9 | D18_3 | 27 | D44_1 | 45 | DW58_2 |
| 10 | D20_1 | 28 | D45_3 | 46 | DW61_1 |
| 11 | D21_1 | 29 | D48_1 | 47 | DW61_2 |
| 12 | D22_1 | 30 | D52_3 | 48 | DW69_1 |
| 13 | D23_3 | 31 | D63_1 | 49 | DW69_2 |
| 14 | D25_1 | 32 | D63_2 | 50 | DW73-1 |
| 15 | D26_1 | 33 | D64_1 | 51 | DW99_1 |
| 16 | D27_1 | 34 | D66_1 | 52 | DW99_2 |
| 17 | D28_1 | 35 | DW16_1 | ||
| 18 | D28_2 | 36 | DW22_1 |
Table 2
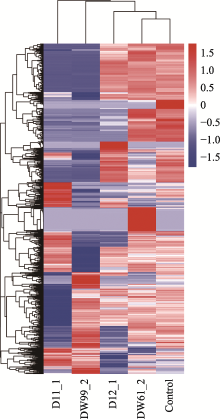
Profiles of the top 50 genes with the most significant differences in D11_1"
| 基因 | 基因全称 | 表达情况 |
|---|---|---|
| TMEM198 | Transmembrane protein 198 | ↑ |
| ALDOB | Aldolase, fructose-bisphosphate B | ↑ |
| UMOD | Uromodulin | ↑ |
| ASS1 | Argininosuccinate synthase 1 | ↑ |
| SLC5A12 | Solute carrier family 5 member 12 | ↑ |
| SLC34A1 | Sus scrofa solute carrier family 34 member 1 (SLC34A1), mRNA | ↑ |
| AGR2 | Anterior gradient protein 2 homolog precursor | ↑ |
| U6 | U6 spliceosomal RNA | ↑ |
| BHMT | Betaine-homocysteine S-methyltransferase 1 | ↑ |
| DDC | Dopa decarboxylase | ↑ |
| DAO | D-amino-acid oxidase | ↑ |
| SLC13A3 | Solute carrier family 13 member 3 | ↑ |
| CDH16 | Cadherin 16 | ↑ |
| CYP2D25 | Vitamin D(3) 25-hydroxylase | ↑ |
| PPARGC1B | PPARG coactivator 1 beta | ↑ |
| FBP1 | Fructose-1,6-bisphosphatase 1 | ↑ |
| G6PC | Glucose-6-phosphatase | ↑ |
| CLDN2 | Claudin-2 | ↑ |
| DMGDH | Dimethylglycine dehydrogenase | ↑ |
| FMO1 | Dimethylaniline monooxygenase [N-oxide-forming] 1 | ↑ |
| UPP2 | Uridine phosphorylase 2 | ↑ |
| CYP4A24 | Sus scrofa cytochrome P450,family 4,subfamily A,polypeptide 21 (CYP4A21), mRNA | ↑ |
| HNF4A | Hepatocyte nuclear factor 4-alpha | ↑ |
| ADSL | Adenylosuccinate lyase | ↓ |
| IGFBP6 | Insulin-like growth factor-binding protein 6 precursor | ↓ |
| ORMDL2 | ORMDL sphingolipid biosynthesis regulator 2 | ↓ |
| MRPS35 | Mitochondrial ribosomal protein S35 | ↓ |
| TM7SF3 | Transmembrane 7 superfamily member 3 | ↓ |
| DERA | Deoxyribose-phosphate aldolase | ↓ |
| LTBR | Tumor necrosis factor receptor superfamily member 3 precursor | ↓ |
| TULP3 | Tubby like protein 3 | ↓ |
| PPHLN1 | Periphilin 1 | ↓ |
| PUS7L | Pseudouridylate synthase 7 like | ↓ |
| SLC38A1 | Solute carrier family 38 member 1 | ↓ |
| NEDD1 | Neural precursor cell expressed, developmentally down-regulated 1 | ↓ |
| SELENOO | Sus scrofa selenoprotein O (SELENOO), mRNA | ↓ |
| SLC35B3 | Solute carrier family 35 member B3 | ↓ |
| FAM8A1 | Family with sequence similarity 8 member A1 | ↓ |
| MBOAT1 | Membrane bound O-acyltransferase domain containing 1 | ↓ |
| novel gene | Lysosomal thioesterase PPT2 precursor | ↓ |
| MAN2A2 | Mannosidase alpha class 2A member 2 | ↓ |
| HMG20A | High mobility group 20A | ↓ |
| CSPG4 | Chondroitin sulfate proteoglycan 4 | ↓ |
| SRP54 | Signal recognition particle 54 | ↓ |
| FOS | Proto-oncogene c-Fos | ↓ |
| SPTLC2 | Serine palmitoyltransferase long chain base subunit 2 | ↓ |
| ATXN3 | Ataxin-3 | ↓ |
Table 3
Profiles of the top 50 genes with the most significant differences in D12_1"
| 基因 | 基因全称 | 表达情况 |
|---|---|---|
| PARVG | Gamma-parvin | ↑ |
| POU3F1 | POU class 3 homeobox 1 | ↑ |
| ASS1 | Argininosuccinate synthase 1 | ↑ |
| UMOD | Uromodulin | ↑ |
| PNMA2 | Paraneoplastic Ma antigen 2 | ↑ |
| ADGRG7 | Adhesion G protein-coupled receptor G7 | ↑ |
| KRT28 | Keratin 28 | ↑ |
| GSDMB | Gasdermin B | ↑ |
| U6 | U6 spliceosomal RNA | ↑ |
| RNF223 | Ring finger protein 223 | ↑ |
| TBX10 | T-box 10 | ↑ |
| TMPRSS2 | Transmembrane protease, serine 12 | ↑ |
| HTR1E | 5-hydroxytryptamine receptor 1E | ↑ |
| HIC2 | HIC ZBTB transcriptional repressor 2 | ↑ |
| SLC34A1 | Sus scrofa solute carrier family 34 member 1 (SLC34A1), mRNA. | ↑ |
| ALDOB | Aldolase, fructose-bisphosphate B | ↑ |
| CSN1S1 | Sus scrofa casein alpha s1 (CSN1S1), mRNA. | ↑ |
| SLC2A12 | Solute carrier family 2 member 12 | ↑ |
| CD53 | CD53 molecule | ↑ |
| NAGA | Alpha-N-acetylgalactosaminidase precursor | ↓ |
| ADSL | Adenylosuccinate lyase | ↓ |
| C12orf4 | Homolog isoform 2 | ↓ |
| SLC35B3 | Solute carrier family 35 member B3 | ↓ |
| LEMD2 | LEM domain containing 2 | ↓ |
| GOLGA5 | Golgin A5 | ↓ |
| GSTA4 | Glutathione S-transferase A4 | ↓ |
| FAM98C | Family with sequence similarity 98 member C | ↓ |
| LDLRAP1 | Low density lipoprotein receptor adaptor protein 1 | ↓ |
| PLK3 | Polo like kinase 3 | ↓ |
| SMOC2 | SPARC related modular calcium binding 2 | ↓ |
| SPG21 | Sus scrofa spastic paraplegia 21 (autosomal recessive, Mast syndrome) (SPG21), mRNA | ↓ |
| PCLAF | Sus scrofa PCNA-associated factor (LOC100514810), mRNA | ↓ |
| SERPIN2 | Serpin family B member 2 | ↓ |
| AEN | Apoptosis enhancing nuclease | ↓ |
| GCNT1 | Glucosaminyl (N-acetyl) transferase 1, core 2 | ↓ |
| PPP6C | Serine/threonine-protein phosphatase 6 catalytic subunit | ↓ |
| PCSK6 | Proprotein convertase subtilisin/kexin type 6 | ↓ |
| BOP1 | Block of proliferation 1 | ↓ |
| FAM49B | Protein FAM49B | ↓ |
| PLAT | Tissue-type plasminogen activator precursor | ↓ |
| SMOX | Spermine oxidase | ↓ |
| ASPN | Asporin precursor | ↓ |
| IL1R1 | Interleukin 1 receptor type 1 | ↓ |
Table 4
Profiles of the top 50 genes with the most significant differences in D61_2"
| 基因 | 基因全称 | 表达情况 |
|---|---|---|
| NPPB | Natriuretic peptides B Brain natriuretic peptide 32 Brain natriuretic peptide 26 | ↑ |
| GRIK2 | Glutamate ionotropic receptor kainate type subunit 2 | ↑ |
| PAX1 | Paired box 1 | ↑ |
| DOK5 | Docking protein 5 | ↑ |
| ANKR2 | Ankyrin repeat domain 2 | ↑ |
| SLC114 | Solute carrier family 16 member 14 | ↑ |
| GPR37 | G protein-coupled receptor 37 | ↑ |
| TRPV2 | Transient receptor potential cation channel subfamily V member 2 | ↑ |
| RHCE | Sus scrofa Rh blood group CcEe antigens (RHCE), mRNA. | ↑ |
| MFNG | MFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase | ↑ |
| UBAPL | Ubiquitin associated protein 1 like | ↑ |
| ASPG | Asparaginase | ↑ |
| CRYBA1 | Crystallin beta A1 | ↑ |
| RECQL | ATP-dependent DNA helicase Q1 | ↓ |
| RIMKLB | Ribosomal modification protein rimK like family member B | ↓ |
| C1R | Complement C1r | ↓ |
| WASHC4 | WASH complex subunit 4 | ↓ |
| GNPTAB | N-acetylglucosamine-1-phosphate transferase alpha and beta subunits | ↓ |
| SELENOO | Sus scrofa selenoprotein O (SELENOO), mRNA. | ↓ |
| MAN2A2 | Mannosidase alpha class 2A member 2 | ↓ |
| STRA6 | Stimulated by retinoic acid 6 | ↓ |
| ISLR | Immunoglobulin superfamily containing leucine rich repeat | ↓ |
| HECTD1 | HECT domain E3 ubiquitin protein ligase 1 | ↓ |
| C14orf119 | Chromosome 14 open reading frame 119 | ↓ |
| NFAT5 | Nuclear factor of activated T-cells 5 | ↓ |
| E2F4 | E2F transcription factor 4 | ↓ |
| INPP5B | Inositol polyphosphate-5-phosphatase B | ↓ |
| SMOC2 | SPARC related modular calcium binding 2 | ↓ |
| MTHFD1L | Methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1 like | ↓ |
| LATS1 | Large tumor suppressor kinase 1 | ↓ |
| ME2 | Malic enzyme 2 | ↓ |
| TRIP4 | Thyroid hormone receptor interactor 4 | ↓ |
| LEO1 | LEO1 homolog, Paf1/RNA polymerase II complex component | ↓ |
| VPS39 | VPS39, HOPS complex subunit | ↓ |
| DPP8 | Dipeptidyl peptidase 8 | ↓ |
| HACD3 | 3-hydroxyacyl-CoA dehydratase 3 | ↓ |
| PRPF39 | Pre-mRNA processing factor 39 | ↓ |
Table 5
Profiles of the top 50 genes with the most significant differences in D99_2"
| 基因 | 基因全称 | 表达情况 |
|---|---|---|
| GBX2 | Gastrulation brain homeobox 2 | ↑ |
| PCDH12 | Protocadherin 12 | ↑ |
| ARHGEF9 | Cdc42 guanine nucleotide exchange factor 9 | ↑ |
| TRAM1L1 | Translocation associated membrane protein 1-like 1 | ↑ |
| U6 | U6 spliceosomal RNA | ↑ |
| DMTN | Dematin actin binding protein | ↑ |
| CEP72 | Centrosomal protein 72 | ↑ |
| YBX2 | Y-box binding protein 2 | ↑ |
| ZNF768 | Zinc finger protein 768 | ↑ |
| NOTCH4 | Neurogenic locus notch homolog protein 4 precursor | ↑ |
| GARNL3 | GTPase activating Rap/RanGAP domain like 3 | ↑ |
| MTBP | MDM2 binding protein | ↑ |
| UHRF1 | Ubiquitin like with PHD and ring finger domains 1 | ↑ |
| PACSIN2 | Protein kinase C and casein kinase substrate in neurons 2 | ↑ |
| EP300 | E1A binding protein p300 | ↓ |
| ADSL | Adenylosuccinate lyase | ↓ |
| PWP1 | PWP1 homolog, endonuclein | ↓ |
| IGFBP6 | Insulin-like growth factor-binding protein 6 precursor | ↓ |
| MMP19 | Matrix metallopeptidase 19 | ↓ |
| ESYT1 | Extended synaptotagmin 1 | ↓ |
| SMARCC2 | SWI/SNF related, matrix associated, actin dependent regulator of chromatin subfamily c member 2 | ↓ |
| PTGES3 | Prostaglandin E synthase 3 | ↓ |
| MON2 | MON2 homolog, regulator of endosome-to-Golgi trafficking | ↓ |
| XPOT | Exportin for tRNA | ↓ |
| TMEM19 | Transmembrane protein 19 | ↓ |
| TBC1D15 | TBC1 domain family member 15 | ↓ |
| DNM1L | Dynamin 1 like | ↓ |
| FAR2 | Fatty acyl-CoA reductase 2 | ↓ |
| ARNTL2 | Aryl hydrocarbon receptor nuclear translocator like 2 | ↓ |
| TM7SF3 | Transmembrane 7 superfamily member 3 | ↓ |
| FGFR1OP2 | FGFR1 oncogene partner 2 | ↓ |
| AEBP2 | AE binding protein 2 | ↓ |
| LRP6 | LDL receptor related protein 6 | ↓ |
| C1R | Complement C1r | ↓ |
| NOP2 | Sus scrofa NOP2 nucleolar protein (NOP2), mRNA | ↓ |
| [1] |
Wilmut I, Schnieke AE, Mcwhir J, Kind AJ, Campbell KH . Viable offspring derived from fetal and adult mammalian cells. Nature, 1997,385(6619):810-813.
doi: 10.1038/385810a0 pmid: 9039911 |
| [2] |
Cibelli JB, Stice SL, Golueke PJ, Kane JJ, Jerry J, Blackwell C, de León FAP, Robl JM. Cloned transgenic calves produced from nonquiescent fetal fibroblasts. Science, 1998,280(5367):1256-1258.
pmid: 9596577 |
| [3] |
Onishi A, Iwamoto M, Akita T, Mikawa S, Takeda K, Awata T, Hanada H, Perry AC . Pig cloning by microinjection of fetal fibroblast nuclei. Science, 2000,289(5482):1188-1190.
pmid: 10947985 |
| [4] |
Liu Z, Cai YJ, Wang Y, Nie YH, Zhang CC, Xu YT, Zhang XT, Lu Y, Wang ZY, Poo M, Sun Q . Cloning of macaque monkeys by somatic cell nuclear transfer. Cell, 2018,172(4):881-887.
doi: 10.1016/j.cell.2018.01.020 pmid: 29395327 |
| [5] |
Beyhan Z, Iager AE, Cibelli JB . Interspecies nuclear transfer: implications for embryonic stem cell biology. Cell Stem Cell, 2007,1(5):502-512.
doi: 10.1016/j.stem.2007.10.009 |
| [6] | Niemann H, Lucas-Hahn A . Somatic cell nuclear transfer cloning: practical applications and current legislation. Reprod Domest Anim, 2012,47(Suppl. 5):2-10. |
| [7] |
Tachibana M, Amato P, Sparman M, Gutierrez N M, Tippner-Hedges R, Ma H, Kang E, Fulati A, Lee H S, Sritanaudomchai H, Masterson K, Larson J, Eaton D, Sadler-Fredd K, Battaglia D, Lee D, Wu D, Jensen J, Patton P, Gokhale S, Stouffer RL, Wolf D, Mitalipov S . Human embryonic stem cells derived by somatic cell nuclear transfer. Cell, 2013,153(6):1228-1238.
doi: 10.1016/j.cell.2013.05.006 |
| [8] |
Ao Z, Wu X, Zhou J, Gu T, Wang XW, Shi JS, Zhao CF, Cai GY, Zheng EQ, Liu DW, Wu ZF, Li ZC . Cloned pig fetuses exhibit fatty acid deficiency from impaired placental transport. Mol Reprod Dev, 2019,86(11):1569-1581.
pmid: 31347235 |
| [9] |
Liu Y, Li J, Løvendahl P, Schmidt M, Larsen K, Callesen H . In vitro manipulation techniques of porcine embryos: a meta-analysis related to transfers, pregnancies and piglets. Reprod Fertil Dev, 2015,27(3):429-439.
doi: 10.1071/RD13329 pmid: 25482653 |
| [10] |
Jang G, Park ES, Cho JK, Bhuiyan MM, Lee BC, Kang SK, Hwang WS . Preimplantational embryo development and incidence of blastomere apoptosis in bovine somatic cell nuclear transfer embryos reconstructed with long-term cultured donor cells. Theriogenology, 2004,62(3-4):512-521.
pmid: 15226007 |
| [11] | Zhang DF, Liu D, Tang LL, Wang Y, Chen Y, Wang K, Wang GL, Schellander K, Cailu L . Effects of different donor cells on the development of nuclear-trans-ferred porcine embryos. Hereditas(Beijing), 2007,29(2):211-217. |
| 张德福, 刘东, 汤琳琳, 王英, 陈茵, 王凯, 王根林 , KARL Schellander, LIN Cailu. 不同供体细胞及其处理对猪核移植重构胚体外发育的影响. 遗传, 2007,29(2):211-217. | |
| [12] |
Ruan ZY, Zhao X, Li ZD, Qin XL, Shao QM, Ruan QY, Deng YF, Jiang JR, Huang B, Lu FH, Shi DS . Effect of sex differences in donor foetal fibroblast on the early development and DNA methylation status of buffalo (Bubalus bubalis) nuclear transfer embryos. Reprod Domest Anim, 2019,54(1):11-22.
doi: 10.1111/rda.13286 pmid: 30051521 |
| [13] |
Rideout WR, Eggan K, Jaenisch R . Nuclear cloning and epigenetic reprogramming of the genome. Science, 2001,293(5532):1093-1098.
doi: 10.1126/science.1063206 pmid: 11498580 |
| [14] | Yang XQ, Wu ZF, Li ZC . Advances in epigenetic reprogramming of somatic cells nuclear transfer in mammals. Hereditas(Beijing), 2019,41(12):1099-1109. |
| 杨旭琼, 吴珍芳, 李紫聪 . 哺乳动物体细胞核移植表观遗传重编程研究进展. 遗传, 2019,41(12):1099-1109. | |
| [15] |
Zhai YH, Li W, Zhang ZR, Cao YQ, Wang ZZ, Zhang S, Li ZY . Epigenetic states of donor cells significantly affect the development of somatic cell nuclear transfer (SCNT) embryos in pigs. Mol Reprod Dev, 2018,85(1):26-37.
doi: 10.1002/mrd.22935 pmid: 29205617 |
| [16] |
Yamanaka S . Elite and stochastic models for induced pluripotent stem cell generation. Nature, 2009,460(7251):49-52.
doi: 10.1038/nature08180 pmid: 19571877 |
| [17] |
Inoue K, Ogonuki N, Mochida K, Yamamoto Y, Takano K, Kohda T, Ishino F, Ogura A . Effects of donor cell type and genotype on the efficiency of mouse somatic cell cloning. Biol Reprod, 2003,69(4):1394-1400.
doi: 10.1095/biolreprod.103.017731 pmid: 12801984 |
| [18] |
Zhou C, Zhang JC, Zhang M, Wang DB, Ma Y, Wang Y, Wang YZ, Huang YM, Zhang Y . Transcriptional memory inherited from donor cells is a developmental defect of bovine cloned embryos. Faseb J, 2020,34(1):1637-1651.
doi: 10.1096/fj.201900578RR pmid: 31914649 |
| [19] |
Picelli S, Faridani OR, Björklund AK, Winberg G, Sagasser S, Sandberg R . Full-length RNA-seq from single cells using Smart-seq2. Nat Protoc, 2014,9(1):171-181.
doi: 10.1038/nprot.2014.006 pmid: 24385147 |
| [20] |
Grubman A, Chew G, Ouyang JF, Sun GZ, Choo XY, Mclean C, Simmons RK, Buckberry S, Vargas-Landin DB, Poppe D, Pflueger J, Lister R, Rackham O, Petretto E, Polo JM . A single-cell atlas of entorhinal cortex from individuals with Alzheimer's disease reveals cell-type- specific gene expression regulation. Nat Neurosci, 2019,22(12):2087-2097
pmid: 31768052 |
| [21] |
Tang FC, Barbacioru C, Wang YZ, Nordman E, Lee C, Xu NN, Wang XH, Bodeau J, Tuch BB, Siddiqui A, Lao K , Surani MA. mRNA-Seq whole-transcriptome analysis of a single cell. Nat Methods, 2009,6(5):377-382.
doi: 10.1038/nmeth.1315 pmid: 19349980 |
| [22] |
Kim D, Langmead B, Salzberg SL . HISAT: a fast spliced aligner with low memory requirements. Nat Methods, 2015,12(4):357-360.
doi: 10.1038/nmeth.3317 pmid: 25751142 |
| [23] |
Wang LK, Feng ZX, Wang X, Wang XW, Zhang XG . DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics, 2010,26(1):136-138.
doi: 10.1093/bioinformatics/btp612 |
| [24] |
Young MD, Wakefield MJ, Smyth GK, Oshlack A . Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol, 2010,11(2):R14.
doi: 10.1186/gb-2010-11-2-r14 pmid: 20132535 |
| [25] |
Robinson MD, Mccarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics, 2010,26(1):139-140.
doi: 10.1093/bioinformatics/btp616 pmid: 19910308 |
| [26] |
Navin N, Hicks J . Future medical applications of single-cell sequencing in cancer. Genome Med, 2011,3(5):31.
pmid: 21631906 |
| [27] |
Novick A, Weiner M . Enzyme induction as an all-or-none phenomenon. Proc Natl Acad Sci USA, 1957,43(7):553-566.
pmid: 16590055 |
| [28] |
Coskun AF, Eser U, Islam S . Cellular identity at the single-cell level. Mol Biosyst, 2016,12(10):2965-2979.
doi: 10.1039/c6mb00388e pmid: 27460751 |
| [29] |
Menon M, Mohammadi S, Davila-Velderrain J, Goods BA, Cadwell TD, Xing Y, Stemmer-Rachamimov A, Shalek AK, Love JC, Kellis M, Hafler BP . Single-cell transcriptomic atlas of the human retina identifies cell types associated with age-related macular degeneration. Nat Commun, 2019,10(1):4902.
pmid: 31653841 |
| [30] |
Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S . Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell, 2007,131(5):861-872.
doi: 10.1016/j.cell.2007.11.019 pmid: 18035408 |
| [31] |
Lowry WE, Richter L, Yachechko R, Pyle AD, Tchieu J, Sridharan R, Clark AT, Plath K . Generation of human induced pluripotent stem cells from dermal fibroblasts. Proc Natl Acad Sci USA, 2008,105(8):2883-2888.
doi: 10.1073/pnas.0711983105 pmid: 18287077 |
| [32] |
Polo JM, Liu S, Figueroa M E, Kulalert W, Eminli S, Tan KY, Apostolou E, Stadtfeld M, Li YS, Shioda T, Natesan S, Wagers AJ, Melnick A, Evans T, Hochedlinger K . Cell type of origin influences the molecular and functional properties of mouse induced pluripotent stem cells. Nat Biotechnol, 2010,28(8):848-855.
doi: 10.1038/nbt.1667 pmid: 20644536 |
| [33] |
Lai L, Tao T, Macháty Z, Kühholzer B, Sun QY, Park KW, Day BN, Prather RS . Feasibility of producing porcine nuclear transfer embryos by using G2/M-stage fetal fibroblasts as donors. Biol Reprod, 2001,65(5):1558-1564.
doi: 10.1095/biolreprod65.5.1558 pmid: 11673275 |
| [34] |
Chesné P, Adenot PG, Viglietta C, Baratte M, Boulanger L, Renard JP . Cloned rabbits produced by nuclear transfer from adult somatic cells. Nat Biotechnol, 2002,20(4):366-369.
doi: 10.1038/nbt0402-366 pmid: 11923842 |
| [35] |
Sutovsky P, Moreno RD, Ramalho-Santos J, Dominko T, Simerly C, Schatten G . Ubiquitin tag for sperm mitochondria. Nature, 1999,402(6760):371-372.
doi: 10.1038/46466 pmid: 10586873 |
| [36] |
Onishi A, Iwamoto M, Akita T, Mikawa S, Takeda K, Awata T, Hanada H, Perry AC . Pig cloning by microinjection of fetal fibroblast nuclei. Science, 2000,289(5482):1188-1190.
doi: 10.1126/science.289.5482.1188 pmid: 10947985 |
| [37] |
Gupta S, Sahu D, Bomalaski JS, Frank I, Boorjian SA, Thapa P, Cheville J C, Hansel DE . Argininosuccinate Synthetase-1 (ASS1) loss in high-grade neuroendocrine carcinomas of the urinary bladder: implications for targeted therapy with ADI-PEG 20. Endocr Pathol, 2018,29(3):236-241.
doi: 10.1007/s12022-018-9516-9 pmid: 29453600 |
| [38] |
Moren L, Perryman R, Crook T, Langer JK, Oneill K, Syed N, Antti H . Metabolomic profiling identifies distinct phenotypes for ASS1 positive and negative GBM. BMC Cancer, 2018,18(1):167.
pmid: 29422017 |
| [39] |
Bang J, Lee E, Lee AR, Il Lee J, Choi SH, Seol D, Park C, Lee DR . The effect of cell penetrating peptide-conjugated coactivator-associated arginine methyltransferase 1 (CPP-CARM1) on the cloned mouse embryonic development. Sci Rep, 2018,8(1):16721.
doi: 10.1038/s41598-018-35077-0 pmid: 30425285 |
| [40] |
Abdel-Hady AE, Beige J, Kreutz R, Bolbrinker J . Effect of UMOD genotype on long-term graft survival after kidney transplantation in patients treated with cyclosporine-based therapy. Pharmacogenomics J, 2018,18(2):227-231.
pmid: 28418009 |
| [41] |
Bailie C, Kilner J, Maxwell AP, Mcknight AJ . Development of next generation sequencing panel for UMOD and association with kidney disease. PLoS One, 2017,12(6):e0178321.
doi: 10.1371/journal.pone.0178321 pmid: 28609449 |
| [42] |
Davis D, Kannan M, Wattenberg B . Orm/ORMDL proteins: Gate guardians and master regulators. Adv Biol Regul, 2018,70:3-18.
doi: 10.1016/j.jbior.2018.08.002 pmid: 30193828 |
| [43] |
Fava RA, Browning JL, Gatumu M, Skarstein K, Bolstad AI . LTBR-pathway in Sjogren's syndrome: CXCL13 levels and B-cell-enriched ectopic lymphoid aggregates in NOD mouse lacrimal glands are dependent on LTBR. Adv Exp Med Biol, 2011,691:383-390.
doi: 10.1007/978-1-4419-6612-4_39 pmid: 21153342 |
| [44] |
Zhu QQ, Li N, Li F, Sang J, Deng H, Han QY, Lv Y, Li CY, Liu ZW . Association of LTBR polymorphisms with chronic hepatitis B virus infection and hepatitis B virus-related hepatocellular carcinoma. Int Immunopharmacol, 2017,49:126-131.
doi: 10.1016/j.intimp.2017.05.031 pmid: 28575727 |
| [45] |
Sia D, Losic B, Moeini A, Cabellos L, Hao K, Revill K, Bonal D, Miltiadous O, Zhang ZY, Hoshida Y, Cornella H, Castillo-Martin M, Pinyol R, Kasai Y, Roayaie S, Thung S N, Fuster J, Schwartz ME, Waxman S, Cordon-Cardo C, Schadt E, Mazzaferro V, Llovet JM . Massive parallel sequencing uncovers actionable FGFR2-PPHLN1 fusion and ARAF mutations in intrahepatic cholangiocarcinoma. Nat Commun, 2015,6:6087.
doi: 10.1038/ncomms7087 pmid: 25608663 |
| [46] |
Chung YG, Matoba S, Liu YT, Eum JH, Lu FL, Jiang W, Lee JE, Sepilian V, Cha KY, Lee DR, Zhang Y . Histone demethylase expression enhances human somatic cell nuclear transfer efficiency and promotes derivation of pluripotent stem cells. Cell Stem Cell, 2015,17(6):758-766.
doi: 10.1016/j.stem.2015.10.001 pmid: 26526725 |
| [47] |
Boone PM, Yuan B, Gu S, Ma ZW, Gambin T, Gonzaga-Jauregui C, Jain M, Murdock TJ, White JJ, Jhangiani SN, Walker K, Wang QY, Muzny DM, Gibbs RA, Hejtmancik JF, Lupski JR, Posey JE, Lewis RA . Hutterite-type cataract maps to chromosome 6p21.32- p21.31, cosegregates with a homozygous mutation in LEMD2, and is associated with sudden cardiac death. Mol Genet Genomic Med, 2016,4(1):77-94.
doi: 10.1002/mgg3.181 pmid: 26788539 |
| [48] | Mcgee LJ, Jiang AL, Lan Y. Golga5 is dispensable for mouse embryonic development and postnatal survival.Genesis, 2017, 55(7): 10. 1002/dvg. 23039. |
| [49] |
O'Grady GL, Best HA, Sztal TE, Schartner V, Sanjuan-Vazquez M, Donkervoort S, Abath NO, Sutton RB, Ilkovski B, Romero NB, Stojkovic T, Dastgir J, Waddell L B, Boland A, Hu Y, Williams C, Ruparelia AA, Maisonobe T, Peduto AJ, Reddel SW, Lek M, Tukiainen T, Cummings BB, Joshi H, Nectoux J, Brammah S, Deleuze JF, Ing VO, Ramm G, Ardicli D, Nowak KJ, Talim B, Topaloglu H, Laing NG, North K N, Macarthur DG, Friant S, Clarke NF, Bryson-Richardson RJ, Bönnemann CG, Laporte J, Cooper ST,. Variants in the oxidoreductase PYROXD1 cause early-onset myopathy with internalized nuclei and myofibrillar disorganization. Am J Hum Genet, 2016,99(5):1086-1105.
doi: 10.1016/j.ajhg.2016.09.005 pmid: 27745833 |
| [50] |
Al-Dabbagh N, Al-Shahrani H, Al-Dohayan N, Mustafa M, Arfin M, Al-Asmari AK . The SPARC-related modular calcium binding protein 2 (SMOC2) gene polymorphism in primary glaucoma: a case-control study. Clin Ophthalmol, 2017,11:549-555.
doi: 10.2147/OPTH.S126459 pmid: 28356709 |
| [51] |
Huang XQ, Zhou ZQ, Zhang XF, Chen CL, Tang Y, Zhu Q, Zhang JH, Xia JC . Overexpression of SMOC2 attenuates the tumorigenicity of hepatocellular carcinoma cells and is associated with a positive postoperative prognosis in human hepatocellular carcinoma. J Cancer, 2017,8(18):3812-3827.
doi: 10.7150/jca.20775 pmid: 29151969 |
| [52] |
Hagan N, Guarente J, Ellisor D, Zervas M . The temporal contribution of the Gbx2 lineage to cerebellar neurons. Front Neuroanat, 2017,11:50.
doi: 10.3389/fnana.2017.00050 pmid: 28785208 |
| [53] |
Mallika C, Guo QX, Li JYH . Gbx2 is essential for maintaining thalamic neuron identity and repressing habenular characters in the developing thalamus. Dev Biol, 2015,407(1):26-39.
doi: 10.1016/j.ydbio.2015.08.010 pmid: 26297811 |
| [54] |
Alber M, Kalscheuer VM, Marco E, Sherr E, Lesca G, Till M, Gradek G, Wiesener A, Korenke C, Mercier S, Becker F, Yamamoto T, Scherer SW, Marshall CR, Walker S, Dutta UR, Dalal A B, Suckow V, Jamali P, Kahrizi K, Najmabadi H, Minassian BA . ARHGEF9 disease: Phenotype clarification and genotype-phenotype correlation. Neurol Genet, 2017,3(3):e148.
doi: 10.1212/NXG.0000000000000148 pmid: 28589176 |
| [55] |
Klein KM, Pendziwiat M, Eilam A, Gilad R, Blatt I, Rosenow F, Kanaan M, Helbig I, Afawi Z . The phenotypic spectrum of ARHGEF9 includes intellectual disability, focal epilepsy and febrile seizures. J Neurol, 2017,264(7):1421-1425.
doi: 10.1007/s00415-017-8539-3 pmid: 28620718 |
| [56] |
Li HS, Song MM, Yang W, Cao P, Zheng L, Zuo YC . A Comparative analysis of single-cell transcriptome identifies reprogramming driver factors for efficiency improvement. Molecular Therapy - Nucleic Acids, 2020,19:1053-1064.
doi: 10.1016/j.omtn.2019.12.035 pmid: 32045876 |
| [57] |
Inoue K, Ogonuki N, Mochida K, Yamamoto Y, Takano K, Kohda T, Ishino F, Ogura A . Effects of donor cell type and genotype on the efficiency of mouse somatic cell cloning. Biol Reprod, 2003,69(4):1394-1400.
doi: 10.1095/biolreprod.103.017731 pmid: 12801984 |
| [58] |
Xie BT, Zhang H, Wei RY, Li QN, Weng XG, Kong QR, Liu ZH . Histone H3 lysine 27 trimethylation acts as an epigenetic barrier in porcine nuclear reprogramming. Reproduction, 2016,151(1):9-16.
doi: 10.1530/REP-15-0338 pmid: 26515777 |
| [59] |
Matoba S, Liu YT, Lu FL, Iwabuchi KA, Shen L, Inoue A, Zhang Y . Embryonic development following somatic cell nuclear transfer impeded by persisting histone methylation. Cell, 2014,159(4):884-895.
doi: 10.1016/j.cell.2014.09.055 |
| [60] |
Liu Y, Wu FR, Zhang L, Wu XQ, Li DK, Xin J, Xie J, Kong F, Wang WY, Wu QQ, Zhang D, Wang R, Gao SR, Li WY . Transcriptional defects and reprogramming barriers in somatic cell nuclear reprogramming as revealed by single-embryo RNA sequencing. BMC Genomics, 2018,19(1):734.
doi: 10.1186/s12864-018-5091-1 pmid: 30305014 |
| [1] | Xi Han, Fucheng Luo. Application of single-cell RNA sequencing in probing oligodendroglia heterogeneity and neurological disorders [J]. Hereditas(Beijing), 2023, 45(3): 198-211. |
| [2] | Beiping Zeng, Hongen Xu, Lu Mao, Wenxue Tang. Molecular diagnosis of hereditary deafness and application of stepwise testing strategy [J]. Hereditas(Beijing), 2023, 45(1): 29-41. |
| [3] |
Zhuo Wang, Xiaohan Shen, Qihui Shi.
|
| [4] | Baosong Xing, Jing Wang, Junfeng Chen, Qiang Ma, Qiaoling Ren, Jiaqing Zhang, Hua Zhang, Liushuai Hua, Jiajie Sun, Hai Cao. Analysis of differentially expressed circRNAs in longissimus muscle between castrated and intact male pigs [J]. Hereditas(Beijing), 2021, 43(11): 1066-1077. |
| [5] | Liang Qu, Su Li, Huaji Qiu. Applications of single-cell RNA sequencing in virology [J]. Hereditas(Beijing), 2020, 42(3): 269-277. |
| [6] | Qiang Zhang, Mingliang Gu. Single-cell sequencing and its application in breast cancer [J]. Hereditas(Beijing), 2020, 42(3): 250-268. |
| [7] | Zheng Ao, Xiang Chen, Zhenfang Wu, Zicong Li. Progress on abnormal development of cloned pigs generated by somatic cell transfer nuclear [J]. Hereditas(Beijing), 2020, 42(10): 993-1003. |
| [8] | Jianxin Mo,Haoqiang Wang,Guangyan Huang,Gengyuan Cai,Zhenfang Wu,Xianwei Zhang. Heterogenous expression and enzymatic property analysis of microbial-derived pectinases in pig PK15 cells [J]. Hereditas(Beijing), 2019, 41(8): 736-745. |
| [9] | Baojun Wu,Zhuo Wang,Yu Dong,Yuliang Deng,Qihui Shi. Identification and single-cell sequencing analysis of rare tumor cells in malignant pleural effusion of lung cancer patients [J]. Hereditas(Beijing), 2019, 41(2): 175-184. |
| [10] | Xuqiong Yang, Zhenfang Wu, Zicong Li. Advances in epigenetic reprogramming of somatic cells nuclear transfer in mammals [J]. Hereditas(Beijing), 2019, 41(12): 1099-1109. |
| [11] | Youwang Lu,Kunhua Wang. Research progress on genetic heterogeneity between primary and paired metastatic colorectal cancer [J]. Hereditas(Beijing), 2017, 39(6): 482-490. |
| [12] | Zheng Ao, Dewu Liu, Gengyuan Cai, Zhenfang Wu, Zicong Li. Placental developmental defects in cloned mammalian animals [J]. HEREDITAS(Beijing), 2016, 38(5): 402-410. |
| [13] | Linqi Wang. Adaptation strategies: how environmental fungi become fatal? [J]. HEREDITAS(Beijing), 2015, 37(5): 436-441. |
| [14] | Jing Liu, Yanan Wang, Yaqi Sun, Hongyang Wang, Chao Wang, Zhongzhen Peng, Bang Liu. Investigation of genes within copy number variation regions in pig chromosome 13 and analysis of the genetic law [J]. HEREDITAS, 2014, 36(4): 354-359. |
| [15] | Ji Huili, Lu Shengsheng, Pan Dengke. Epigenetic reprogramming by somatic cell nuclear transfer: questions and potential solutions [J]. HEREDITAS(Beijing), 2014, 36(12): 1211-1218. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||